A Sensitive Aptamer Fluorescence Anisotropy Sensor for Cd2+ Using Affinity-Enhanced Aptamers with Phosphorothioate Modification
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemical and Reagents
2.2. Isothermal Titration Calorimetry Measurement
2.3. Fluorescence Anisotropy Measurement
2.4. Detection of Cd2+ in Complex Sample Matrix
3. Results and Discussion
3.1. FA Sensor for Cd2+ Using TMR-Labeled Aptamers
3.2. Enhancing Aptamer Affinity with Phosphorothioate Modification
3.3. Optimization of Aptamer FA Sensor for Cd2+
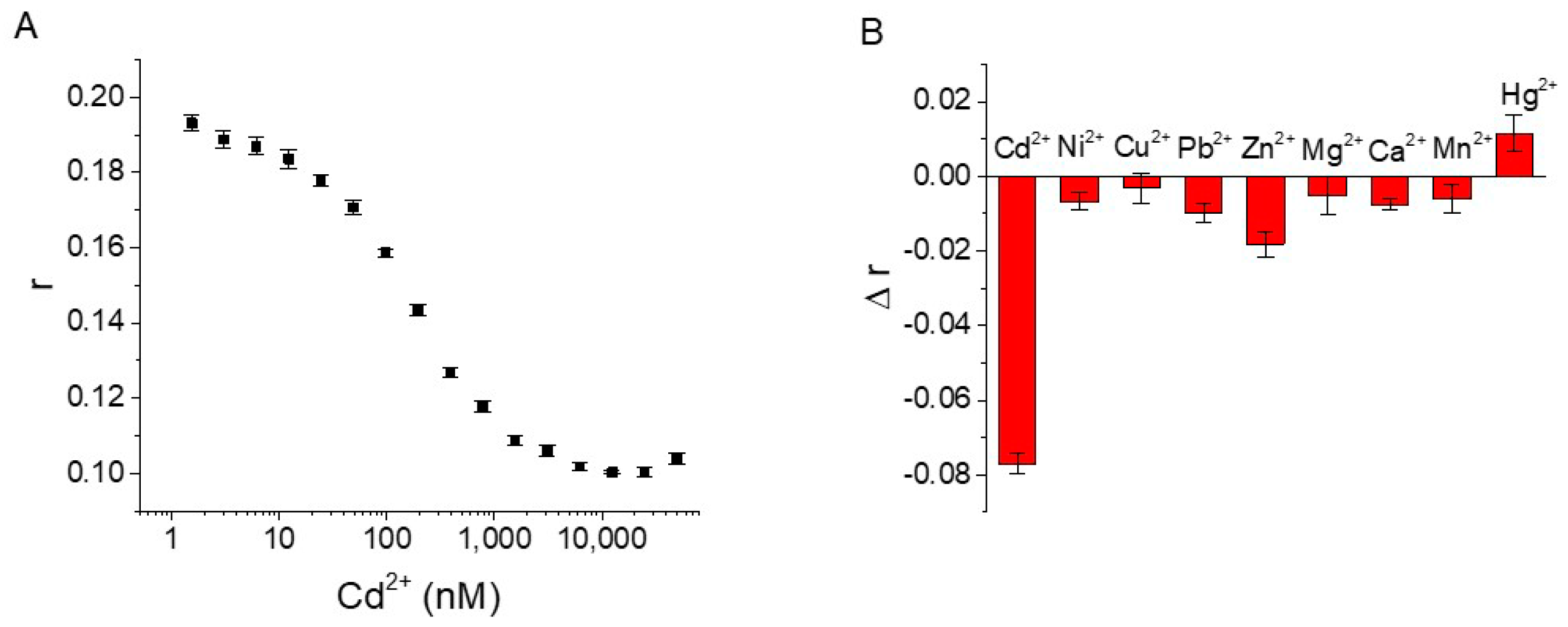
3.4. Detection of Cd2+ with Aptamer FA Sensor
3.5. Selectivity Test and Practical Sample Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Newbigging, A.M.; Yan, X.W.; Le, X.C. Cadmium in soybeans and the relevance to human exposure. J. Environ. Sci. 2015, 37, 157–162. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, P.; Zhao, F.J. Dietary cadmium exposure, risks to human health and mitigation strategies. Crit. Rev. Environ. Sci. Technol. 2022, 7, 1–25. [Google Scholar] [CrossRef]
- Mahajan, M.; Gupta, P.K.; Singh, A.; Vaish, B.; Singh, P.; Kothari, R.; Singh, R.P. A comprehensive study on aquatic chemistry, health risk and remediation techniques of cadmium in groundwater. Sci. Total Environ. 2022, 818, 151784. [Google Scholar] [CrossRef]
- Balali-Mood, M.; Naseri, K.; Tahergorabi, Z.; Khazdair, M.R.; Sadeghi, M. Toxic mechanisms of five heavy metals: Mercury, lead, chromium, cadmium, and arsenic. Front. Pharmacol. 2021, 12, 643972. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.H.; Chen, G.L.; Xu, K.; Wang, J. Cadmium in cereal crops: Uptake and transport mechanisms and minimizing strategies. J. Agric. Food Chem. 2022, 70, 5961–5974. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, S.; Dahiya, D.; Sharma, V.; Khan, N.; Chaurasia, D.; Nadda, A.K.; Varjani, S.; Pandey, A.; Bhargava, P.C. Advances from conventional to real time detection of heavy metal(loid)s for water monitoring: An overview of biosensing applications. Chemosphere 2022, 307, 136124. [Google Scholar] [CrossRef] [PubMed]
- Maghsoudi, A.S.; Hassani, S.; Mirnia, K.; Abdollahi, M. Recent advances in nanotechnology-based biosensors development for detection of arsenic, lead, mercury, and cadmium. Int. J. Nanomed. 2021, 16, 803–832. [Google Scholar] [CrossRef]
- Cao, Y.P.; Deng, B.Y.; Yan, L.Z.; Huang, H.L. An environmentally-friendly, highly efficient, gas pressure-assisted sample introduction system for ICP-MS and its application to detection of cadmium and lead in human plasma. Talanta 2017, 167, 520–525. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment-RNA ligands to bacteriophage-T4 DNA-polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, H.Q.; Wang, Z.X.; Newbigging, A.M.; Reid, M.S.; Li, X.F.; Le, X.C. Aptamers facilitating amplified detection of biomolecules. Anal. Chem. 2015, 87, 274–292. [Google Scholar] [CrossRef]
- Prante, M.; Segal, E.; Scheper, T.; Bahnemann, J.; Walter, J. Aptasensors for Point-of-Care Detection of Small Molecules. Biosensors 2020, 10, 108. [Google Scholar] [CrossRef]
- Farzin, L.; Shamsipur, M.; Sheibani, S. A review: Aptamer-based analytical strategies using the nanomaterials for environmental and human monitoring of toxic heavy metals. Talanta 2017, 174, 619–627. [Google Scholar] [CrossRef]
- Chang, C.C. Recent advancements in aptamer-based surface plasmon resonance biosensing strategies. Biosensors 2021, 11, 233. [Google Scholar] [CrossRef]
- Wang, H.Y.; Cheng, H.; Wang, J.; Xu, L.J.; Chen, H.X.; Pei, R.J. Selection and characterization of DNA aptamers for the development of light-up biosensor to detect Cd(II). Talanta 2016, 154, 498–503. [Google Scholar] [CrossRef]
- Wu, Y.G.; Zhan, S.S.; Wang, L.M.; Zhou, P. Selection of a DNA aptamer for cadmium detection based on cationic polymer mediated aggregation of gold nanoparticles. Analyst 2014, 139, 1550–1561. [Google Scholar] [CrossRef]
- Fakude, C.T.; Arotiba, O.A.; Mabuba, N. Electrochemical aptasensing of cadmium (II) on a carbon black-gold nanoplatform. J. Electroanal. Chem. 2020, 858, 113796. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Liu, C.; Su, X.Y.; Zhang, W.; Zou, X.B. Signal on-off ratiometric electrochemical sensor based on semi-complementary aptamer couple for sensitive cadmium detection in mussel. Sens. Actuator B Chem. 2021, 346, 130506. [Google Scholar] [CrossRef]
- Gan, Y.; Liang, T.; Hu, Q.W.; Zhong, L.J.; Wang, X.Y.; Wan, H.; Wang, P. In-situ detection of cadmium with aptamer functionalized gold nanoparticles based on smartphone-based colorimetric system. Talanta 2020, 208, 120231. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, D.W.; Ding, J.N.; Hayat, K.; Yang, X.J.; Zhan, X.J.; Zhang, D.; Lu, Y.T.; Zhou, P. A facile aptasensor for instantaneous determination of cadmium ions based on fluorescence amplification effect of MOPS on FAM-labeled aptamer. Biosensors 2021, 11, 133. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Q. Rapid sensitive fluorescence detection of cadmium (II) with pyrene excimer switching aptasensor. J. Environ. Sci. 2022, in press. [Google Scholar] [CrossRef]
- Hall, M.D.; Yasgar, A.; Peryea, T.; Braisted, J.C.; Jadhav, A.; Simeonov, A.; Coussens, N.P. Fluorescence polarization assays in high-throughput screening and drug discovery: A review. Methods Appl. Fluores. 2016, 4, 022001. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Tao, J.; Uppal, J.S.; Peng, H.Y.; Wang, H.L.; Le, X.C. Nucleic acid aptamers improving fluorescence anisotropy and fluorescence polarization assays for small molecules. Trac-Trends Anal. Chem. 2019, 110, 401–409. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, S.; De Ruyck, K.; Beloglazova, N.V.; Eremin, S.A.; De Saeger, S.; Zhang, S.; Shen, J.; Wang, Z. Fluorescence polarization assays for chemical contaminants in food and environmental analyses. Trac-Trends Anal. Chem. 2019, 114, 293–313. [Google Scholar] [CrossRef]
- Li, C.L.; Mi, T.J.; Conti, G.O.; Yu, Q.; Wen, K.; Shen, J.Z.; Ferrante, M.; Wang, Z.H. Development of a screening fluorescence polarization immunoassay for the simultaneous detection of fumonisins B-1 and B-2 in maize. J. Agric. Food Chem. 2015, 63, 4940–4946. [Google Scholar] [CrossRef]
- Li, Y.P.; Zhao, Q. Aptamer structure switch fluorescence anisotropy assay for aflatoxin B1 using tetramethylrhodamine-guanine interaction to enhance signal change. Chin. Chem. Lett. 2020, 31, 1982–1985. [Google Scholar] [CrossRef]
- Zhao, Q.; Lv, Q.; Wang, H.L. Identification of allosteric nucleotide sites of tetramethylrhodamine-labeled aptamer for noncompetitive aptamer-based fluorescence anisotropy detection of a small molecule, ochratoxin A. Anal. Chem. 2014, 86, 1238–1245. [Google Scholar] [CrossRef]
- Johnson, D.K. A fluorescence polarization immunoassay for cadmium(II). Anal. Chim. Acta 1999, 399, 161–172. [Google Scholar] [CrossRef]
- Zhu, X.; Xu, L.; Lou, Y.; Yu, H.; Li, X.; Blake, D.A.; Liu, F. Preparation of specific monoclonal antibodies (MAbs) against heavy metals: MAbs that recognize chelated cadmium ions. J. Agric. Food Chem. 2007, 55, 7648–7653. [Google Scholar] [CrossRef]
- Huang, P.J.J.; Liu, J.W. Rational evolution of Cd2+-specific DNAzymes with phosphorothioate modified cleavage junction and Cd2+ sensing. Nucleic Acids Res. 2015, 43, 6125–6133. [Google Scholar] [CrossRef]
- Zhang, D.P.; Shen, H.J.; Li, G.H.; Zhao, B.L.; Yu, A.C.; Zhao, Q.; Wang, H.L. Specific and sensitive fluorescence anisotropy sensing of guanine-quadruplex structures via a photoinduced electron transfer mechanism. Anal. Chem. 2012, 84, 8088–8094. [Google Scholar] [CrossRef]
- Zhou, B.; Yang, X.Y.; Wang, Y.S.; Yi, J.C.; Zeng, Z.; Zhang, H.; Chen, Y.T.; Hu, X.J.; Suo, Q.L. Label-free fluorescent aptasensor of Cd2+ detection based on the conformational switching of aptamer probe and SYBR green I. Microchem. J. 2019, 144, 377–382. [Google Scholar] [CrossRef]
- Zhu, Y.F.; Wang, Y.S.; Zhou, B.; Yu, J.H.; Peng, L.L.; Huang, Y.Q.; Li, X.J.; Chen, S.H.; Tang, X.; Wang, X.F. A multifunctional fluorescent aptamer probe for highly sensitive and selective detection of cadmium(II). Anal. Bioanal. Chem. 2017, 409, 4951–4958. [Google Scholar] [CrossRef]
- Lai, B.; Wang, H.T.; Su, W.T.; Wang, Z.P.; Zhu, B.W.; Yu, C.X.; Tan, M.Q. A phosphorescence resonance energy transfer-based “off-on” long afterglow aptasensor for cadmium detection in food samples. Talanta 2021, 232, 122409. [Google Scholar] [CrossRef]
- Zhou, D.H.; Wu, W.; Li, Q.; Pan, J.F.; Chen, J.H. A label-free and enzyme-free aptasensor for visual Cd2+ detection based on split DNAzyme fragments. Anal. Methods 2019, 11, 3546–3551. [Google Scholar] [CrossRef]
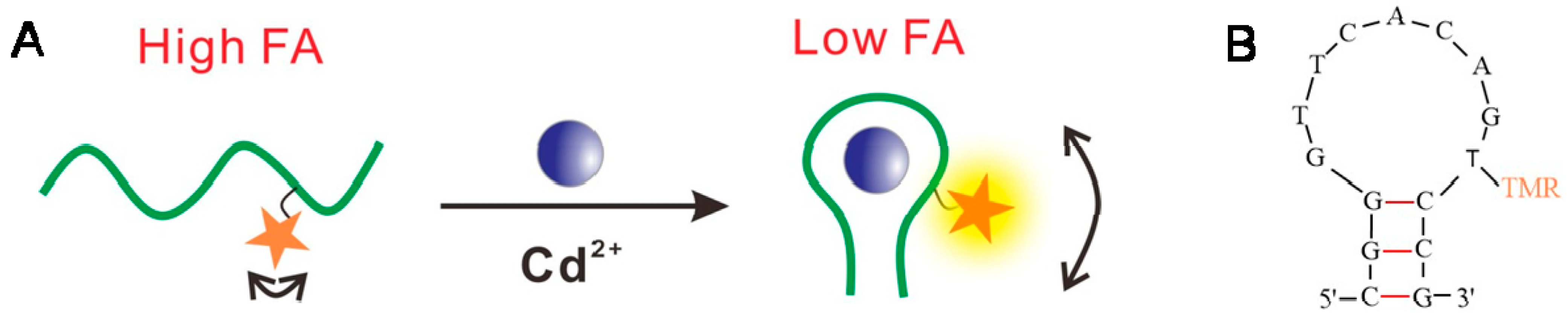
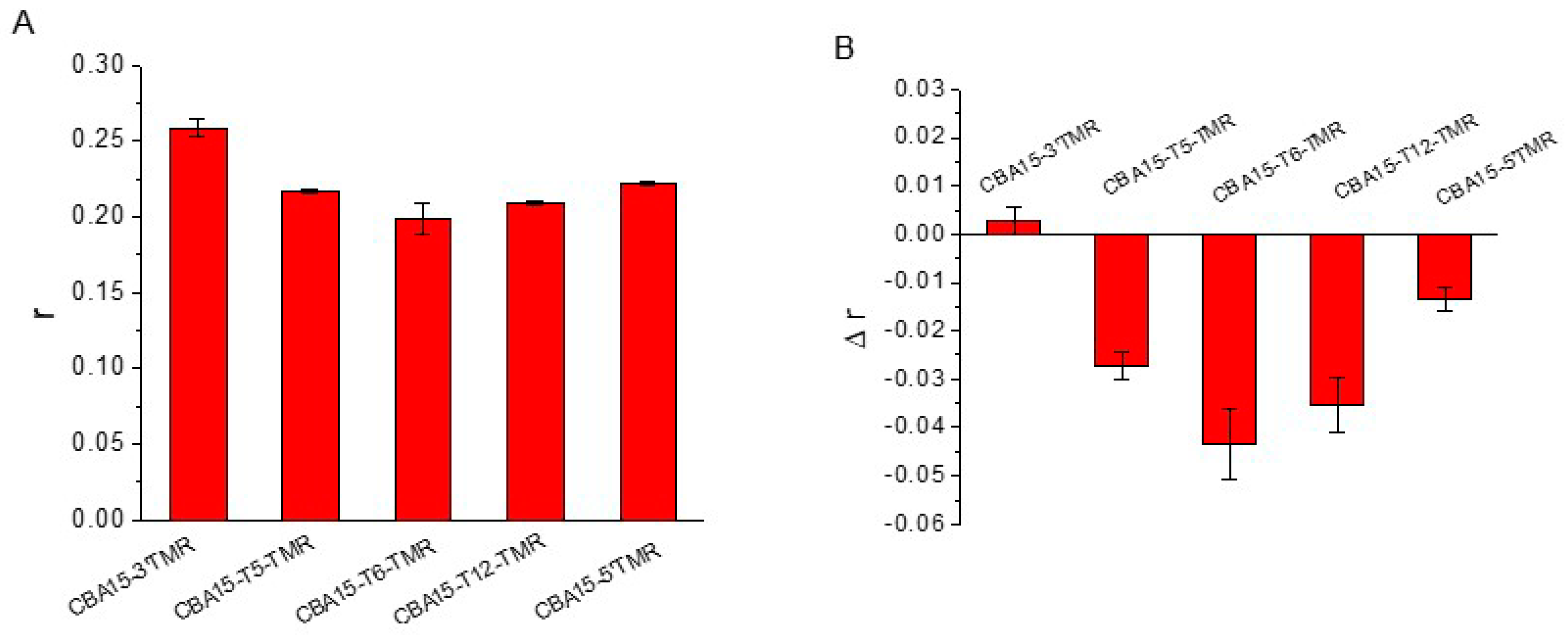
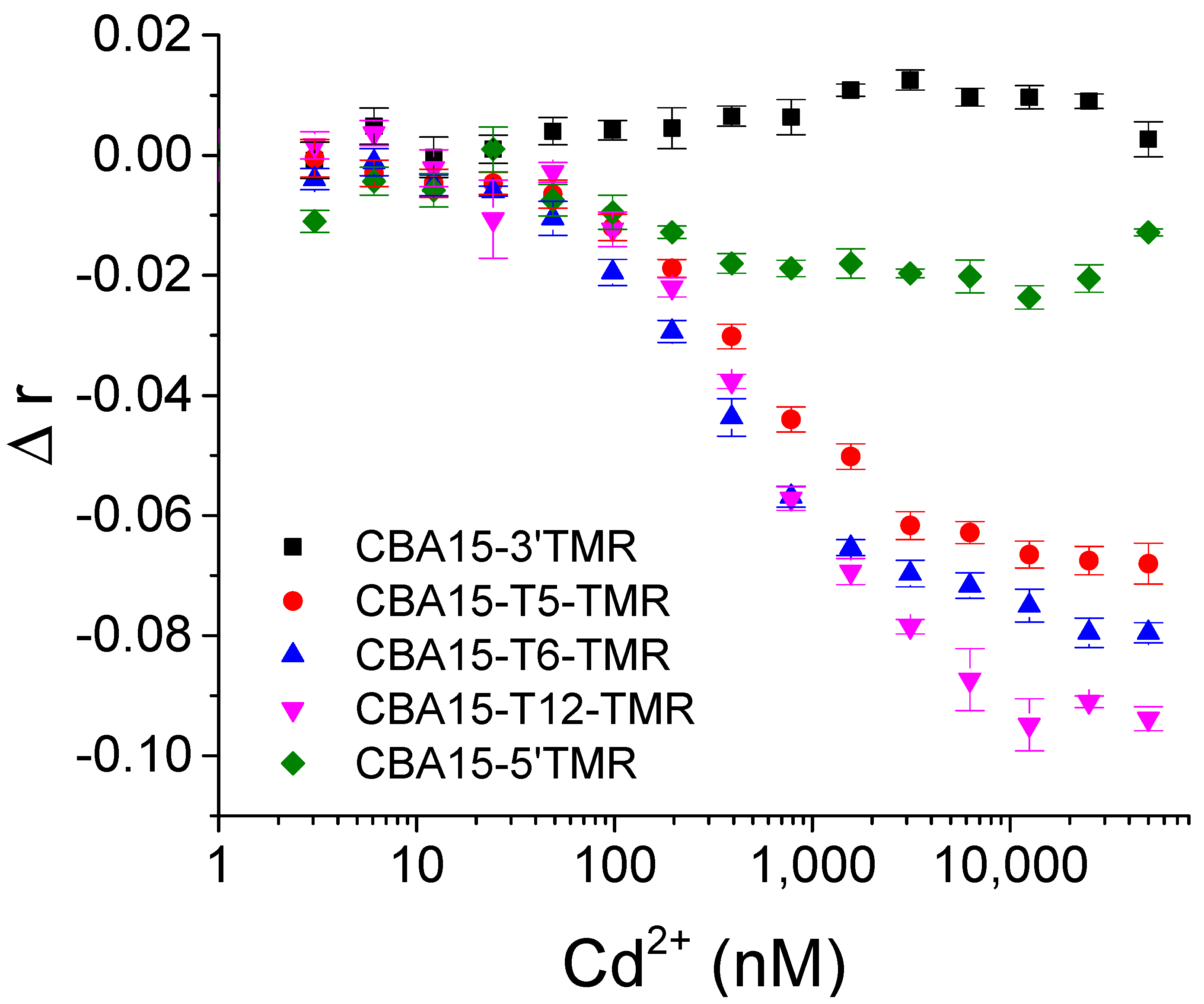

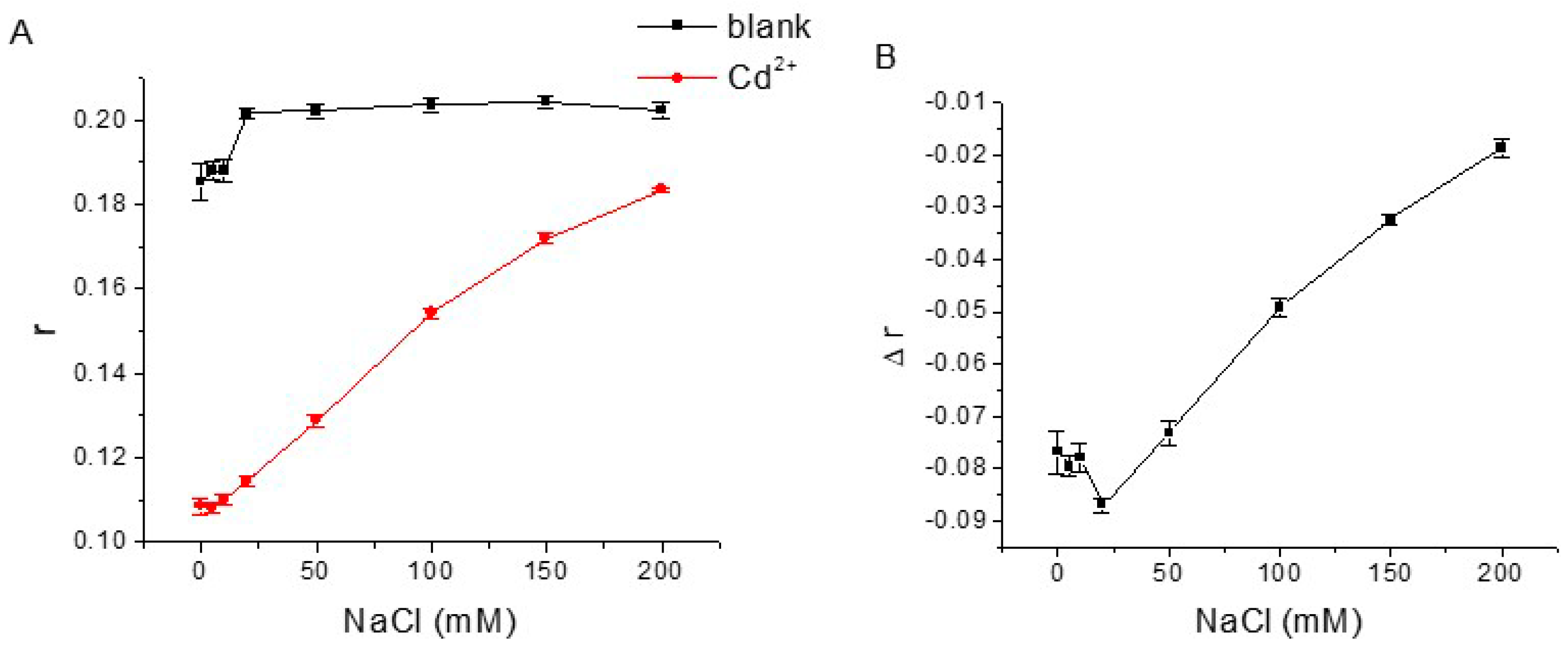
| Name | Sequences |
|---|---|
| CBA15 | 5′-CGG GTT CAC AGT CCG-3′ |
| CBA15-3′-TMR | 5′-CGG GTT CAC AGT CCG-TMR-3′ a |
| CBA15-T5-TMR | 5′-CGG GT(TMR)T CAC AGT CCG-3′ |
| CBA15-T6-TMR | 5′-CGG GTT(TMR) CAC AGT CCG-3′ |
| CBA15-T12-TMR | 5′-CGG GTT CAC AGT(TMR) CCG-3′ |
| CBA15-5′-TMR | 5′-TMR-CGG GTT CAC AGT CCG-3′ |
| CBA15-G3S-T12-TMR | 5′-CGGPS GTT CAC AGT(TMR) CCG-3′ b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, H.; Zhao, Q. A Sensitive Aptamer Fluorescence Anisotropy Sensor for Cd2+ Using Affinity-Enhanced Aptamers with Phosphorothioate Modification. Biosensors 2022, 12, 887. https://doi.org/10.3390/bios12100887
Yu H, Zhao Q. A Sensitive Aptamer Fluorescence Anisotropy Sensor for Cd2+ Using Affinity-Enhanced Aptamers with Phosphorothioate Modification. Biosensors. 2022; 12(10):887. https://doi.org/10.3390/bios12100887
Chicago/Turabian StyleYu, Hao, and Qiang Zhao. 2022. "A Sensitive Aptamer Fluorescence Anisotropy Sensor for Cd2+ Using Affinity-Enhanced Aptamers with Phosphorothioate Modification" Biosensors 12, no. 10: 887. https://doi.org/10.3390/bios12100887
APA StyleYu, H., & Zhao, Q. (2022). A Sensitive Aptamer Fluorescence Anisotropy Sensor for Cd2+ Using Affinity-Enhanced Aptamers with Phosphorothioate Modification. Biosensors, 12(10), 887. https://doi.org/10.3390/bios12100887





