Integration of Power-Free and Self-Contained Microfluidic Chip with Fiber Optic Particle Plasmon Resonance Aptasensor for Rapid Detection of SARS-CoV-2 Nucleocapsid Protein
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
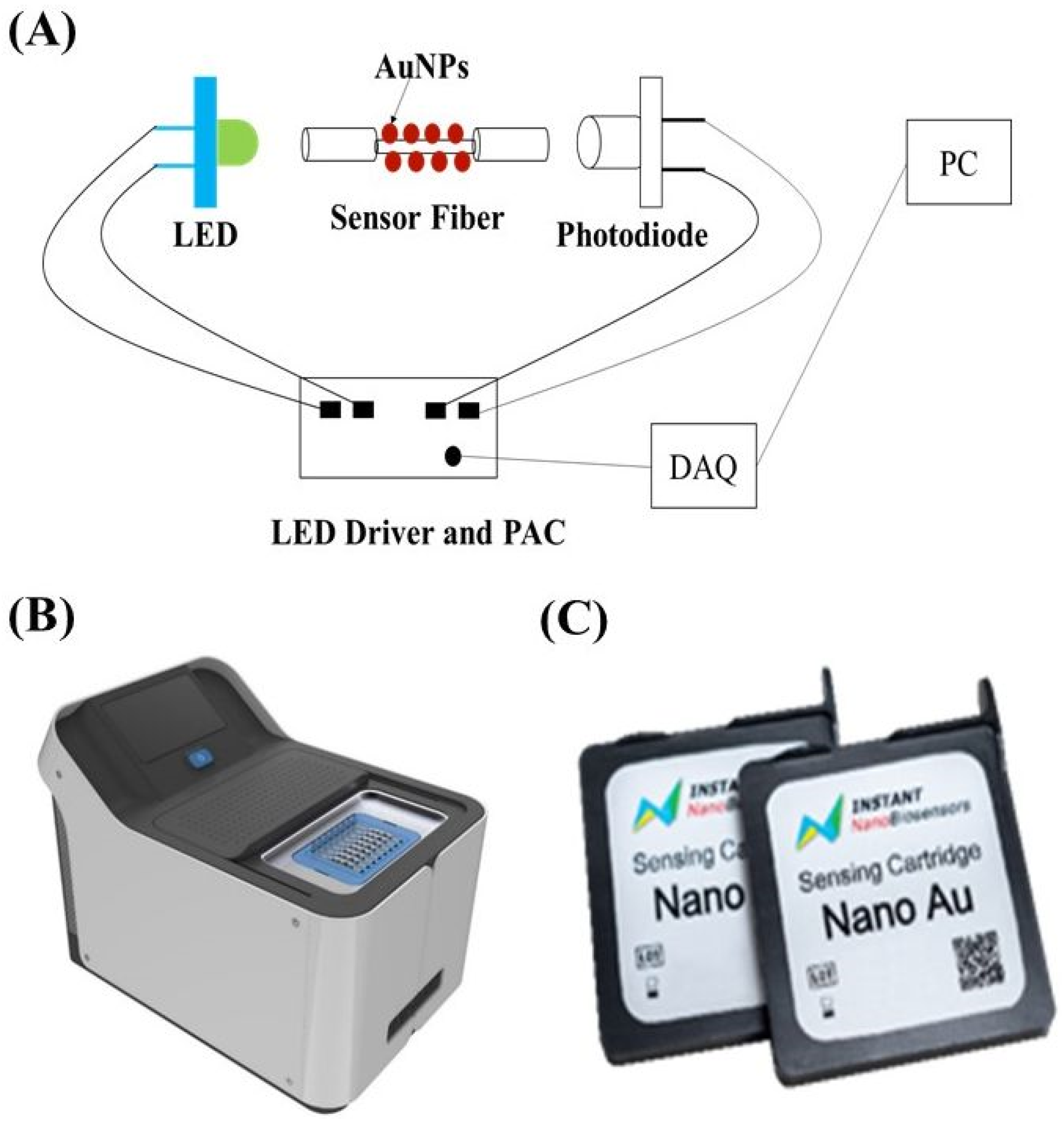
2.2. Biosensing System
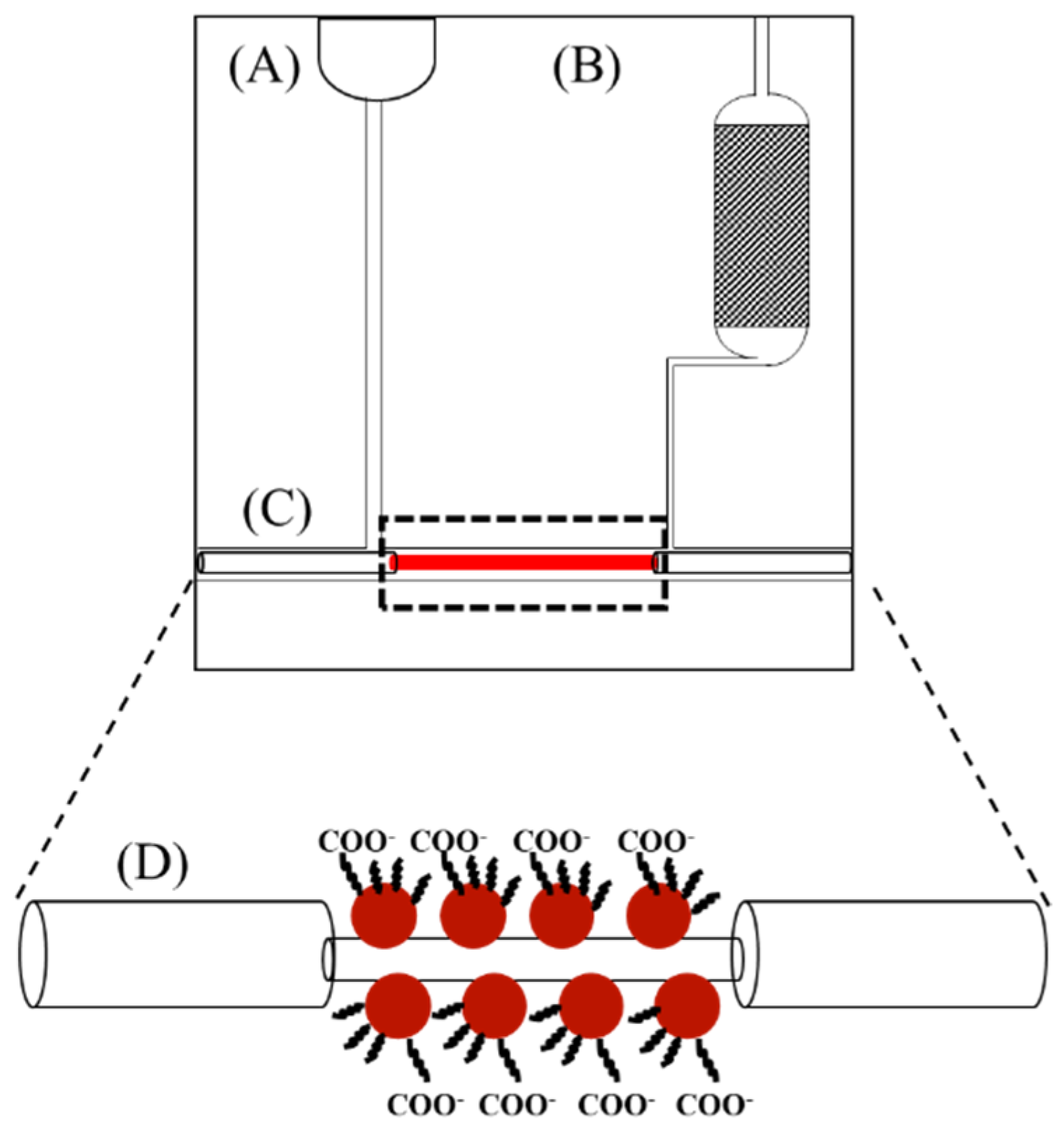
2.3. Sensor Chip and Test Protocol
2.4. Method to Calculate Binding Kinetic Constants
3. Results and Discussion
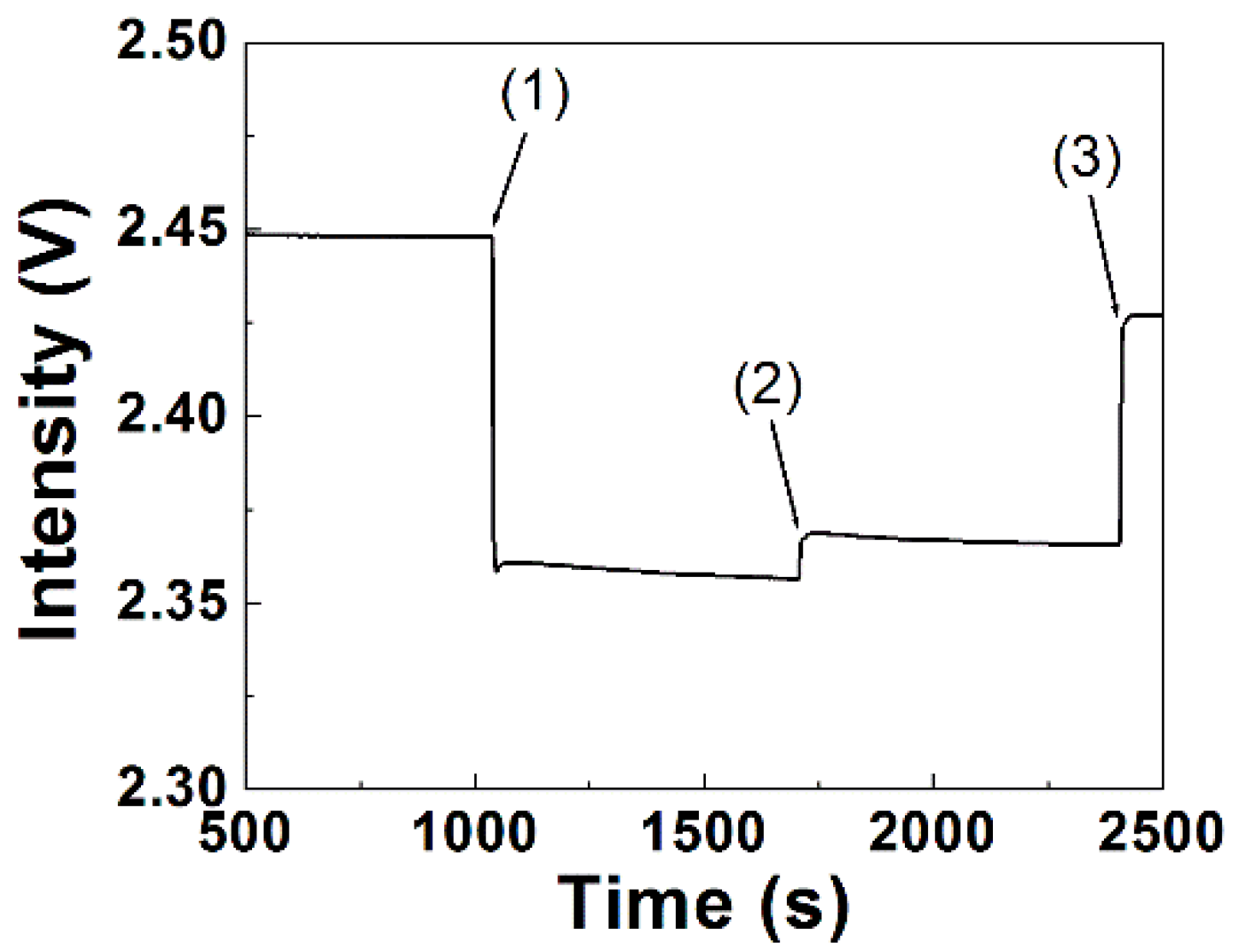
3.1. Principle of Label-Free Detection by FOPPR Biosensor
3.2. Power-Free Microfluidic Chip
3.3. Reproducibility in Bioconjugation of ssDNA Aptamer
3.4. Estimation of Binding Affinity and Kinetic Constants
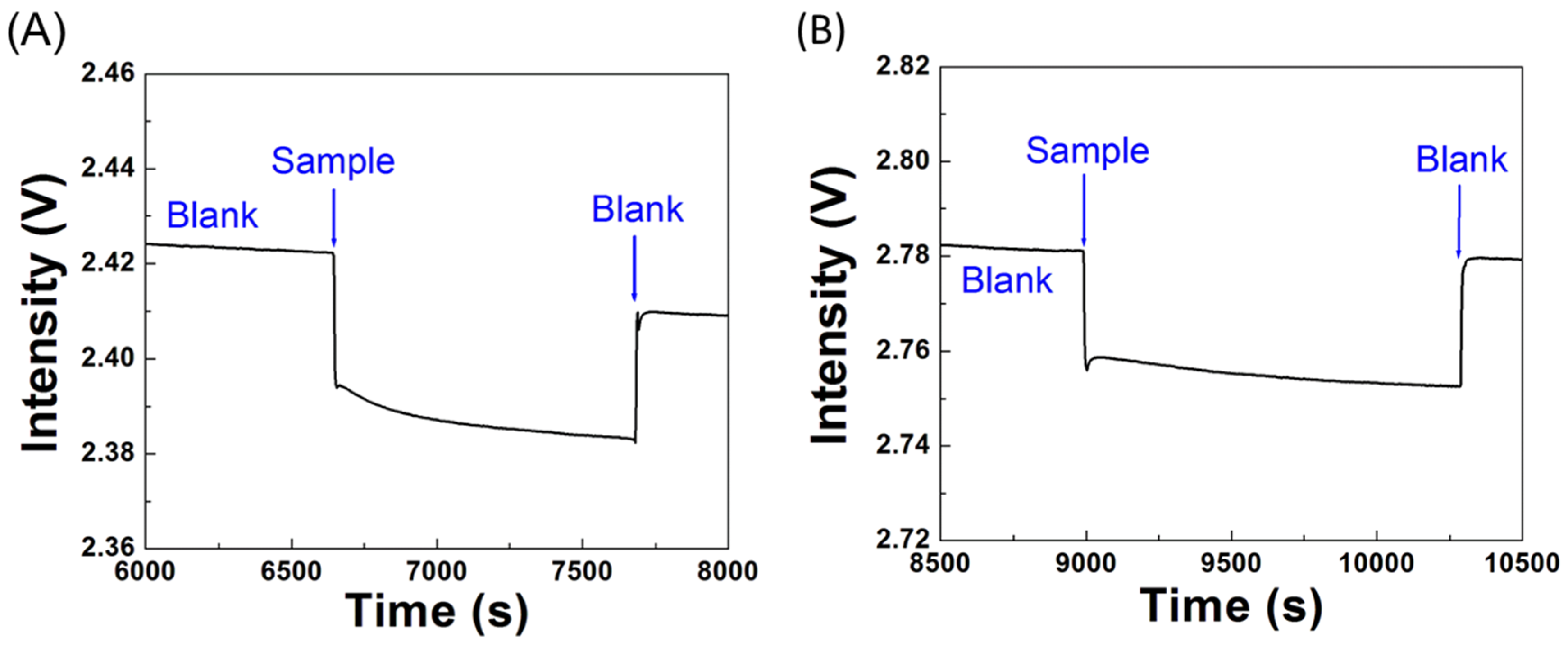
3.5. Specificity Tests
3.6. Sensitivity of the Biosensor
3.7. Real Sample Tests
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, L.; Lee, H.; Jetta, D.; Oh, K.W. Vacuum-driven power-free microfluidics utilizing the gas solubility or permeability of polydimethylsiloxane (PDMS). Lab Chip 2015, 15, 3962–3979. [Google Scholar] [CrossRef] [PubMed]
- Arata, H.; Komatsu, H.; Han, A.; Hosokawa, K.; Maeda, M. Rapid microRNA detection using power-free microfluidic chip: Coaxial stacking effect enhances the sandwich hybridization. Analyst 2012, 137, 3234–3237. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, H.; Azzazy, H.M.E. Power-free chip enzyme immunoassay for detection of prostate specific antigen (PSA) in serum. Biosens. Bioelectron. 2013, 49, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Gao, N.; Chang, J.; Dai, P.; Zhu, Z.; You, H. One-sampling and rapid analysis of cancer biomarker on a power-free and low-cost microfluidic chip. Anal. Sci. 2021, 37, 1695–1700. [Google Scholar] [CrossRef]
- Chen, H.; Kou, X.; Yang, Z.; Wang, W.N.J. Shape- and size-dependent refractive index sensitivity of gold nanoparticles. Langmuir 2008, 24, 5233–5237. [Google Scholar] [CrossRef]
- Jensen, T.R.; Duval, M.L.; Kelly, K.L.; Lazarides, A.A.; Schatz, G.C.; Duyne, R.P.V. Nanosphere lithography: effect of the external dielectric medium on the surface plasmon resonance spectrum of a periodic array of silver nanoparticles. J. Phys. Chem. B 1999, 103, 9846–9853. [Google Scholar] [CrossRef]
- Cheng, S.-F.; Chau, L.-K. Colloidal gold-modified optical fiber for chemical and biochemical sensing. Anal. Chem. 2003, 75, 16–21. [Google Scholar] [CrossRef]
- Chau, L.-K.; Lin, Y.-F.; Cheng, S.-F.; Lin, T.-J. Fiber-optic chemical and biochemical probes based on localized surface plasmon resonance. Sens. Actuators B 2006, 113, 100–105. [Google Scholar] [CrossRef]
- Wu, W.-T.; Chen, C.-H.; Chiang, C.-Y.; Chau, L.-K. Effect of surface coverage of gold nanoparticles on refractive index sensitivity in fiber-optic nanoplasmonic sensing. Sensors 2018, 18, 1759. [Google Scholar] [CrossRef]
- Lin, T.-J.; Chung, M.-F. Detection of cadmium by a fiber-optic biosensor based on localized surface plasmon resonance. Biosens. Bioelectron. 2009, 24, 1213–1218. [Google Scholar] [CrossRef]
- Fan, S.-M.; Chiang, C.-Y.; Tseng, Y.-T.; Wu, T.-Y.; Chen, Y.-L.; Huang, C.-J.; Chau, L.-K. Detection of Hg(II) at part-per-quadrillion levels by fiber optic plasmonic absorption using DNA hairpin and DNA-gold nanoparticle conjugates. ACS Appl. Nano Mater. 2021, 4, 10128–10135. [Google Scholar] [CrossRef]
- Lai, N.-S.; Wang, C.-C.; Chiang, H.-L.; Chau, L.-K. Detection of antinuclear antibodies by a colloidal gold modified optical fiber: Comparison with ELISA. Anal. Bioanl. Chem. 2007, 388, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.-Y.; Hsieh, M.-L.; Huang, K.-W.; Chau, L.-K.; Chang, C.-M.; Lyu, S.-R. Fiber-optic particle plasmon resonance sensor for detection of interleukin-1β in synovial fluids. Biosens. Bioelectron. 2010, 26, 1036–1042. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-C.; Chiang, C.-Y.; Li, C.-H.; Chang, T.-C.; Chiang, C.-S.; Chau, L.-K.; Huang, K.-W.; Wu, C.-W.; Wang, S.-C.; Lyu, S.-R. Quantification of tumor necrosis factor-α and matrix metalloproteinases-3 in synovial fluid by a fiber-optic particle plasmon resonance sensor. Analyst 2013, 138, 4599–4606. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.-Y.; Huang, T.-T.; Wang, C.-H.; Huang, C.-J.; Tsai, T.-H.; Yu, S.-N.; Chen, Y.-T.; Hong, S.-W.; Hsu, C.-W.; Chang, T.-C.; et al. Fiber optic nanogold-linked immunosorbent assay for rapid detection of procalcitonin at femtomolar concentration level. Biosens. Bioelectron. 2020, 151, 111871. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.-T.; Li, W.-Y.; Yu, Y.-W.; Chiang, C.-Y.; Liu, S.-Q.; Chau, L.-K.; Lai, N.-S.; Chou, C.-C. Fiber optic particle plasmon resonance biosensor for label-free detection of nucleic acids and its application to HLA-B27 mRNA detection in patients with ankylosing spondylitis. Sensors 2020, 20, 3137. [Google Scholar] [CrossRef]
- Ngo, L.T.; Wang, W.-K.; Tseng, Y.-T.; Chang, T.-C.; Kuo, P.-L.; Chau, L.-K.; Huang, T.-T. MutS protein-based fiber optic particle plasmon resonance biosensor for detecting single nucleotide polymorphisms. Anal. Bioanal. Chem. 2021, 413, 3329–3337. [Google Scholar] [CrossRef]
- Lin, T.-J.; Huang, K.-T.; Liu, C.-Y. Determination of organophosphorous pesticides by a novel biosensor based on localized surface plasmon resonance. Biosens. Bioelectron. 2006, 22, 513–518. [Google Scholar] [CrossRef]
- Chaudhari, P.P.; Chau, L.-K.; Tseng, Y.-T.; Huang, C.-J.; Chen, Y.-L. A fiber optic nanoplasmonic biosensor for the sensitive detection of ampicillin and its analogs. Microchim. Acta 2020, 187, 396. [Google Scholar] [CrossRef]
- Chang, T.-C.; Chiang, C.-Y.; Lin, M.-H.; Chen, I.-K.; Chau, L.-K.; Hsu, D.-S.; Shieh, S.-S.; Kuo, C.-J.; Wang, S.-C.; Chen, Y.-f. Fiber optic particle plasmon resonance immunosensor for rapid and sensitive detection of methamphetamine based on competitive inhibition. Microchem. J. 2020, 157, 105026. [Google Scholar] [CrossRef]
- Lin, H.-Y.; Huang, C.-H.; Lu, S.-H.; Kuo, I.-T.; Chau, L.-K. Direct detection of orchid viruses using nanorod-based fiber optic particle plasmon resonance immunosensor. Biosens. Bioelectron. 2014, 51, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.N. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Xiong, X.; Park, Y.-J.; Tortorici, M.A.; Snijder, J.; Quispe, J.; Cameroni, E.; Gopal, R.; Dai, M.; Lanzavecchia, A.; et al. Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 2019, 176, 1026–1039. [Google Scholar] [CrossRef] [PubMed]
- Savastano, A.; Opakua, A.I.; Rankovic, M.; Zweckstetter, M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat. Commun. 2020, 11, 6041. [Google Scholar] [CrossRef]
- Li, D.; Li, J. Immunologic testing for SARS-CoV-2 infection from the antigen perspective. J. Clin. Microbiol. 2021, 59, e02160-20. [Google Scholar] [CrossRef]
- Ou, J.; Zhou, Z.; Dai, R.; Zhao, S.; Wu, X.; Zhang, J.; Lan, W.; Cui, L.; Wu, J.; Seto, D.; et al. Emergence of SARS-CoV-2 spike RBD mutants that enhance viral infectivity through increased human ACE2 receptor binding affinity. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lan, L.; Xu, D.; Ye, G.; Xia, C.; Wang, S.; Li, Y.; Xu, H. Positive RT-PCR test results in patients recovered from COVID-19. JAMA 2020, 323, 1502–1503. [Google Scholar] [CrossRef]
- Singanayagam, A.; Patel, M.; Charlett, A.; Bernal, J.L.; Saliba, V.; Ellis, J.; Ladhani, S.; Zambon, M.; Gopal, R. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Eurosurveillance 2020, 25, 2001483. [Google Scholar] [CrossRef]
- Yam, W.C.; Chan, K.H.; Poon, L.L.M.; Guan, Y.; Yuen, K.Y.; Seto, W.H.; Peiris, J.S.M. Evaluation of reverse transcription-PCR assays for rapid diagnosis of severe acute respiratory syndrome associated with a novel coronavirus. J. Clin. Microbiol. 2003, 41, 4521–4524. [Google Scholar] [CrossRef]
- Lau, E.H.Y.; Tsang, O.T.Y.; Hui, D.S.C.; Kwan, M.Y.W.; Chan, W.-H.; Chiu, S.S.; Ko, R.L.W.; Chan, K.H.; Cheng, S.M.S.; Perera, R.A.P.M.; et al. Neutralizing antibody titres in SARS-CoV-2 infections. Nat. Commun. 2021, 12, 63. [Google Scholar] [CrossRef]
- Toropov, N.; Osborne, E.; Joshi, L.T.; Davidson, J.; Morgan, C.; Page, J.; Pepperell, J.; Vollmer, F. SARS-CoV-2 tests: Bridging the gap between laboratory sensors and clinical applications. ACS Sens. 2021, 6, 2815–2837. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, C.; Liu, R.; Liang, C.; Lu, J.; Li, J.; Tang, X.; Li, C.; Li, T. Smartphone-based high-throughput fiber-integrated immunosensing system for point-of-care testing of the SARS-CoV-2 nucleocapsid protein. ACS Sens. 2022, 7, 1985–1995. [Google Scholar] [CrossRef] [PubMed]
- Shan, D.; Johnson, J.M.; Fernandes, S.C.; Suib, H.; Hwang, S.; Wuelfing, D.; Mendes, M.; Holdridge, M.; Burke, E.M.; Beauregard, K.; et al. N-protein presents early in blood, dried blood and saliva during asymptomatic and symptomatic SARS-CoV-2 infection. Nat. Commun. 2021, 12, 1931. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, D.K.; Nandwana, V.; Henrich, S.E.; Josyula, V.P.V.; Thaxton, C.S.; Qi, C.; Simons, L.M.; Hultquist, J.F.; Ozer, E.A.; Shekhawat, G.S.; et al. Highly sensitive and ultra-rapid antigen-based detection of SARS-CoV-2 using nanomechanical sensor platform. Biosens. Bioelectron. 2022, 195, 113647. [Google Scholar] [CrossRef]
- Grant, B.D.; Anderson, C.E.; Williford, J.R.; Alonzo, L.F.; Glukhova, V.A.; Boyle, D.S.; Weigl, B.H.; Nichols, K.P. SARS-CoV-2 coronavirus nucleocapsid antigen-detecting half-strip lateral flow assay toward the development of point of care tests using commercially available reagents. Anal. Chem. 2020, 92, 11305–11309. [Google Scholar] [CrossRef]
- Kim, H.-Y.; Lee, J.-H.; Kim, M.J.; Park, S.C.; Choi, M.; Lee, W.; Ku, K.B.; Kim, B.T.; Park, E.C.; Kim, H.G.; et al. Development of a SARS-CoV-2-specific biosensor for antigen detection using scFv-Fc fusion proteins. Biosens. Bioelectron. 2021, 175, 112868. [Google Scholar] [CrossRef]
- Guo, J.; Chen, S.; Tian, S.; Liu, K.; Ni, J.; Zhao, M.; Kang, Y.; Ma, X.; Guo, J. 5G-enabled ultra-sensitive fluorescence sensor for proactive prognosis of COVID-19. Biosens. Bioelectron. 2021, 181, 113160. [Google Scholar] [CrossRef]
- Raziq, A.; Kidakova, A.; Boroznjak, R.; Reut, J.; Opik, A.; Syritski, V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021, 178, 113029. [Google Scholar] [CrossRef]
- Li, J.; Lillehoj, P.B. Microfluidic magneto immunosensor for rapid, high sensitivity measurements of SARS-CoV-2 nucleocapsid protein in serum. ACS Sens. 2021, 6, 1270–1278. [Google Scholar] [CrossRef]
- Tian, J.; Liang, Z.; Hu, O.; He, Q.; Sun, D.; Chen, Z. An electrochemical dual-aptamer biosensor based on metal-organic frameworks MIL-53 decorated with Au@Pt nanoparticles and enzymes for detection of COVID-19 nucleocapsid protein. Electrochim. Acta 2021, 387, 138553. [Google Scholar] [CrossRef]
- Shao, W.; Shurin, M.R.; Wheeler, S.E.; He, X.; Star, A. Rapid detection of SARS-CoV-2 antigens using high-purity semiconducting single-walled carbon nanotube-based field-effect transistors. ACS Appl. Mater. Interfaces 2021, 13, 10321–10327. [Google Scholar] [CrossRef] [PubMed]
- Salahandish, R.; Haghayegh, F.; Ayala-Charca, G.; Hyun, J.E.; Khalghollah, M.; Zare, A.; Far, B.; Berenger, B.M.; Niu, Y.D.; Ghafar-Zadeh, E.; et al. Bi-ECDAQ: An electrochemical dual-immuno-biosensor accompanied by a customized bi-potentiostat for clinical detection of SARS-CoV-2 nucleocapsid proteins. Biosens. Bioelectron. 2022, 203, 114018. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, D.W.; Trinh, J.T.; Ryan, M.J.; Cantu, C.M.; Lu, J.; Nicklen, F.D.; Du, Y.; Sun, R.; Wu, B.M.; Kamei, D.T. On-demand nanozyme signal enhancement at the push of a button for the improved detection of SARS-CoV-2 nucleocapsid protein in serum. Analyst 2021, 146, 7483–7490. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zhang, X.; Zhang, X.; ul ain Zahra, Q.; Huang, Z.; Chen, Y.; Song, C.; Song, M.; Jiang, H.; Luo, Z.; et al. An electrochemical aptasensor with N protein binding aptamer-complementary oligonucleotide as probe for ultra-sensitive detection of COVID-19. Biosens. Bioelectron. 2022, 213, 114436. [Google Scholar] [CrossRef] [PubMed]
- Janata, J. Ion optodes. Anal. Chem. 1992, 64, 921A–927A. [Google Scholar] [CrossRef]
- Etienne, E.E.; Nunna, B.B.; Talukder, N.; Wang, Y.; Lee, E.S. COVID-19 biomarkers and advanced sensing technologies for point-of-care (POC) diagnosis. Bioengineering 2021, 8, 98. [Google Scholar] [CrossRef]
- Valera, E.; Jankelow, A.; Lim, J.; Kindratenko, V.; Ganguli, A.; White, K.; Kumar, J.; Bashir, R. COVID-19 point-of-care diagnostics: Present and future. ACS Nano 2021, 15, 7899–7906. [Google Scholar] [CrossRef]
- Famulok, M.; Mayer, G. Aptamers and SELEX in chemistry & biology. Chem. Biol. 2014, 21, 1055–1058. [Google Scholar]
- Zhang, L.; Fang, X.; Liu, X.; Ou, H.; Zhang, H.; Wang, J.; Li, Q.; Cheng, W.; Zhang, W.; Luo, Z. Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers. Chem. Commun. 2020, 56, 10235–10238. [Google Scholar] [CrossRef]
- Chang, T.-C.; Wu, C.-C.; Wang, S.-C.; Chau, L.-K.; Hsieh, W.-H. Using a fiber optic particle plasmon resonance biosensor to determine kinetic constants of antigen-antibody binding reaction. Anal. Chem. 2013, 85, 245–250. [Google Scholar] [CrossRef]
- Chau, L.-K.; Chiang, C.-Y.; Hsu, W.-T.; Chen, S.-H.; Cheng, S.-F. Optical waveguide-based biosensors using noble metal nanoparticles. In From Bioimaging to Biosensors—Noble Metal Nanoparticles in Biodetection; Chau, L.-K., Chang, H.-T., Eds.; Pan Stanford Publishing: Singapore, 2013; Chapter 5; pp. 77–102. [Google Scholar]
- Wu, C.-W.; Chiang, C.-Y.; Chen, C.-H.; Chiang, C.-S.; Wang, C.-T.; Chau, L.-K. Self-referencing fiber optic particle plasmon resonance sensing system for real-time biological monitoring. Talanta 2016, 146, 291–298. [Google Scholar] [CrossRef] [PubMed]
- Schuck, P. Kinetics of ligand binding to receptor immobilized in a polymer matrix, as detected with an evanescent wave biosensor. 1. A computer simulation of the influence of mass transport. Biophys. J. 1996, 70, 1230–1249. [Google Scholar] [CrossRef]
- Goldstein, B.; Coombs, D.; He, X.; Pineda, A.R.; Wolsy, C. The influence of transport on the kinetics of binding to surface receptors: Application to cells and BIAcore. J. Mol. Recognit. 1999, 12, 293–299. [Google Scholar] [CrossRef]
- World Health Organization. COVID-19 Target Product Profiles for Priority Diagnostics to Support Response to the COVID-19 Pandemic. v.1.0. Available online: https://www.who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1 (accessed on 4 April 2022).
- Cennamo, N.; Pasquardini, L.; Arcadio, F.; Lunelli, L.; Vanzetti, L.; Carafa, V.; Altucci, L.; Zeni, L. SARS-CoV-2 spike protein detection through a plasmonic D-shaped plastic optical fiber aptasensor. Talanta 2021, 233, 122532. [Google Scholar] [CrossRef] [PubMed]
- Idili, A.; Parolo, C.; Alvarez-Diduk, R.; MerkoçI, A. Rapid and efficient detection of the SARS-CoV-2 spike protein using an electrochemical aptamer-based sensor. ACS Sens. 2021, 6, 3093–3101. [Google Scholar] [CrossRef]
- Abrego-Martinez, J.C.; Jafari, M.; Chergui, S.; Pavel, C.; Che, D.; Siaj, M. Aptamer-based electrochemical biosensor for rapid detection of SARS-CoV-2: Nanoscale electrode-aptamer-SARS-CoV-2 imaging by photo-induced force microscopy. Biosens. Bioelectron. 2022, 195, 113595. [Google Scholar] [CrossRef]





| Aptamer | Sequence (5′ → 3′) |
|---|---|
| NP-A48 | GCTGGATGTCGCTTACGACAATATTCCTTAGGGGCACCGCTACATTGACACATCCAGC |
| NP-A58 | GCTGGATGTCACCGGATTGTCGGACATCGGATTGTCTGAGTCATATGACACATCCAGC |
| NP-A61 | GCTGGATGTTGACCTTTACAGATCGGATTCTGTGGGGCGTTAAACTGACACATCCAGC |
| NP-A15 | GCTGGATGTTCATGCTGGCAAAATTCCTTAGGGGCACCGTTACTTTGACACATCCAGC |
| NP-A48 | NP-A58 | NP-A61 | NP-A15 | |
|---|---|---|---|---|
| Average | 0.00517 | 0.00145 | 0.00251 | 0.00440 |
| SD | 1.38 × 10−4 | 7.79 × 10−5 | 6.42 × 10−5 | 3.37 × 10−4 |
| CV | 2.7% | 5.4% | 2.6% | 7.7% |
| Aptamer | ka (M−1s−1) [FOPPR] | ka (M−1s−1) [SPR] | kd (s−1) [FOPPR] | kd (s−1) [SPR] | KD (nM) [FOPPR] | KD (nM) [SPR] |
|---|---|---|---|---|---|---|
| NP-A48 | 5.14 × 104 | 8.80 × 105 | 1.35 × 10−4 | 3.85 × 10−4 | 2.63 | 0.49 |
| NP-A58 | 1.89 × 104 | 1.92 × 104 | 7.89 × 10−5 | 1.34 × 10−4 | 4.17 | 0.70 |
| NP-A61 | 2.03 × 104 | 4.58 × 104 | 1.02 × 10−4 | 1.18 × 10−4 | 5.02 | 2.74 |
| NP-A15 | 3.24 × 104 | 2.84 × 105 | 4.44 × 10−4 | 1.16 × 10−3 | 13.70 | 4.38 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, T.-C.; Sun, A.Y.; Huang, Y.-C.; Wang, C.-H.; Wang, S.-C.; Chau, L.-K. Integration of Power-Free and Self-Contained Microfluidic Chip with Fiber Optic Particle Plasmon Resonance Aptasensor for Rapid Detection of SARS-CoV-2 Nucleocapsid Protein. Biosensors 2022, 12, 785. https://doi.org/10.3390/bios12100785
Chang T-C, Sun AY, Huang Y-C, Wang C-H, Wang S-C, Chau L-K. Integration of Power-Free and Self-Contained Microfluidic Chip with Fiber Optic Particle Plasmon Resonance Aptasensor for Rapid Detection of SARS-CoV-2 Nucleocapsid Protein. Biosensors. 2022; 12(10):785. https://doi.org/10.3390/bios12100785
Chicago/Turabian StyleChang, Ting-Chou, Aileen Y. Sun, Yu-Chung Huang, Chih-Hui Wang, Shau-Chun Wang, and Lai-Kwan Chau. 2022. "Integration of Power-Free and Self-Contained Microfluidic Chip with Fiber Optic Particle Plasmon Resonance Aptasensor for Rapid Detection of SARS-CoV-2 Nucleocapsid Protein" Biosensors 12, no. 10: 785. https://doi.org/10.3390/bios12100785
APA StyleChang, T.-C., Sun, A. Y., Huang, Y.-C., Wang, C.-H., Wang, S.-C., & Chau, L.-K. (2022). Integration of Power-Free and Self-Contained Microfluidic Chip with Fiber Optic Particle Plasmon Resonance Aptasensor for Rapid Detection of SARS-CoV-2 Nucleocapsid Protein. Biosensors, 12(10), 785. https://doi.org/10.3390/bios12100785






