Breast Mass Classification Using Diverse Contextual Information and Convolutional Neural Network
Abstract
:1. Introduction
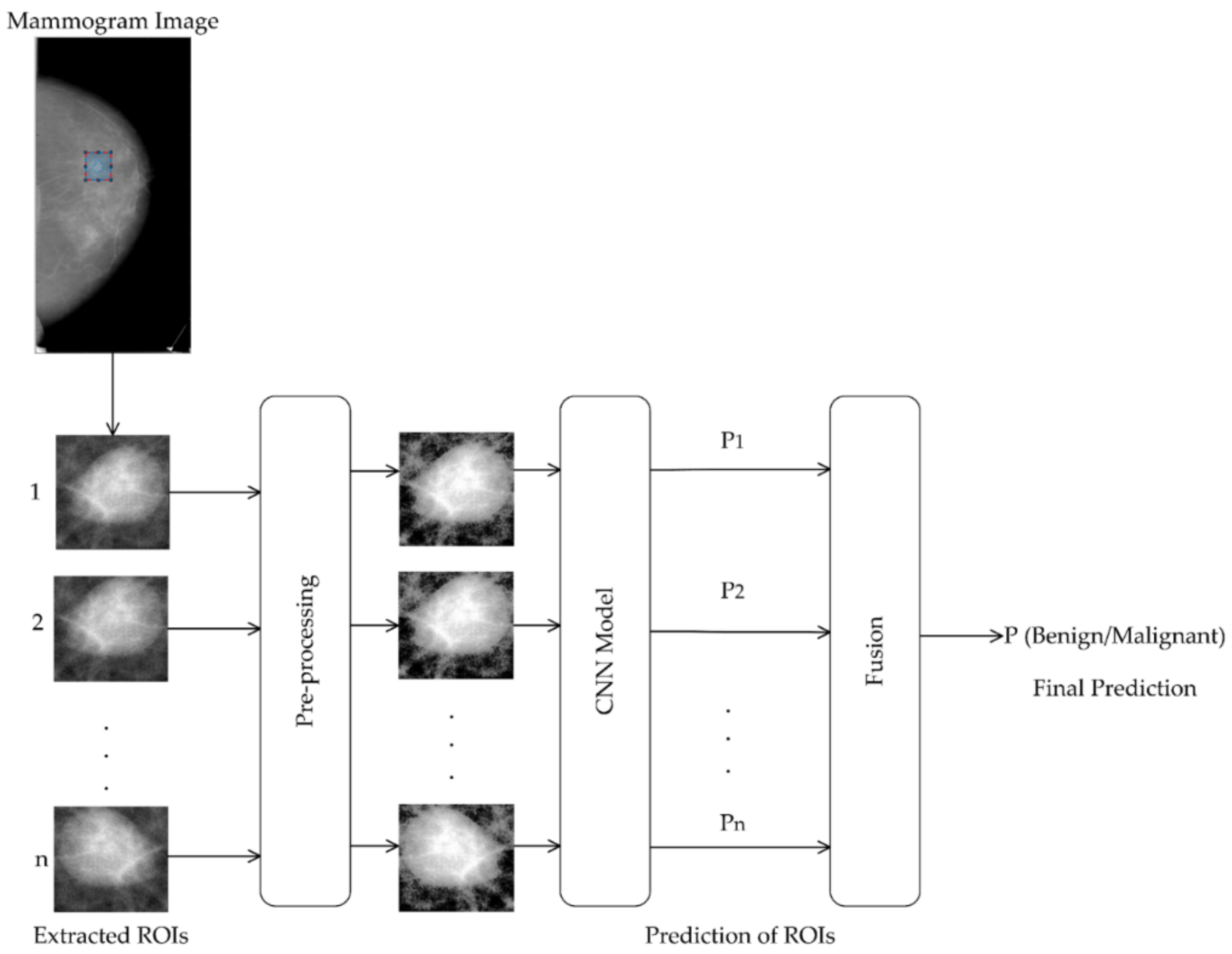
- We proposed a system for the classification of masses into benign and malignant using contextual information. It is based on the idea of an ensemble classifier and uses ResNet-50 as the backbone CNN model. It introduces diversity in decision-making using contextual information with multiple ROIs, not using multiple CNN models.
- For the system, we employed two schemes to extract multiple ROIs from a mass region for modeling diverse contextual information. For fusing the diverse contextual information from different ROIs, we used different fusing techniques to find the best one. Stacking gives the best results.
- We employed the ResNet-50 model as a backbone model. To examine the effect of breast density in the discrimination of benign and malignant masses, we introduced density-specific modifications of ResNet-50 based on the idea of fusing local and global features, knowing that when the density-specific model is used, the breast density must be known. The density-specific models result in better performance than ResNet-50. Finally, the impact of different BI-RADS types on the classification was evaluated.
- The mass regions of each type are not enough to fine-tune even a pre-trained model. We introduced a data augmentation approach for fine-tuning the pre-trained models.
Related Work
2. Proposed Method
2.1. Modeling the Context Information
2.1.1. Scale-Based Multi-Context Regions of Interest (ROIs) Extraction
2.1.2. Translation-Based Multi-Context ROIs Extraction
2.2. Preprocessing
2.3. Backbone Convolutional Neural Network (CNN) Model
2.4. Fine-Tuned ResNet-50
2.5. Density Specific Modification of ResNet-50–DResNet-50
2.5.1. DIResNet-50 for BI-RADS I
2.5.2. DIIResNet-50 for BI-RADS II
2.5.3. DIIIResNet-50 for BI-RADS III
2.5.4. DIVResNet-50 for BI-RADS IV
2.6. Fusion Techniques
2.6.1. Majority of the Decisions
2.6.2. Soft Voting
2.6.3. Max Voting
2.6.4. Stacking
2.7. Training of CNN Models
2.7.1. Datasets
- CBIS-DDSM [47]. The Curated Breast Imaging Subset of DDSM (CBIS-DDSM) is a challenging dataset that contains digitized film images of 753 calcifications and 891 masses converted to Digital Imaging and Communications in Medicine (DICOM) format. This dataset has the updated annotations of mass regions on mediolateral oblique (MLO) and bilateral craniocaudal (CC) views. The database size and ground truth verification make the DDSM a useful tool in developing and testing support systems without any bias. Using the annotations, we extracted benign and malignant mass regions. We only considered the mass abnormality with breast density. For evaluation, we used the protocol provided for this dataset; the dataset is separated into train and test datasets, as shown in Table 2. In the sequel, DT, D1T, D2T, D3T, D4T stand for the complete training data set (D1T∪D2T∪D3T∪D4T), and the training datasets for BI-RADS.I, BI-RADS.II, BI-RADS.III, BI-RADS.IV, respectively. Similarly, DTs, D1Ts, D2Ts, D3Ts, D4Ts represent the complete test data set (D1Ts∪D2Ts∪D3Ts∪D4Ts), and the test datasets for BI-RADS.I, BI-RADS.II, BI-RADS.III, BI-RADS.IV, respectively.
- INbreast [48]. It is the largest public dataset that contains 410 full-field digital mammographic (FFDM) images provided in Digital Imaging and Communications in Medicine (DICOM) format. Each case consists of mediolateral oblique (MLO) and bilateral craniocaudal (CC) views. According to the database size and ground truth verification, the INbreast provides useful data for building and testing support systems without bias. According to the annotation, the statistics of this dataset are given in Table 3.
2.7.2. Data Augmentation
2.7.3. Fine-Tuning the Backbone Models
3. Evaluation Protocol
- We used the evaluation protocol provided for CBIS-DDSM. The training set was used to fine-tune the backbone models; it was divided into training and validation sets with a ratio of 90:10. The new training set was utilized to fit the model independently, and the validation set was employed to control the training process. After completing a model’s training, its performance was evaluated on the test set of CBIS-DDSM without bias.
- For INbreast the cases are randomly divided into 80% for training,10% for validation, and 10% for testing, which allows us to run five-fold cross-validation. Cross-validation provides a less biased estimate of the model’s ability for unseen data.
4. Result
4.1. Why ResNet-50 as a Backbone Model?
4.2. The Effect of Multi-Scale and Multi-Context Schemes for a Test Region
4.3. The Effect of Different Fusion Techniques
4.4. The Effect of Density-Specific Models
4.5. Comparison with State-of-the-Art Methods
5. Discussion
- The system uses ResNet-50 and its modified versions as the backbone model. It would be better if a new data-dependent model is designed which is adaptive to mammogram images.
- The method fails when the mass appears in extremely dense breast tissue because the characteristic similarity between the dense tissue and masses makes breast mass classification difficult. Figure 16 shows mass regions of test images that are difficult to classify accurately.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 5 October 2021).
- Coleman, C. Early Detection and Screening for Breast Cancer. Semin. Oncol. Nurs. 2017, 33, 141–155. [Google Scholar] [CrossRef]
- Autier, P.; Boniol, M. Mammography screening: A major issue in medicine. Eur. J. Cancer 2018, 90, 34–62. [Google Scholar] [CrossRef]
- Chaira, T. Intuitionistic fuzzy approach for enhancement of low contrast mammogram images. Int. J. Imaging Syst. Technol. 2020, 30, 1162–1172. [Google Scholar] [CrossRef]
- Qiu, Y.; Yan, S.; Gundreddy, R.R.; Wang, Y.; Cheng, S.; Liu, H.; Bin Zheng, B. A new approach to develop computer-aided diagnosis scheme of breast mass classification using deep learning technology. J. X-ray Sci. Technol. 2017, 25, 751–763. [Google Scholar] [CrossRef] [Green Version]
- Falconí, L.G.; Pérez, M.; Aguilar, W.G. Transfer Learning in Breast Mammogram Abnormalities Classification with Mobilenet and Nasnet. In Proceedings of the 2019 International Conference on Systems, Signals, and Image Processing (IWSSIP), Osijek, Croatia, 5–7 June, 2019; pp. 109–114. [Google Scholar]
- Perre, A.C.; Alexandre, L.A.; Freire, L.C. Lesion Classification in Mammograms Using Convolutional Neural Networks and Transfer Learning. Comput. Methods Biomech. Biomed. Eng. Imaging Vis. 2019, 7, 550–556. [Google Scholar] [CrossRef]
- Khan, H.N.; Shahid, A.R.; Raza, B.; Dar, A.H.; Alquhayz, H. Multi-View Feature Fusion Based Four Views Model for Mammogram Classification Using Convolutional Neural Network. IEEE Access 2019, 7, 165724–165733. [Google Scholar] [CrossRef]
- Li, H.; Chen, D.; Nailon, W.H.; Davies, M.E.; Laurenson, D. A Deep Dual-path Network for Improved Mammogram Image Processing. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal, Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1224–1228. [Google Scholar]
- Tsochatzidis, L.; Costaridou, L.; Pratikakis, I. Deep Learning for Breast Cancer Diagnosis from Mammograms—A Comparative Study. J. Imaging 2019, 5, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duggento, A.; Aiello, M.; Cavaliere, C.; Cascella, G.L.; Cascella, D.; Conte, G.; Guerrisi, M.; Toschi, N. An Ad Hoc Random Initialization Deep Neural Network Architecture for Discriminating Malignant Breast Cancer Lesions in Mammographic Images. Contrast Media Mol. Imaging 2019, 2019, 5982834. [Google Scholar] [CrossRef]
- Shu, X.; Zhang, L.; Wang, Z.; Lv, Q.; Yi, Z. Deep Neural Networks with Region-Based Pooling Structures for Mammographic Image Classification. IEEE Trans. Med. Imaging 2020, 39, 2246–2255. [Google Scholar] [CrossRef]
- Alhakeem, Z.; Jang, S.I. LBP-HOG Descriptor Based on Matrix Projection for Mammogram Classification. arXiv 2021, arXiv:1904.00187. [Google Scholar]
- Chougrad, H.; Zouaki, H.; Alheyane, O. Deep Convolutional Neural Networks for Breast Cancer Screening. Comput. Methods Programs Biomed. 2018, 157, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Al-Antari, M.A.; Al-Masni, M.A.; Choi, M.T.; Han, S.M.; Kim, T.S. A Fully Integrated Computer-Aided Diagnosis System for Digital X-Ray Mammograms via Deep Learning Detection, Segmentation, and Classification. Int. J. Med. Inform. 2018, 117, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Gou, C.; Wang, J.; Wang, F.-Y. Simultaneous Segmentation and Classification of Mass Region from Mammograms Using a Mixed-Supervision Guided Deep Model. IEEE Signal Process. Lett. 2019, 27, 196–200. [Google Scholar] [CrossRef]
- Aly, G.H.; Marey, M.A.E.-R.; Amin, S.E.-S.; Tolba, M.F. YOLO V3 and YOLO V4 for Masses Detection in Mammograms with ResNet and Inception for Masses Classification. In Proceedings of the International Conference on Advanced Machine Learning Technologies and Applications, Cairo, Egypt, 20–22 March 2021; pp. 145–153. [Google Scholar] [CrossRef]
- Lou, M.; Wang, R.; Qi, Y.; Zhao, W.; Xu, C.; Meng, J.; Deng, X.; Ma, Y. MGBN: Convolutional neural networks for automated benign and malignant breast masses classification. Multimedia Tools Appl. 2021, 80, 1–20. [Google Scholar] [CrossRef]
- Polikar, R. Ensemble Machine Learning; Springer: Boston, MA, USA, 2012; pp. 1–34. [Google Scholar]
- Shen, R.; Zhou, K.; Yan, K.; Tian, K.; Zhang, J. Multi-Context Multi-Task Learning Networks for Mass Detection in Mammogram. In Medical Physics; Springer: Boston, MA, USA, 2019. [Google Scholar]
- Luo, S.; Cheng, B. Diagnosing Breast Masses in Digital Mammography Using Feature Selection and Ensemble Methods. J. Med. Syst. 2010, 36, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Dietterich, T.G. Ensemble Methods in Machine Learning. In Multiple Classifier Systems; Springer: Berlin/Heidelberg, Germany, 2000. [Google Scholar]
- Nguyen, Q.H.; Do, T.T.T.; Wang, Y.; Heng, S.S.; Chen, K.; Ang, W.H.M.; Philip, C.E.; Singh, M.; Pham, H.N.; Nguyen, B.P.; et al. Breast Cancer Prediction using Feature Selection and Ensemble Voting. In Proceedings of the 2019 International Conference on System Science and Engineering (ICSSE), Dong Hoi, Vietnam, 20–21 July 2019; pp. 250–254. [Google Scholar]
- Swiderski, B.; Osowski, S.; Kurek, J.; Kruk, M.; Lugowska, I.; Rutkowski, P.; Barhoumi, W. Novel methods of image description and ensemble of classifiers in application to mammogram analysis. Expert Syst. Appl. 2017, 81, 67–78. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, L.; Zhang, W.; Huang, X.; Iosifidis, V.; Liu, Z.; Zhang, M.; Messina, E.; Zhang, J. Using Machine Learning to Automate Mammogram Images Analysis. In Proceedings of the 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Seoul, South Korea, 16–19 December 2020; pp. 757–764. [Google Scholar]
- Arora, R.; Rai, P.K.; Raman, B. Deep feature–based automatic classification of mammograms. Med. Biol. Eng. Comput. 2020, 58, 1199–1211. [Google Scholar] [CrossRef]
- Fezza, S.A.; Bakhti, Y.; Hamidouche, W.; Deforges, O. Perceptual Evaluation of Adversarial Attacks for CNN-based Image Classification. In Proceedings of the 2019 Eleventh International Conference on Quality of Multimedia Experience (QoMEX), Berlin, Germany, 5–7 June 2019; pp. 1–6. [Google Scholar]
- Savelli, B.; Bria, A.; Molinara, M.; Marrocco, C.; Tortorella, F. A multi-context CNN ensemble for small lesion detection. Artif. Intell. Med. 2020, 103, 101749. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet Classification with Deep Convolutional Neural Networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Abdelhafiz, D.; Yang, C.; Ammar, R.; Nabavi, S. Deep convolutional neural networks for mammography: Advances, challenges and applications. BMC Bioinform. 2019, 20, 1–20. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Z.; Gao, X.; Wang, Y.; Li, J. A deep feature based framework for breast masses classification. Neurocomputing 2016, 197, 221–231. [Google Scholar] [CrossRef]
- Kabbai, L.; Abdellaoui, M.; Douik, A. Image Classification by Combining Local and Global Features. Vis. Comput. 2019, 35, 679–693. [Google Scholar] [CrossRef]
- Zou, J.; Li, W.; Chen, C.; Du, Q. Scene classification using local and global features with collaborative representation fusion. Inf. Sci. 2016, 348, 209–226. [Google Scholar] [CrossRef]
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2016, 39, 640–651. [Google Scholar] [CrossRef]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Chu, B.; Yang, D.; Tadinada, R. Visualizing Residual Networks. arXiv 2017, arXiv:1701.02362. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); IEEE: Piscataway, NJ, USA, 2016. [Google Scholar]
- Rokach, L. Ensemble-based classifiers. Artif. Intell. Rev. 2009, 33, 1–39. [Google Scholar] [CrossRef]
- Wolpert, D.H. Stacked Generalization. Neural Netw. 1992, 5, 241–259. [Google Scholar] [CrossRef]
- Aroef, C.; Rivan, Y.; Rustam, Z. Comparing random forest and support vector machines for breast cancer classification. Telkomnika Telecommun. Comput. Electron. Control. 2020, 18, 815–821. [Google Scholar] [CrossRef]
- Sarosa, S.J.A.; Utaminingrum, F.; Bachtiar, F.A. Mammogram Breast Cancer Classification Using Gray-Level Co-Occurrence Matrix and Support Vector Machine. In Proceedings of the 2018 International Conference on Sustainable Information Engineering and Technology (SIET), Malang, Indonesia, 10–12 November 2018; pp. 54–59. [Google Scholar]
- Gunn, S. Support Vector Machines for Classification and Regression. ISIS Tech. Rep. 1998, 14, 5–16. [Google Scholar]
- Cristianini, N.; Shawe-Taylor, J. An Introduction to Support Vector Machines and Other Kernel-Based Learning Methods; Cambridge University Press: Cambridge, UK, 2000. [Google Scholar]
- Lee, R.S.; Gimenez, F.; Hoogi, A.; Miyake, K.K.; Gorovoy, M.; Rubin, D. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci. Data 2017, 4, 1–9. [Google Scholar] [CrossRef]
- Moreira, I.C.; Amaral, I.; Domingues, I.; Cardoso, A.; Cardoso, M.J.; Cardoso, J. INbreast: Toward a Full-field Digital Mammographic Database. Acad. Radiol. 2012, 19, 236–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, S.; Gatt, A.; Stamatescu, V.; McDonnell, M.D. Understanding Data Augmentation for Classification: When to Warp? In Proceedings of the 2016 International Conference on Digital Image Computing: Techniques and Applications (DICTA), Gold Coast, QLD, Australia, 30 November–2 December 2016; pp. 1–6. [Google Scholar]
- Ranganathan, P.; Pramesh, C.S.; Aggarwal, R. Common pitfalls in statistical analysis: Measures of agreement. Perspect. Clin. Res. 2017, 8, 187–191. [Google Scholar] [CrossRef]
- Zhu, W.; Zeng, N.; Wang, N. Sensitivity, specificity, accuracy, associated confidence interval and ROC analysis with practical SAS implementations. NESUGProc. Health Care Life Sci. Baltim. Md. 2010, 19, 67. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A.A. Inception-v4, Inception-Resnet and the Impact of Residual Connections on Learning. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning Transferable Architectures for Scalable Image Recognition. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018; pp. 8697–8710. [Google Scholar]















| Group | OUTPUT SIZE | 50-Layer |
|---|---|---|
| 112 × 112 | 7 × 7, 64, stride 2 | |
| 56 × 56 | 3 × 3 max pool, stride 2 | |
| 28 × 28 | ||
| 14 × 14 | ||
| 7 × 7 | ||
| 1 × 1 | average pool, 1000-d fc, SoftMax |
| Dataset | Dataset Set Pathology | Train | Test |
|---|---|---|---|
| D1 (BI-RADS.I) | Benign | 103 | 24 |
| Malignant | 128 | 21 | |
| D2 (BI-RADS.II) | Benign | 257 | 74 |
| Malignant | 248 | 70 | |
| D3 (BI-RADS.III) | Benign | 145 | 63 |
| Malignant | 125 | 29 | |
| D4 (BI-RADS.IV) | Benign | 53 | 21 |
| Malignant | 38 | 12 | |
| D (D1+D2+D3+D4) | Benign | 558 | 182 |
| Malignant | 539 | 132 |
| Dataset | Dataset Set Pathology | Number of Masses |
|---|---|---|
| D1 (BI-RADS.I) | Benign | 12 |
| Malignant | 30 | |
| D2 (BI-RADS.II) | Benign | 6 |
| Malignant | 32 | |
| D3 (BI-RADS.III) | Benign | 13 |
| Malignant | 8 | |
| D4 (BI-RADS.IV) | Benign | 6 |
| Malignant | 1 | |
| D (D1+D2+D3+D4) | Benign | 37 |
| Malignant | 71 |
| Model | Sen (%) | SP (%) | ACC (%) | Kappa (%) | F1-Score |
|---|---|---|---|---|---|
| ResNet-50 [32] | 99.24 | 87.36 | 92.36 | 84.70 | 91.61 |
| DensNet-201 [52] | 81.01 | 97.44 | 89.17 | 78.40 | 88.28 |
| InceptionResNetV2 [53] | 81.53 | 97.45 | 89.49 | 79 | 88.58 |
| NasNetLarge [54] | 81.25 | 98.70 | 89.81 | 79.70 | 89.04 |
| Scheme | Sen (%) | SP (%) | ACC (%) | Kappa (%) | F1-Score |
|---|---|---|---|---|---|
| single | 67.42 | 78.02 | 73.57 | 45.60 | 68.20 |
| {5,10,15,20,25}-MS1 | 81.06 | 91.76 | 87.26 | 73.60 | 84.25 |
| {10,20,30,40,50}-MS2 | 82.58 | 90.66 | 87.26 | 73.70 | 84.50 |
| {50,60,70,80,100}-MS3 | 83.33 | 92.31 | 88.54 | 76.30 | 85.94 |
| Scheme | Sen (%) | SP (%) | ACC (%) | Kappa (%) | F1-Score |
|---|---|---|---|---|---|
| single | 67.42 | 78.02 | 73.57 | 45.60 | 68.20 |
| 256 with 3 contexts-MC1 | 73.48 | 86.26 | 80.89 | 60.40 | 76.38 |
| 256 with 5 contexts-MC2 | 87.12 | 93.96 | 91.08 | 81.60 | 89.15 |
| Fusion Technique | Sen (%) | SP (%) | Acc (%) | Kappa (%) | F1-Score | |
|---|---|---|---|---|---|---|
| Stacking | Random Forest | 87.88 | 93.96 | 91.40 | 82.30 | 89.59 |
| SVM with RBF | 99.24 | 87.36 | 92.36 | 84.70 | 91.61 | |
| SVM with Linear | 92.42 | 92.31 | 92.36 | 84.40 | 91.04 | |
| SVM with Polynomial | 87.12 | 93.96 | 91.08 | 81.60 | 89.15 | |
| Majority voting | 77.27 | 83.52 | 80.89 | 60.80 | 77.27 | |
| Soft Voting | 78.79 | 84.62 | 82.17 | 63.10 | 78.79 | |
| Max voting | 81.06 | 84.07 | 82.80 | 64.40 | 79.85 | |
| Models | Sen (%) | SP (%) | ACC (%) | Kappa (%) | F1-Score |
|---|---|---|---|---|---|
| DIRresNet-50_D1T_D1Ts | 100 | 91.67 | 95.56 | 91.12 | 95.45 |
| DII RresNet-50_D2T_D2Ts | 92.86 | 87.84 | 90.28 | 80.57 | 90.28 |
| DIII RresNet-50D3T_D3Ts | 96.55 | 96.83 | 96.74 | 92.50 | 94.92 |
| DIVRresNet-50_D4T_D4Ts | 91.67 | 100 | 96.97 | 93.30 | 95.65 |
| ResNet-50_D1T _D1Ts | 90.48 | 87.50 | 88.89 | 77.70 | 88.37 |
| ResNet-50_D2T _D2Ts | 90 | 75.68 | 82.64 | 65.40 | 83.44 |
| ResNet-50_D3T _D3Ts | 82.76 | 84.13 | 83.70 | 63.90 | 76.19 |
| ResNet-50_D4T _ D4Ts | 66.67 | 100 | 87.88 | 71.80 | 80 |
| Datasets | Models | Sen (%) | SP (%) | ACC (%) | Kappa (%) | F1-Score |
|---|---|---|---|---|---|---|
| CBIS-DDSM | DIRresNet-50_ DT_DTs | 98.48 | 92.31 | 94.90 | 89.65 | 94.20 |
| DIIRresNet-50_DT_DTs | 96.97 | 92.31 | 94.27 | 88.36 | 93.43 | |
| DIIIRresNet-50_DT_DTs | 97.73 | 91.21 | 93.95 | 87.75 | 93.14 | |
| DIVRresNet-50_DT_DTs | 91.67 | 96.70 | 94.59 | 88.83 | 93.44 | |
| RresNet-50_DT_ DTs | 99.24 | 87.36 | 92.36 | 84.67 | 91.61 | |
| INbreast | DIRresNet-50_ DT_DTs | 100 | 97.5 | 99.09 | 98.07 | 99.31 |
| DIIRresNet-50_DT_DTs | 100 | 100 | 100 | 100 | 100 | |
| DIIIRresNet-50_DT_DTs | 97.14 | 97.5 | 97.19 | 94.31 | 97.77 | |
| DIVRresNet-50_DT_DTs | 100 | 97.14 | 99 | 97.73 | 99.26 | |
| RresNet-50_DT_ DTs | 98.57 | 97.5 | 98.18 | 96.14 | 98.61 |
| References | Models\Descriptors | Sen (%) | SP (%) | AUC (%) | ACC (%) | |
|---|---|---|---|---|---|---|
| CBIS-DDSM | Khan et el. [8] | ResNet-50 | 75.46 | 62.75 | 69.10 | 69.98 |
| MVFF | 81.82 | 72.02 | 76.90 | 77.66 | ||
| Tsochatzidis et al. [10] | ResNet-50 from scratch | - | - | 80.40 | 74.90 | |
| Fine-tuning ResNet-50 | - | - | 63.70 | 62.70 | ||
| Duggento et al. [11] | AlexNet | 84.40 | 62.44 | - | 71.19 | |
| AL Hakeem and Jang [13] | LBP-HOG | - | - | - | 64.35 | |
| Li et al. [9] | Dual-core Net | - | - | 85 | - | |
| Shu et al. [12] | Region-based Group-max Pooling | - | - | 83.3 | 76.2 | |
| Global Group-max Pooling | - | - | 82.3 | 76.7 | ||
| Proposed system | Multi-context ResNet-50 | 99.24 | 87.36 | 97.17 | 92.36 | |
| Multi-context DIRresNet-50 | 98.48 | 92.31 | 94.38 | 94.90 | ||
| Multi-context DIIRresNet-50 | 96.97 | 92.31 | 93.59 | 94.27 | ||
| Multi-context DIIIRresNet-50 | 97.73 | 91.21 | 96.55 | 93.95 | ||
| Multi-context DIVRresNet-50 | 91.67 | 96.70 | 94.83 | 94.59 | ||
| INbreast | Chougrad et al. [14] | Resnet-50 | - | - | - | 92.50 |
| Al-antari et al. [15] | Alex Net | 97.14 | 92.41 | 94.78 | 95.64 | |
| Shen et al. [16] | ResCU-Net | - | - | 96.16 | 94.12 | |
| Ghada et al. [17] | ResNet | - | - | - | 90 | |
| Inception | - | - | - | 95 | ||
| Lou et al. [18] | ResNet-50 | 69.23 | 74 | 84.96 | 72.37 | |
| MGBN-50 | 77.16 | 88.24 | 93.11 | 84.50 | ||
| Proposed system | Multi-context ResNet-50 | 98.57 | 97.500 | 99.02 | 98.18 | |
| Multi-context DIRresNet-50 | 100 | 97.5 | 99.64 | 99.09 | ||
| Multi-context DIIRresNet-50 | 100 | 100 | 100 | 100 | ||
| Multi-context DIIIRresNet-50 | 97.14 | 97.50 | 97.35 | 97.19 | ||
| Multi-context DIVRresNet-50 | 100 | 97.14 | 99.56 | 99 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Busaleh, M.; Hussain, M.; Aboalsamh, H.A.; Amin, F.-e.-. Breast Mass Classification Using Diverse Contextual Information and Convolutional Neural Network. Biosensors 2021, 11, 419. https://doi.org/10.3390/bios11110419
Busaleh M, Hussain M, Aboalsamh HA, Amin F-e-. Breast Mass Classification Using Diverse Contextual Information and Convolutional Neural Network. Biosensors. 2021; 11(11):419. https://doi.org/10.3390/bios11110419
Chicago/Turabian StyleBusaleh, Mariam, Muhammad Hussain, Hatim A. Aboalsamh, and Fazal-e- Amin. 2021. "Breast Mass Classification Using Diverse Contextual Information and Convolutional Neural Network" Biosensors 11, no. 11: 419. https://doi.org/10.3390/bios11110419
APA StyleBusaleh, M., Hussain, M., Aboalsamh, H. A., & Amin, F.-e.-. (2021). Breast Mass Classification Using Diverse Contextual Information and Convolutional Neural Network. Biosensors, 11(11), 419. https://doi.org/10.3390/bios11110419





