Limited Substrate Specificity of PS/γ-Secretase Is Supported by Novel Multiplexed FRET Analysis in Live Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Constructs
2.2. Cell Culture and Plasmid Transfection
2.3. Confocal Microscopy and FRET Analysis
2.4. Statistical Analysis
3. Results
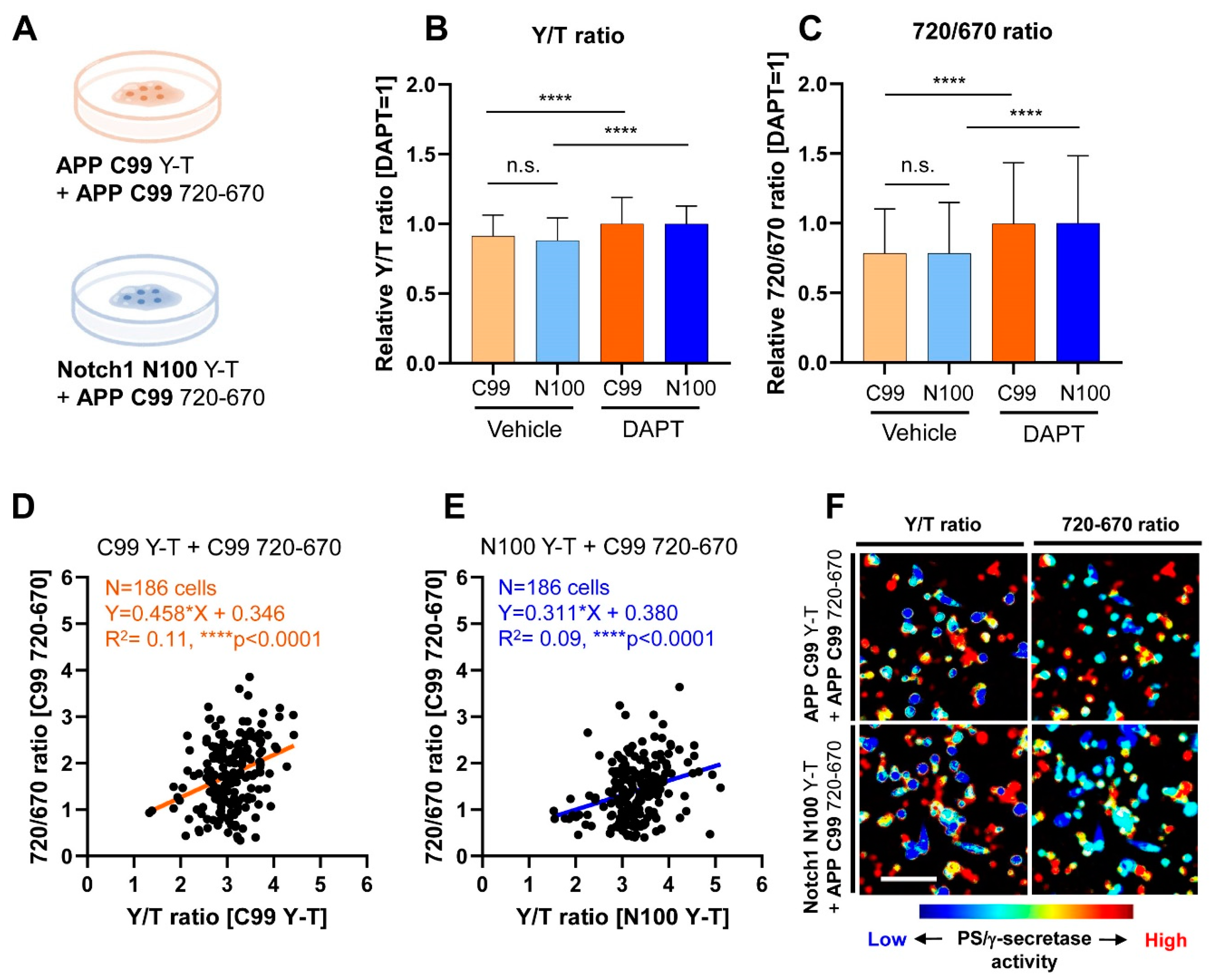
3.1. Co-Recording of APP C99 and Notch 1 N100 Processing by PS/γ-Secretase on a Cell-by-Cell Basis in Live Cells
3.2. Simultaneous Monitoring of WT and FAD-Linked Mutant APP C99 Processing by PS/γ-Secretase
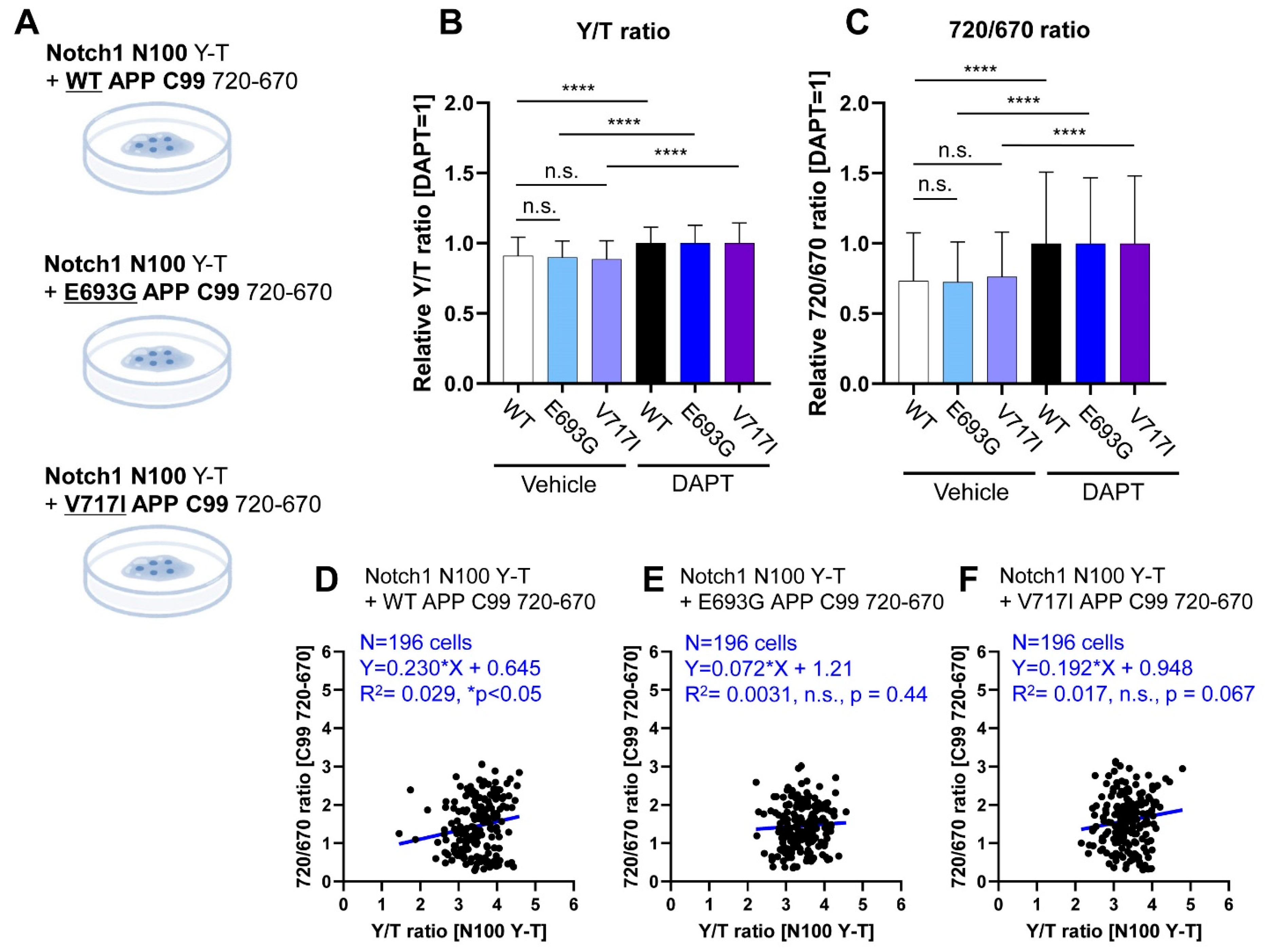
3.3. Co-Monitoring of Notch1 N100 and FAD-Linked Mutant APP C99 Processing by PS/γ-Secretase
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Strooper, B.; Saftig, P.; Craessaerts, K.; Vanderstichele, H.; Guhde, G.; Annaert, W.; Von Figura, K.; Van Leuven, F. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature 1998, 391, 387–390. [Google Scholar] [CrossRef]
- De Strooper, B.; Annaert, W.; Cupers, P.; Saftig, P.; Craessaerts, K.; Mumm, J.S.; Schroeter, E.H.; Schrijvers, V.; Wolfe, M.S.; Ray, W.J.; et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 1999, 398, 518–522. [Google Scholar] [CrossRef]
- Wolfe, M.S.; Xia, W.; Ostaszewski, B.L.; Diehl, T.S.; Kimberly, W.T.; Selkoe, D.J. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature 1999, 398, 513–517. [Google Scholar] [CrossRef] [PubMed]
- Sherrington, R.; Rogaev, E.I.; Liang, Y.; Rogaeva, E.A.; Levesque, G.; Ikeda, M.; Chi, H.; Lin, C.; Li, G.; Holman, K.; et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature 1995, 375, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Levy-Lahad, E.; Wasco, W.; Poorkaj, P.; Romano, D.M.; Oshima, J.; Pettingell, W.H.; Yu, C.E.; Jondro, P.D.; Schmidt, S.D.; Wang, K.; et al. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science 1995, 269, 973–977. [Google Scholar] [CrossRef]
- George-Hyslop, P.H.S.; E Tanzi, R.; Polinsky, R.J.; Haines, J.L.; Nee, L.; Watkins, P.C.; Myers, R.H.; Feldman, R.G.; Pollen, D.; Drachman, D.; et al. The genetic defect causing familial Alzheimer’s disease maps on chromosome 21. Science 1987, 235, 885–890. [Google Scholar] [CrossRef]
- Haapasalo, A.; Kovacs, D.M. The many substrates of presenilin/g-secretase. J. Alzheimer’s Dis. 2011, 25, 3–28. [Google Scholar] [CrossRef] [PubMed]
- Güner, G.; Lichtenthaler, S.F. The substrate repertoire of g-secretase/presenilin. Semin. Cell Dev. Biol. 2020, 105, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Bronson, R.T.; Chen, D.F.; Xia, W.; Selkoe, D.J.; Tonegawa, S. Skeletal and CNS defects in Presenilin-1-deficient mice. Cell 1997, 89, 629–639. [Google Scholar] [CrossRef]
- Wong, P.C.; Zheng, H.; Chen, H.; Becher, M.W.; Sirinathsinghji, D.J.; Trumbauer, M.E.; Chen, H.Y.; Price, D.L.; Van der Ploeg, L.H.; Sisodia, S.S. Presenilin 1 is required for Notch1 andDII1 expression in the paraxial mesoderm. Nature 1997, 387, 288–292. [Google Scholar] [CrossRef]
- Huppert, S.S.; Le, A.; Schroeter, E.H.; Mumm, J.S.; Saxena, M.T.; Milner, L.A.; Kopan, R. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature 2000, 405, 966–970. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yang, W.; Wen, W.; Sun, J.; Su, B.; Liu, B.; Ma, D.; Lv, D.; Wen, Y.; Qu, T.; et al. Gamma-secretase gene mutations in familial acne inversa. Science 2010, 330, 1065. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yan, Y.; Wang, B. γ-Secretase Genetics of Hidradenitis Suppurativa: A Systematic Literature Review. Dermatology 2020, 1–7. [Google Scholar]
- Doody, R.S.; Raman, R.; Farlow, M.; Iwatsubo, T.; Vellas, B.; Joffe, S.; Kieburtz, K.; He, F.; Sun, X.; Thomas, R.G.; et al. A phase 3 trial of semagacestat for treatment of Alzheimer’s disease. N. Engl. J. Med. 2013, 369, 341–350. [Google Scholar] [CrossRef]
- Struhl, G.; Adachi, A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol. Cell 2000, 6, 625–636. [Google Scholar] [CrossRef]
- Zhou, R.; Yang, G.; Guo, X.; Zhou, Q.; Lei, J.; Shi, Y. Recognition of the amyloid precursor protein by human gamma-secretase. Science 2019, 363, eaaw0930. [Google Scholar] [CrossRef]
- Yang, G.; Zhou, R.; Zhou, Q.; Guo, X.; Yan, C.; Ke, M.; Lei, J.; Shi, Y. Structural basis of Notch recognition by human gamma-secretase. Nature 2019, 565, 192–197. [Google Scholar] [CrossRef]
- Maesako, M.; Sekula, N.M.; Aristarkhova, A.; Feschenko, P.; Anderson, L.C.; Berezovska, O. Visualization of PS/γ-Secretase Activity in Living Cells. iScience 2020, 23, 101139. [Google Scholar] [CrossRef]
- Houser, M.C.Q.; Hou, S.S.; Perrin, F.; Turchyna, Y.; Bacskai, B.J.; Berezovska, O.; Maesako, M. A novel NIR FRET biosensor for reporting PS/γ-secretase activity in living cells. Sensors 2020, 20, 5980. [Google Scholar] [CrossRef]
- Shcherbakova, D.M.; Cammer, N.C.; Huisman, T.M.; Verkhusha, V.V.; Hodgson, L. Direct multiplex imaging and optogenetics of Rho GTPases enabled by near-infrared FRET. Nat. Chem. Biol. 2018, 14, 591–600. [Google Scholar] [CrossRef]
- Nilsberth, C.; Westlind-Danielsson, A.; Eckman, C.B.; Condron, M.M.; Axelman, K.; Forsell, C.; Stenh, C.; Luthman, J.; Teplow, D.B.; Younkin, S.G.; et al. The ‘Arctic’ APP mutation (E693G) causes Alzheimer’s disease by enhanced Abeta protofibril formation. Nat. Neurosci. 2001, 4, 887–893. [Google Scholar] [CrossRef]
- Goate, A.; Chartier-Harlin, M.C.; Mullan, M.; Brown, J.; Crawford, F.; Fidani, L.; Giuffra, L.; Haynes, A.; Irving, N.; James, L.; et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature 1991, 349, 704–706. [Google Scholar] [CrossRef] [PubMed]
- Koo, E.H.; Squazzo, S.L. Evidence That Production and Release of Amyloid Beta-Protein Involves the Endocytic Pathway. J. Biol. Chem. 1994, 269, 17386–17389. [Google Scholar] [CrossRef]
- Xia, W.; Zhang, J.; Kholodenko, D.; Citron, M.; Podlisny, M.B.; Teplow, D.B.; Haass, C.; Seubert, P.; Koo, E.H.; Selkoe, D.J. Enhanced production and oligomerization of the 42-residue amyloid beta-protein by Chinese hamster ovary cells stably expressing mutant presenilins. J. Biol. Chem. 1997, 272, 7977–7982. [Google Scholar] [CrossRef] [PubMed]
- Dovey, H.F.; John, V.; Anderson, J.P.; Chen, L.Z.; de Saint Andrieu, P.; Fang, L.Y.; Freedman, S.B.; Folmer, B.; Goldbach, E.; Holsztynska, E.J.; et al. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J. Neurochem. 2001, 76, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Mayer, S.C.; Kreft, A.F.; Harrison, B.; Abou-Gharbia, M.; Antane, M.; Aschmies, S.; Atchison, K.; Chlenov, M.; Cole, D.C.; Comery, T.; et al. Discovery of begacestat, a Notch-1-sparing gamma-secretase inhibitor for the treatment of Alzheimer’s disease. J. Med. Chem. 2008, 51, 7348–7351. [Google Scholar] [CrossRef] [PubMed]
- Gillman, K.W.; Starrett, J.E., Jr.; Parker, M.F.; Xie, K.; Bronson, J.J.; Marcin, L.R.; McElhone, K.E.; Bergstrom, C.P.; Mate, R.A.; Williams, R.; et al. Discovery and Evaluation of BMS-708163, a Potent, Selective and Orally Bioavailable γ-Secretase Inhibitor. ACS Med. Chem. Lett. 2010, 1, 120–124. [Google Scholar] [CrossRef]
- Lleó, A.; Berezovska, O.; Ramdya, P.; Fukumoto, H.; Raju, S.; Shah, T.; Hyman, B.T. Notch1 competes with the amyloid precursor protein for gamma-secretase and down-regulates presenilin-1 gene expression. J. Biol. Chem. 2003, 278, 47370–47375. [Google Scholar] [CrossRef]
- Lleó, A.; Waldron, E.; von Arnim, C.A.; Herl, L.; Tangredi, M.M.; Peltan, I.D.; Strickland, D.K.; Koo, E.H.; Hyman, B.T.; Pietrzik, C.U.; et al. Low density lipoprotein receptor-related protein (LRP) interacts with presenilin 1 and is a competitive substrate of the amyloid precursor protein (APP) for gamma-secretase. J. Biol. Chem. 2005, 280, 27303–27309. [Google Scholar] [CrossRef]
- Saito, T.; Suemoto, T.; Brouwers, N.; Sleegers, K.; Funamoto, S.; Mihira, N.; Matsuba, Y.; Yamada, K.; Nilsson, P.; Takano, J.; et al. Potent amyloidogenicity and pathogenicity of Aβ43. Nat. Neurosci. 2011, 14, 1023–1032. [Google Scholar] [CrossRef]
- Xia, D.; Watanabe, H.; Wu, B.; Lee, S.H.; Li, Y.; Tsvetkov, E.; Bolshakov, V.Y.; Shen, J.; Kelleher, R.J., 3rd. Presenilin-1 knockin mice reveal loss-of-function mechanism for familial Alzheimer’s disease. Neuron 2015, 85, 967–981. [Google Scholar] [CrossRef]
- Mehta, S.; Zhang, Y.; Roth, R.H.; Zhang, J.F.; Mo, A.; Tenner, B.; Huganir, R.L.; Zhang, J. Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat. Cell. Biol. 2018, 20, 1215–1225. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.F.; Liu, B.; Hong, I.; Mo, A.; Roth, R.H.; Tenner, B.; Lin, W.; Zhang, J.Z.; Molina, R.S.; Drobizhev, M.; et al. An ultrasensitive biosensor for high-resolution kinase activity imaging in awake mice. Nat. Chem. Biol. 2021, 17, 39–46. [Google Scholar] [CrossRef]
- Qian, Y.; Piatkevich, K.D.; Mc Larney, B.; Abdelfattah, A.S.; Mehta, S.; Murdock, M.H.; Gottschalk, S.; Molina, R.S.; Zhang, W.; Chen, Y.; et al. A genetically encoded near-infrared fluorescent calcium ion indicator. Nat. Methods 2019, 16, 171–174. [Google Scholar] [CrossRef]
- Subach, O.M.; Kunitsyna, T.A.; Mineyeva, O.A.; Lazutkin, A.A.; Bezryadnov, D.V.; Barykina, N.V.; Piatkevich, K.D.; Ermakova, Y.G.; Bilan, D.S.; Belousov, V.V.; et al. Slowly Reducible Genetically Encoded Green Fluorescent Indicator for In Vivo and Ex Vivo Visualization of Hydrogen Peroxide. Int. J. Mol. Sci. 2019, 20, 3138. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Cosio, D.M.O.; Piatkevich, K.D.; Aufmkolk, S.; Su, W.C.; Celiker, O.T.; Schohl, A.; Murdock, M.H.; Aggarwal, A.; Chang, Y.F.; et al. Improved genetically encoded near-infrared fluorescent calcium ion indicators for in vivo imaging. PLoS Biol. 2020, 18, e3000965. [Google Scholar] [CrossRef] [PubMed]
- Shemetov, A.A.; Monakhov, M.V.; Zhang, Q.; Canton-Josh, J.E.; Kumar, M.; Chen, M.; Matlashov, M.E.; Li, X.; Yang, W.; Nie, L.; et al. A near-infrared genetically encoded calcium indicator for in vivo imaging. Nat. Biotechnol. 2021, 39, 368–377. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Houser, M.C.Q.; Turchyna, Y.; Perrin, F.; Chibnik, L.; Berezovska, O.; Maesako, M. Limited Substrate Specificity of PS/γ-Secretase Is Supported by Novel Multiplexed FRET Analysis in Live Cells. Biosensors 2021, 11, 169. https://doi.org/10.3390/bios11060169
Houser MCQ, Turchyna Y, Perrin F, Chibnik L, Berezovska O, Maesako M. Limited Substrate Specificity of PS/γ-Secretase Is Supported by Novel Multiplexed FRET Analysis in Live Cells. Biosensors. 2021; 11(6):169. https://doi.org/10.3390/bios11060169
Chicago/Turabian StyleHouser, Mei C. Q., Yuliia Turchyna, Florian Perrin, Lori Chibnik, Oksana Berezovska, and Masato Maesako. 2021. "Limited Substrate Specificity of PS/γ-Secretase Is Supported by Novel Multiplexed FRET Analysis in Live Cells" Biosensors 11, no. 6: 169. https://doi.org/10.3390/bios11060169
APA StyleHouser, M. C. Q., Turchyna, Y., Perrin, F., Chibnik, L., Berezovska, O., & Maesako, M. (2021). Limited Substrate Specificity of PS/γ-Secretase Is Supported by Novel Multiplexed FRET Analysis in Live Cells. Biosensors, 11(6), 169. https://doi.org/10.3390/bios11060169





