An Exosomal miRNA Biomarker for the Detection of Pancreatic Ductal Adenocarcinoma
Abstract
1. Introduction
2. Materials and Methods
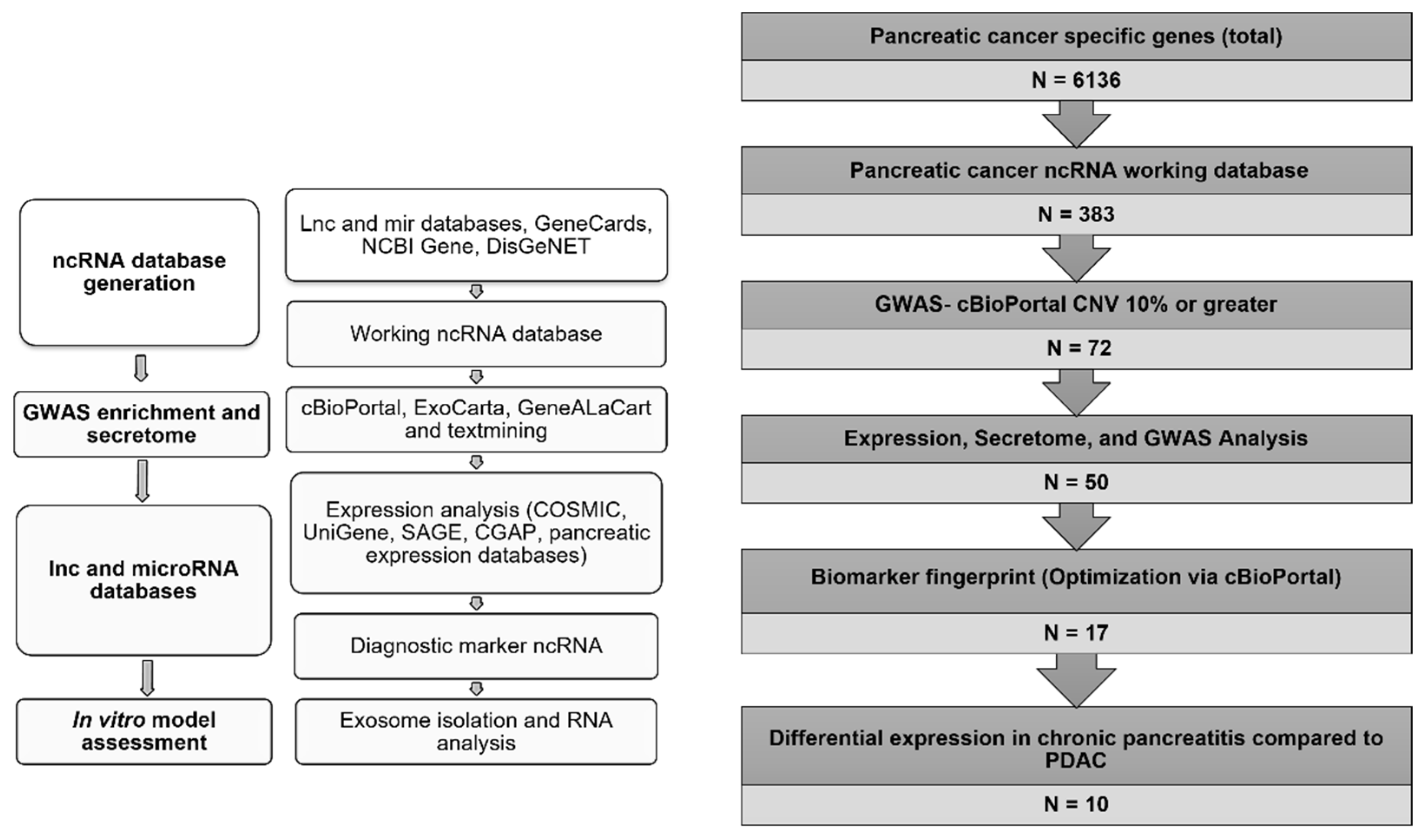
2.1. Database Generation and Biomarkers Identification
2.2. Cell Lines
2.3. Exosome Isolation from Cell Culture Media
2.4. Isolation and Profiling of Exosomal miRNA
2.5. microRNA Nomenclature
3. Results
3.1. Establishment of a Noncoding RNA Biomarker
3.1.1. Database Generation and Lead Identification
3.1.2. Selection of Biomarker Panel for Diagnostics and Screening
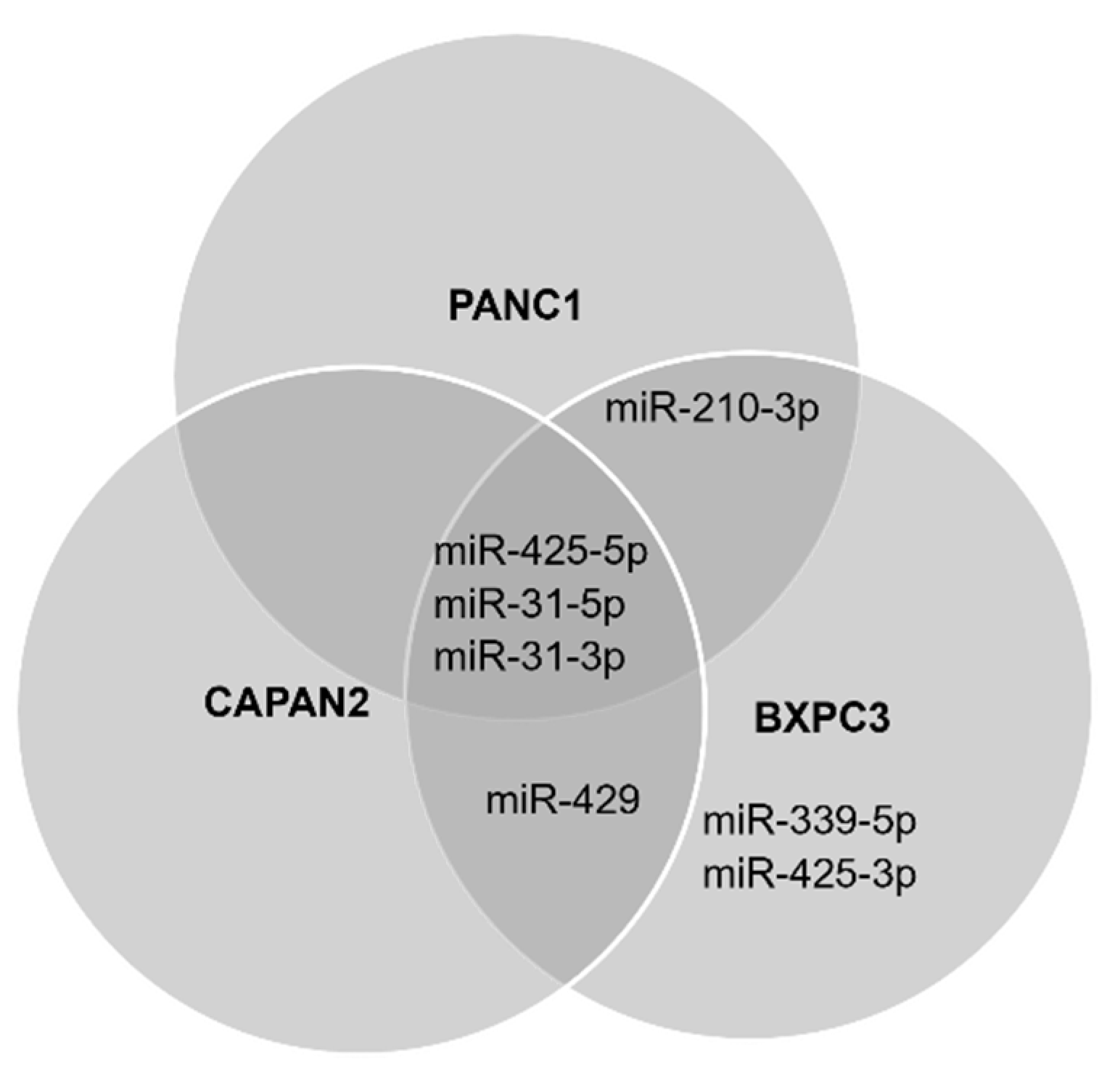
3.2. Detection of the miRNA Panel in Pancreatic Cancer In Vitro Models
3.3. Cell Preferences for Mature miRNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balachandran, V.P.; Beatty, G.L.; Dougan, S.K. Broadening the Impact of Immunotherapy to Pancreatic Cancer: Challenges and Opportunities. Gastroenterology 2019, 156, 2056–2072. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting Cancer Incidence and Deaths to 2030: The Unexpected Burden of Thyroid, Liver, and Pancreas Cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.R.; Cannon, A.; Atri, P.; Wichman, C.S.; Smith, L.M.; Ganti, A.K.; Are, C.; Sasson, A.R.; Kumar, S.; Batra, S.K. Advanced Pancreatic Cancer: A Meta-Analysis of Clinical Trials over Thirty Years. Oncotarget 2018, 9, 19396–19405. [Google Scholar] [CrossRef] [PubMed]
- Guillén-Ponce, C.; Blázquez, J.; González, I.; De-Madaria, E.; Montáns, J.; Carrato, A. Diagnosis and Staging of Pancreatic Ductal Adenocarcinoma. Clin. Transl. Oncol. 2017, 19, 1205–1216. [Google Scholar] [CrossRef] [PubMed]
- Gbolahan, O.B.; Tong, Y.; Sehdev, A.; O’Neil, B.; Shahda, S. Overall Survival of Patients with Recurrent Pancreatic Cancer Treated with Systemic Therapy: A Retrospective Study. BMC Cancer 2019, 19, 468. [Google Scholar] [CrossRef]
- Cid-Arregui, A.; Juarez, V. Perspectives in the Treatment of Pancreatic Adenocarcinoma. World J. Gastroenterol. 2015, 21, 9297–9316. [Google Scholar] [CrossRef]
- Scarà, S.; Bottoni, P.; Scatena, R. CA 19-9: Biochemical and Clinical Aspects. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2015; Volume 867, pp. 247–260. [Google Scholar]
- Thomsen, M.; Skovlund, E.; Sorbye, H.; Bolstad, N.; Nustad, K.J.; Glimelius, B.; Pfeiffer, P.; Kure, E.H.; Johansen, J.S.; Tveit, K.M.; et al. Prognostic Role of Carcinoembryonic Antigen and Carbohydrate Antigen 19-9 in Metastatic Colorectal Cancer: A BRAF-Mutant Subset with High CA 19-9 Level and Poor Outcome. Br. J. Cancer 2018, 118, 1609–1616. [Google Scholar] [CrossRef]
- Yang, G.-B. Clinical Value of Serum Cancer Antigen 19-9 as a Tumor Screening Marker among Healthy Individuals. JBUON 2015, 20, 1612–1616. [Google Scholar]
- Takaori, K.; Bassi, C.; Biankin, A.; Brunner, T.B.; Cataldo, I.; Campbell, F.; Cunningham, D.; Falconi, M.; Frampton, A.E.; Furuse, J.; et al. International Association of Pancreatology (IAP)/European Pancreatic Club (EPC) Consensus Review of Guidelines for the Treatment of Pancreatic Cancer. Pancreatology 2016, 16, 14–27. [Google Scholar] [CrossRef]
- Bekkali, N.L.H.; Oppong, K.W. Pancreatic Ductal Adenocarcinoma Epidemiology and Risk Assessment: Could We Prevent? Possibility for an Early Diagnosis. Endosc. Ultrasound 2017, 6, S58–S61. [Google Scholar]
- Principe, D.R.; Rana, A. Updated Risk Factors to Inform Early Pancreatic Cancer Screening and Identify High Risk Patients. Cancer Lett. 2020, 485, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Kirkegård, J.; Mortensen, F.V.; Cronin-Fenton, D. Chronic Pancreatitis and Pancreatic Cancer Risk: A Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 2017, 112, 1366–1372. [Google Scholar] [CrossRef] [PubMed]
- Paroder, V.; Flusberg, M.; Kobi, M.; Rozenblit, A.M.; Chernyak, V. Pancreatic Cysts: What Imaging Characteristics Are Associated with Development of Pancreatic Ductal Adenocarcinoma? Eur. J. Radiol. 2016, 85, 1622–1626. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zamboni, G.; Hirabayashi, K.; Castelli, P.; Lennon, A.M. Precancerous Lesions of the Pancreas. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 299–322. [Google Scholar] [CrossRef] [PubMed]
- Zaccari, P.; Cardinale, V.; Severi, C.; Pedica, F.; Carpino, G.; Gaudio, E.; Doglioni, C.; Petrone, M.C.; Alvaro, D.; Arcidiacono, P.G.; et al. Common Features between Neoplastic and Preneoplastic Lesions of the Biliary Tract and the Pancreas. World J. Gastroenterol. 2019, 25, 4343–4359. [Google Scholar] [CrossRef]
- Duggan, S.N.; Ní Chonchubhair, H.M.; Lawal, O.; O’Connor, D.B.; Conlon, K.C. Chronic Pancreatitis: A Diagnostic Dilemma. World J. Gastroenterol. 2016, 22, 2304–2313. [Google Scholar] [CrossRef]
- Brugge, W.R. Diagnosis and Management of Cystic Lesions of the Pancreas. J. Gastrointest. Oncol. 2015, 6, 375–388. [Google Scholar]
- Kahlert, C.; Kalluri, R. Exosomes in Tumor Microenvironment Influence Cancer Progression and Metastasis. J. Mol. Med. 2013, 91, 431–437. [Google Scholar] [CrossRef]
- Makler, A.; Narayanan, R. Mining Exosomal Genes for Pancreatic Cancer Targets. Cancer Genom. Proteom. 2017, 14, 161–172. [Google Scholar] [CrossRef]
- Caivano, A.; Laurenzana, I.; De Luca, L.; La Rocca, F.; Simeon, V.; Trino, S.; D’Auria, F.; Traficante, A.; Maietti, M.; Izzo, T.; et al. High Serum Levels of Extracellular Vesicles Expressing Malignancy-Related Markers Are Released in Patients with Various Types of Hematological Neoplastic Disorders. Tumor Biol. 2015, 36, 9739–9752. [Google Scholar] [CrossRef]
- Cappello, F.; Logozzi, M.; Campanella, C.; Bavisotto, C.C.; Marcilla, A.; Properzi, F.; Fais, S. Exosome Levels in Human Body Fluids: A Tumor Marker by Themselves? Eur. J. Pharm. Sci. 2017, 96, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Patel, T.; Freedman, J.E. Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med. 2018, 379, 958–966. [Google Scholar] [CrossRef] [PubMed]
- Makler, A.; Asghar, W. Exosomal Biomarkers for Cancer Diagnosis and Patient Monitoring. Expert Rev. Mol. Diagn. 2020, 20, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.-Y.; Li, M.-X.; Pan, W.-L.; Chen, Y.; Li, M.-M.; Pang, J.-X.; Zheng, L.; Chen, J.-X.; Duan, W.-J. In Situ Detection of Plasma Exosomal MicroRNA-1246 for Breast Cancer Diagnostics by a Au Nanoflare Probe. ACS Appl. Mater. Interfaces 2018, 10, 39478–39486. [Google Scholar] [CrossRef]
- Hydbring, P.; De Petris, L.; Zhang, Y.; Brandén, E.; Koyi, H.; Novak, M.; Kanter, L.; Hååg, P.; Hurley, J.; Tadigotla, V.; et al. Exosomal RNA-Profiling of Pleural Effusions Identifies Adenocarcinoma Patients through Elevated MiR-200 and LCN2 Expression. Lung Cancer 2018, 124, 45–52. [Google Scholar] [CrossRef]
- Jin, D.; Yang, F.; Zhang, Y.; Liu, L.; Zhou, Y.; Wang, F.; Zhang, G.J. ExoAPP: Exosome-Oriented, Aptamer Nanoprobe-Enabled Surface Proteins Profiling and Detection. Anal. Chem. 2018, 90, 14402–14411. [Google Scholar] [CrossRef]
- Erb, U.; Zoller, M. Progress and Potential of Exosome Analysis for Early Pancreatic Cancer Detection. Expert Rev. Mol. Diagn. 2016, 16, 757–767. [Google Scholar] [CrossRef]
- Nuzhat, Z.; Kinhal, V.; Sharma, S.; Rice, G.E.; Joshi, V.; Salomon, C. Tumour-Derived Exosomes as a Signature of Pancreatic Cancer-Liquid Biopsies as Indicators of Tumour Progression. Oncotarget 2017, 8, 17279–17291. [Google Scholar] [CrossRef]
- Li, A.; Zhang, T.; Zheng, M.; Liu, Y.; Chen, Z. Exosomal Proteins as Potential Markers of Tumor Diagnosis. J. Hematol. Oncol. 2017, 10, 175. [Google Scholar] [CrossRef]
- Sharma, R.; Huang, X.; Brekken, R.A.; Schroit, A.J. Detection of Phosphatidylserine-Positive Exosomes for the Diagnosis of Early-Stage Malignancies. Br. J. Cancer 2017, 117, 545–552. [Google Scholar] [CrossRef]
- Xue, M.; Zhuo, Y.; Shan, B. MicroRNAs, Long Noncoding RNAs, and Their Functions in Human Disease. Methods Mol. Biol. 2017, 1617, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-Coding Rnas in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-Coding RNAs as Regulators in Epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.-C.; Ni, J.-J.; Cui, W.-Y.; Wang, B.-Y.; Zhuo, W. Emerging Roles of LncRNA in Cancer and Therapeutic Opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366. [Google Scholar] [PubMed]
- Dai, X.; Kaushik, A.C.; Zhang, J. The Emerging Role of Major Regulatory RNAs in Cancer Control. Front. Oncol. 2019, 9, 920. [Google Scholar] [CrossRef] [PubMed]
- Carlevaro-Fita, J.; Lanzós, A.; Feuerbach, L.; Hong, C.; Mas-Ponte, D.; Pedersen, J.S.; Abascal, F.; Amin, S.B.; Bader, G.D.; Barenboim, J.; et al. Cancer LncRNA Census Reveals Evidence for Deep Functional Conservation of Long Noncoding RNAs in Tumorigenesis. Commun. Biol. 2020, 3, 56. [Google Scholar] [CrossRef]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A Comprehensive Platform Integrating Information on Human Disease-Associated Genes and Variants. Nucleic Acids Res. 2017, 45, D833–D839. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Thomas, J.K.; Kim, M.S.; Balakrishnan, L.; Nanjappa, V.; Raju, R.; Marimuthu, A.; Radhakrishnan, A.; Muthusamy, B.; Khan, A.A.; Sakamuri, S.; et al. Pancreatic Cancer Database: An Integrative Resource for Pancreatic Cancer. Cancer Biol. Ther. 2014, 15, 963–967. [Google Scholar] [CrossRef]
- Jiang, Q.; Wang, Y.; Hao, Y.; Juan, L.; Teng, M.; Zhang, X.; Li, M.; Wang, G.; Liu, Y. MiR2Disease: A Manually Curated Database for MicroRNA Deregulation in Human Disease. Nucleic Acids Res. 2009, 37, D98–D104. [Google Scholar] [CrossRef]
- Xie, B.; Ding, Q.; Han, H.; Wu, D. MiRCancer: A MicroRNA-Cancer Association Database Constructed by Text Mining on Literature. Bioinformatics 2013, 29, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Ning, S.; Zhang, J.; Wang, P.; Zhi, H.; Wang, J.; Liu, Y.; Gao, Y.; Guo, M.; Yue, M.; Wang, L.; et al. Lnc2Cancer: A Manually Curated Database of Experimentally Supported LncRNAs Associated with Various Human Cancers. Nucleic Acids Res. 2016, 44, D980–D985. [Google Scholar] [CrossRef] [PubMed]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Binns, D.; Dimmer, E.; Huntley, R.; Barrell, D.; O’Donovan, C.; Apweiler, R. QuickGO: A Web-Based Tool for Gene Ontology Searching. Bioinformatics 2009, 25, 3045–3046. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Marzec, J.; Dayem Ullah, A.Z.; Pirrò, S.; Gadaleta, E.; Crnogorac-Jurcevic, T.; Lemoine, N.R.; Kocher, H.M.; Chelala, C. The Pancreatic Expression Database: 2018 Update. Nucleic Acids Res. 2018, 46, D1107–D1110. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.M.; Groehler, A.L.; Lee, K.M.; Ouellette, M.M.; Khazak, V.; Der, C.J. K-Ras Promotes Growth Transformation and Invasion of Immortalized Human Pancreatic Cells by Raf and Phosphatidylinositol 3-Kinase Signaling. Cancer Res. 2007, 67, 2098–2106. [Google Scholar] [CrossRef]
- Gevaert, A.B.; Witvrouwen, I.; Vrints, C.J.; Heidbuchel, H.; van Craenenbroeck, E.M.; van Laere, S.J.; van Craenenbroeck, A.H. MicroRNA Profiling in Plasma Samples Using QPCR Arrays: Recommendations for Correct Analysis and Interpretation. PLoS ONE 2018, 13, e0193173. [Google Scholar] [CrossRef]
- Budak, H.; Bulut, R.; Kantar, M.; Alptekin, B. MicroRNA Nomenclature and the Need for a Revised Naming Prescription. Brief. Funct. Genom. 2016, 15, 65–71. [Google Scholar] [CrossRef]
- Kent, O.A.; Mendell, J.T.; Rottapel, R. Transcriptional Regulation of MiR-31 by Oncogenic KRAS Mediates Metastatic Phenotypes by Repressing RASA1. Mol. Cancer Res. 2016, 14, 267–277. [Google Scholar] [CrossRef]
- Lundberg, I.V.; Wikberg, M.L.; Ljuslinder, I.; Li, X.; Myte, R.; Zingmark, C.; Löfgren-Burström, A.; Edin, S.; Palmqvist, R. MicroRNA Expression in KRAS- and BRAF-Mutated Colorectal Cancers. Anticancer Res. 2018, 38, 677–683. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Ma, P.; Wu, D.; Shu, Y.; Gao, W. Functions and Mechanisms of MicroRNA-31 in Human Cancers. Biomed. Pharmacother. 2018, 108, 1162–1169. [Google Scholar] [CrossRef] [PubMed]
- Edmonds, M.D.; Boyd, K.L.; Moyo, T.; Mitra, R.; Duszynski, R.; Arrate, M.P.; Chen, X.; Zhao, Z.; Blackwell, T.S.; Andl, T.; et al. MicroRNA-31 Initiates Lung Tumorigenesis and Promotes Mutant KRAS-Driven Lung Cancer. J. Clin. Investig. 2016, 126, 349–364. [Google Scholar] [CrossRef]
- Diaz-Riascos, Z.V.; Ginesta, M.M.; Fabregat, J.; Serrano, T.; Busquets, J.; Buscail, L.; Cordelier, P.; Capellá, G. Expression and Role of MicroRNAs from the MiR-200 Family in the Tumor Formation and Metastatic Propensity of Pancreatic Cancer. Mol. Ther.-Nucleic Acids 2019, 17, 491–503. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Jia, B.; Cheng, Y.; Zhou, L.; Qian, B.; Liu, Z.; Wang, Y. MicroRNA-429 Sensitizes Pancreatic Cancer Cells to Gemcitabine through Regulation of PDCD4. Am. J. Transl. Res. 2017, 9, 5048–5055. [Google Scholar]
- Liu, D.; Song, L.; Dai, Z.; Guan, H.; Kang, H.; Zhang, Y.; Yan, W.; Zhao, X.; Zhang, S. MiR-429 Suppresses Neurotrophin-3 to Alleviate Perineural Invasion of Pancreatic Cancer. Biochem. Biophys. Res. Commun. 2018, 505, 1077–1083. [Google Scholar] [CrossRef]
- Guo, C.; Zhao, D.; Zhang, Q.; Liu, S.; Sun, M.Z. MiR-429 Suppresses Tumor Migration and Invasion by Targeting CRKL in Hepatocellular Carcinoma via Inhibiting Raf/MEK/ERK Pathway and Epithelial-Mesenchymal Transition. Sci. Rep. 2018, 8, 2375. [Google Scholar] [CrossRef]
- Lin, M.H.; Chen, Y.Z.; Lee, M.Y.; Weng, K.P.; Chang, H.T.; Yu, S.Y.; Dong, B.J.; Kuo, F.R.; Hung, L.T.; Liu, L.F.; et al. Comprehensive Identification of MicroRNA Arm Selection Preference in Lung Cancer: MiR-324-5p and -3p Serve Oncogenic Functions in Lung Cancer. Oncol. Letters 2018, 15, 9818–9826. [Google Scholar] [CrossRef]
- Li, S.C.; Tsai, K.W.; Pan, H.W.; Jeng, Y.M.; Ho, M.R.; Li, W.H. MicroRNA 3’ End Nucleotide Modification Patterns and Arm Selection Preference in Liver Tissues. BMC Syst. Biol. 2012, 6, S14. [Google Scholar] [CrossRef]
- Tsai, K.W.; Leung, C.M.; Lo, Y.H.; Chen, T.W.; Chan, W.C.; Yu, S.Y.; Tu, Y.T.; Lam, H.C.; Li, S.C.; Ger, L.P.; et al. Arm Selection Preference of MicroRNA-193a Varies in Breast Cancer. Sci. Rep. 2016, 6, 28176. [Google Scholar] [CrossRef]
- Chen, L.; Sun, H.; Wang, C.; Yang, Y.; Zhang, M.; Wong, G. MiRNA Arm Switching Identifies Novel Tumour Biomarkers. EBioMedicine 2018, 38, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of Secretion and Uptake of Exosomes and Other Extracellular Vesicles for Cell-to-Cell Communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, O.; Russo, A.; Scarpa, A.; Santini, D.; Reni, M.; Bittoni, A.; Azzariti, A.; Aprile, G.; Delcuratolo, S.; Signorile, M.; et al. MicroRNA in Pancreatic Adenocarcinoma: Predictive/Prognostic Biomarkers or Therapeutic Targets? Oncotarget 2015, 6, 23323–23341. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.F.; Zhuang, Y.Y.; Huang, F.T.; Zhang, S.N. Noncoding RNAs and Pancreatic Cancer. World J. Gastroenterol. 2016, 22, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, X.; Su, X. Noncoding RNAs in Cancer Immunity: Functions, Regulatory Mechanisms, and Clinical Application. Mol. Cancer 2020, 19, 48. [Google Scholar] [CrossRef]
- Cheetham, S.W.; Gruhl, F.; Mattick, J.S.; Dinger, M.E. Long Noncoding RNAs and the Genetics of Cancer. Br. J. Cancer 2013, 108, 2419–2425. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Chen, Y. Insight into the Role of Long Noncoding RNA in Cancer Development and Progression. Int. Rev. Cell Mol. Biol. 2016, 326, 33–65. [Google Scholar]
- Yu, X.; Zheng, H.; Tse, G.; Zhang, L.; Wu, W.K.K. CASC2: An Emerging Tumour-Suppressing Long Noncoding RNA in Human Cancers and Melanoma. Cell Prolif. 2018, 51, e12506. [Google Scholar] [CrossRef]
- Baradaran, B.; Shahbazi, R.; Khordadmehr, M. Dysregulation of Key MicroRNAs in Pancreatic Cancer Development. Biomed. Pharmacother. 2019, 109, 1008–1015. [Google Scholar] [CrossRef]
- Slack, F.J.; Chinnaiyan, A.M. The Role of Non-Coding RNAs in Oncology. Cell 2019, 179, 1033–1055. [Google Scholar] [CrossRef]
- Qin, Y.; Dang, X.; Li, W.; Ma, Q. MiR-133a Functions as a Tumor Suppressor and Directly Targets FSCN1 in Pancreatic Cancer. Oncol. Res. 2014, 21, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Liu, T.; Huang, P.; Yan, W.; Guo, C.; Xiong, L.; Liu, A. USP39, a Direct Target of MicroRNA-133a, Promotes Progression of Pancreatic Cancer via the AKT Pathway. Biochem. Biophys. Res. Commun. 2017, 486, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Li, Q.; Zhou, Y.; Wang, X.; Zhang, Q.; Wang, Y.; Zhuang, H.; Jiang, X.; Xiong, W. The Long Coding RNA AFAP1-AS1 Promotes Tumor Cell Growth and Invasion in Pancreatic Cancer through Upregulating the IGF1R Oncogene via Sequestration of MiR-133a. Cell Cycle 2018, 17, 1949–1966. [Google Scholar] [CrossRef]
- Kim, E.A.; Jang, J.H.; Sung, E.G.; Song, I.H.; Kim, J.Y.; Lee, T.J. MiR-1208 Increases the Sensitivity to Cisplatin by Targeting TBCK in Renal Cancer Cells. Int. J. Mol. Sci. 2019, 20, 3540. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.X.; Sun, J.X.; Zhao, J.H.; Yang, Y.C.; Shi, J.X.; Wu, Z.H.; Chen, X.W.; Gao, P.; Miao, Z.F.; Wang, Z.N. Non-Coding RNAs Participate in the Regulatory Network of CLDN4 via CeRNA Mediated MiRNA Evasion. Nat. Commun. 2017, 8, 289. [Google Scholar] [CrossRef]
- Feng, S.; He, A.; Wang, D.; Kang, B. Diagnostic Significance of MiR-210 as a Potential Tumor Biomarker of Human Cancer Detection: An Updated Pooled Analysis of 30 Articles. OncoTargets Ther. 2019, 12, 479–493. [Google Scholar] [CrossRef]
- Tréhoux, S.; Lahdaoui, F.; Delpu, Y.; Renaud, F.; Leteurtre, E.; Torrisani, J.; Jonckheere, N.; Van Seuningen, I. Micro-RNAs MiR-29a and MiR-330-5p Function as Tumor Suppressors by Targeting the MUC1 Mucin in Pancreatic Cancer Cells. Biochim. Biophys. Acta-Mol. Cell Res. 2015, 1853, 2392–2403. [Google Scholar] [CrossRef]
- Xu, S.; Lei, S.L.; Liu, K.J.; Yi, S.G.; Yang, Z.L.; Yao, H.L. CircSFMBT1 Promotes Pancreatic Cancer Growth and Metastasis via Targeting MiR-330-5p/PAK1 Axis. Cancer Gene Ther. 2020, 28, 234–249. [Google Scholar] [CrossRef]
- Xiong, X.; Shi, Q.; Yang, X.; Wang, W.; Tao, J. LINC00052 Functions as a Tumor Suppressor through Negatively Modulating MiR-330-3p in Pancreatic Cancer. J. Cell. Physiol. 2019, 234, 15619–15626. [Google Scholar] [CrossRef]
- Jin, Z.H.E.; Jia, B.; Tan, L.; Liu, Y. MiR-330-3p Suppresses Liver Cancer Cell Migration by Targeting MAP2K1. Oncol. Lett. 2019, 18, 314–320. [Google Scholar] [CrossRef]
- Cai, L.; Ye, L.; Hu, X.; He, W.; Zhuang, D.; Guo, Q.; Shu, K.; Jie, Y. MicroRNA MiR-330-3p Suppresses the Progression of Ovarian Cancer by Targeting RIPK4. Bioengineered 2021, 12, 440–449. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Zhou, J.; Gu, J.; Xu, M.; Xu, X.; Yuan, G. Probing the G quadruplex from Hsa-MiR-3620-5p and Inhibition of Its Interaction with the Target Sequence. Talanta 2016, 154, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Kumar, D.; Jaggi, M.; Chauhan, S.C. Targeting MicroRNAs in Pancreatic Cancer: Microplayers in the Big Game. Cancer Res. 2013, 73, 6541–6547. [Google Scholar] [CrossRef] [PubMed]
- Słotwiński, R.; Lech, G.; Słotwińska, S.M. MicroRNAs in Pancreatic Cancer Diagnosis and Therapy. Cent. Eur. J. Immunol. 2018, 43, 314–324. [Google Scholar] [CrossRef]
- Waters, A.M.; Der, C.J. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 2018, 8, a031435. [Google Scholar] [CrossRef]
- Zeitouni, D.; Pylayeva-Gupta, Y.; Der, C.J.; Bryant, K.L. KRAS Mutant Pancreatic Cancer: No Lone Path to an Effective Treatment. Cancers 2016, 8, 45. [Google Scholar] [CrossRef]
- Monticone, M.; Biollo, E.; Maffei, M.; Donadini, A.; Romeo, F.; Storlazzi, C.T.; Giaretti, W.; Castagnola, P. Gene Expression Deregulation by KRAS G12D and G12V in a BRAF V600E Context. Mol. Cancer 2008, 7, 92. [Google Scholar] [CrossRef]
- Bournet, B.; Muscari, F.; Buscail, C.; Assenat, E.; Barthet, M.; Hammel, P.; Selves, J.; Guimbaud, R.; Cordelier, P.; Buscail, L. KRAS G12D Mutation Subtype Is A Prognostic Factor for Advanced Pancreatic Adenocarcinoma. Clin. Transl. Gastroenterol. 2016, 7, e157. [Google Scholar] [CrossRef]
- Abrams, S.L.; Lertpiriyapong, K.; Yang, L.V.; Martelli, A.M.; Cocco, L.; Ratti, S.; Falasca, M.; Murata, R.M.; Rosalen, P.L.; Lombardi, P.; et al. Introduction of WT-TP53 into Pancreatic Cancer Cells Alters Sensitivity to Chemotherapeutic Drugs, Targeted Therapeutics and Nutraceuticals. Adv. Biol. Regul. 2018, 69, 16–34. [Google Scholar] [CrossRef]
- Bernard, V.; Kim, D.U.; San Lucas, F.A.; Castillo, J.; Allenson, K.; Mulu, F.C.; Stephens, B.M.; Huang, J.; Semaan, A.; Guerrero, P.A.; et al. Circulating Nucleic Acids Are Associated with Outcomes of Patients with Pancreatic Cancer. Gastroenterology 2019, 156, 108–118.e4. [Google Scholar] [CrossRef]
- Ying, H.; Dey, P.; Yao, W.; Kimmelman, A.C.; Draetta, G.F.; Maitra, A.; Depinho, R.A. Genetics and Biology of Pancreatic Ductal Adenocarcinoma. Genes Dev. 2016, 30, 355–385. [Google Scholar] [CrossRef] [PubMed]
- Sauter, E.R. Exosomes in Blood and Cancer. Transl. Cancer Res. 2017, 6, S1316–S1320. [Google Scholar] [CrossRef]



| Stage I (N = 7) | Stage II (N = 94) | Stage III (N = 6) | Stage IV (IV = 2) |
|---|---|---|---|
| MIR27A | MIR27A | ||
| MIR31 | MIR31 | MIR27A | |
| MIR93 | MIR93 | MIR31 | |
| MIR96 | MIR96 | MIR93 | |
| MIR122 | MIR122 | MIR96 | |
| MIR130B | MIR130B | ||
| MIR133A1 | MIR130B | ||
| MIR203A | MIR203A | MIR133A1 | |
| MIR210 | MIR210 | MIR203A | |
| MIR330 | MIR330 | MIR210 | MIR210 |
| MIR339 | MIR339 | MIR330 | |
| MIR425 | MIR425 | ||
| MIR429 | MIR429 | ||
| MIR522 | MIR429 | MIR429 | |
| MIR590 | MIR522 | ||
| MIR664A | MIR590 | ||
| MIR1208 | MIR664A | ||
| MIR3620 | MIR3620 | MIR1208 | MIR1208 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makler, A.; Narayanan, R.; Asghar, W. An Exosomal miRNA Biomarker for the Detection of Pancreatic Ductal Adenocarcinoma. Biosensors 2022, 12, 831. https://doi.org/10.3390/bios12100831
Makler A, Narayanan R, Asghar W. An Exosomal miRNA Biomarker for the Detection of Pancreatic Ductal Adenocarcinoma. Biosensors. 2022; 12(10):831. https://doi.org/10.3390/bios12100831
Chicago/Turabian StyleMakler, Amy, Ramaswamy Narayanan, and Waseem Asghar. 2022. "An Exosomal miRNA Biomarker for the Detection of Pancreatic Ductal Adenocarcinoma" Biosensors 12, no. 10: 831. https://doi.org/10.3390/bios12100831
APA StyleMakler, A., Narayanan, R., & Asghar, W. (2022). An Exosomal miRNA Biomarker for the Detection of Pancreatic Ductal Adenocarcinoma. Biosensors, 12(10), 831. https://doi.org/10.3390/bios12100831






