Novel Biorecognition Elements against Pathogens in the Design of State-of-the-Art Diagnostics
Abstract
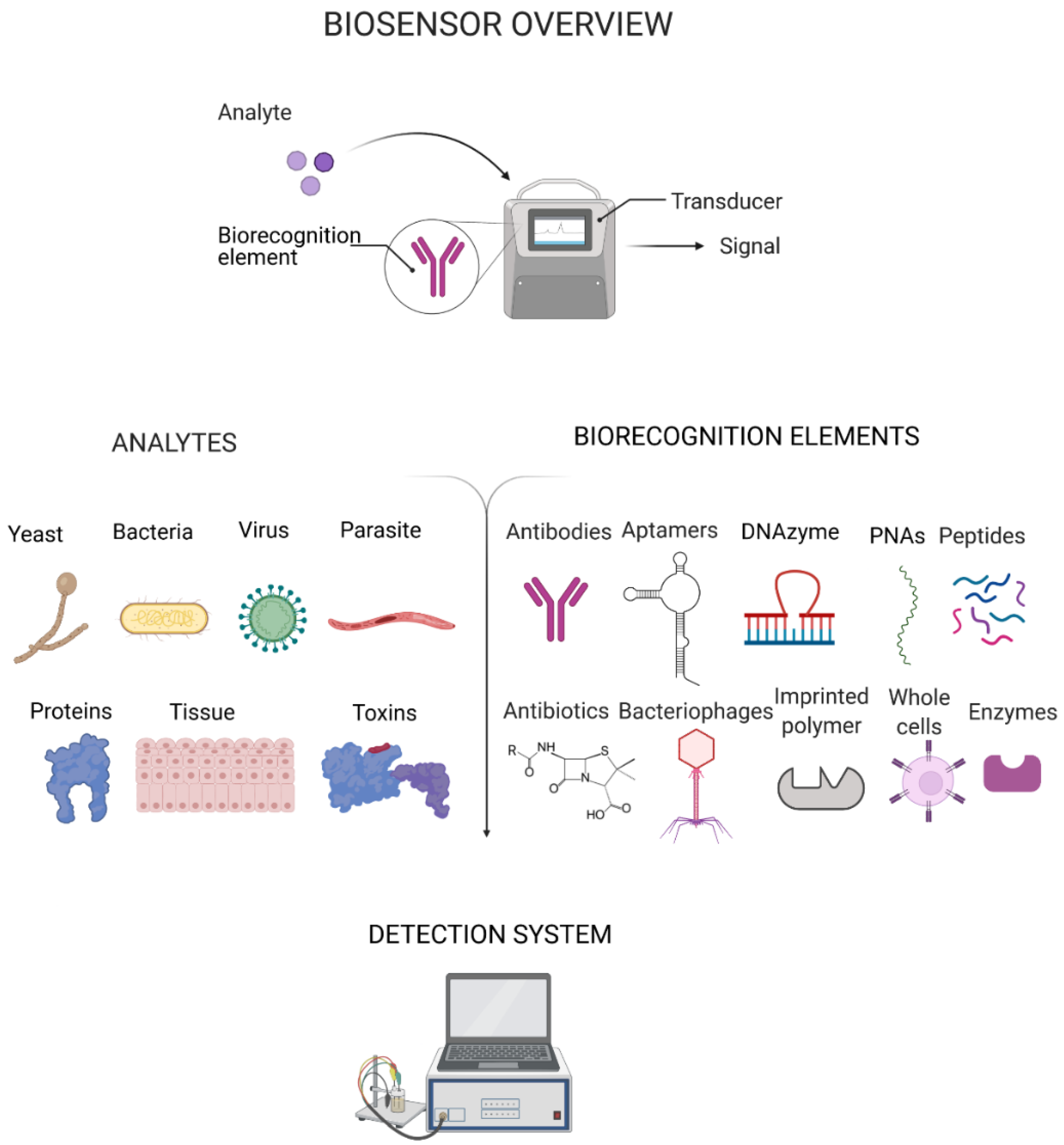
:1. Introduction
2. Recent Progress in Platforms for Pathogen Detection
3. Biorecognition Elements
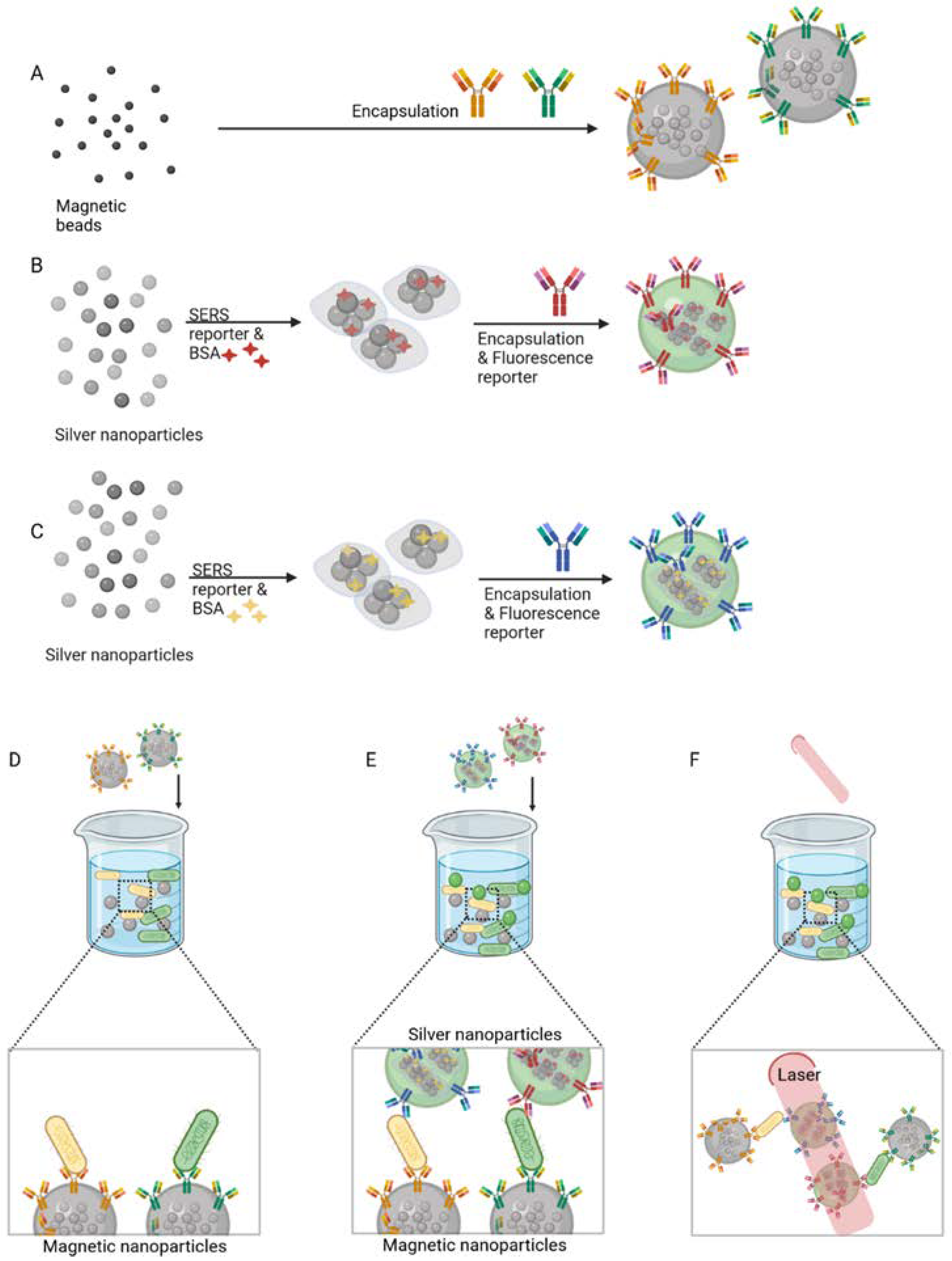
3.1. Antibodies
3.2. Enzymes
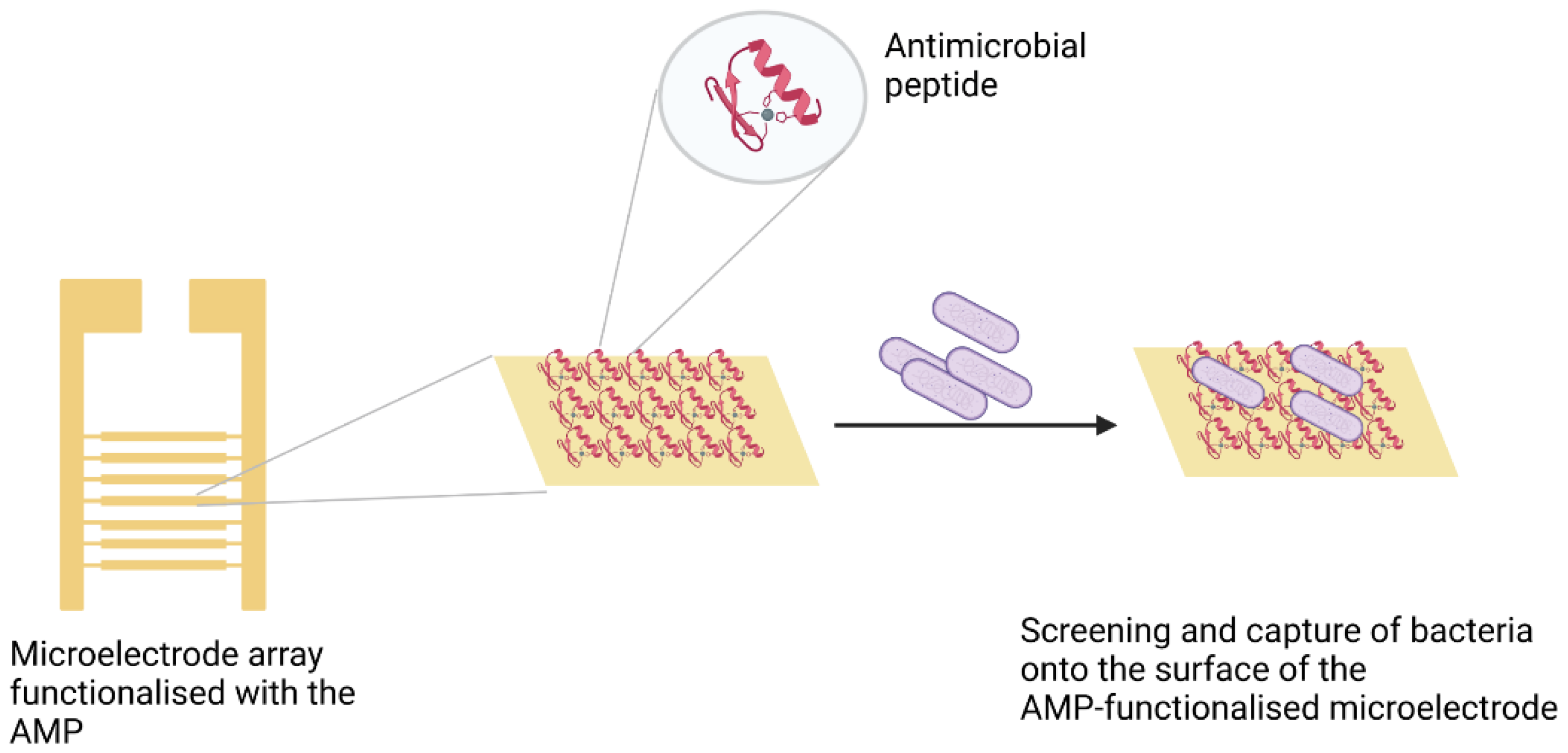
3.3. Peptides
3.4. Nucleic Acid Derivatives
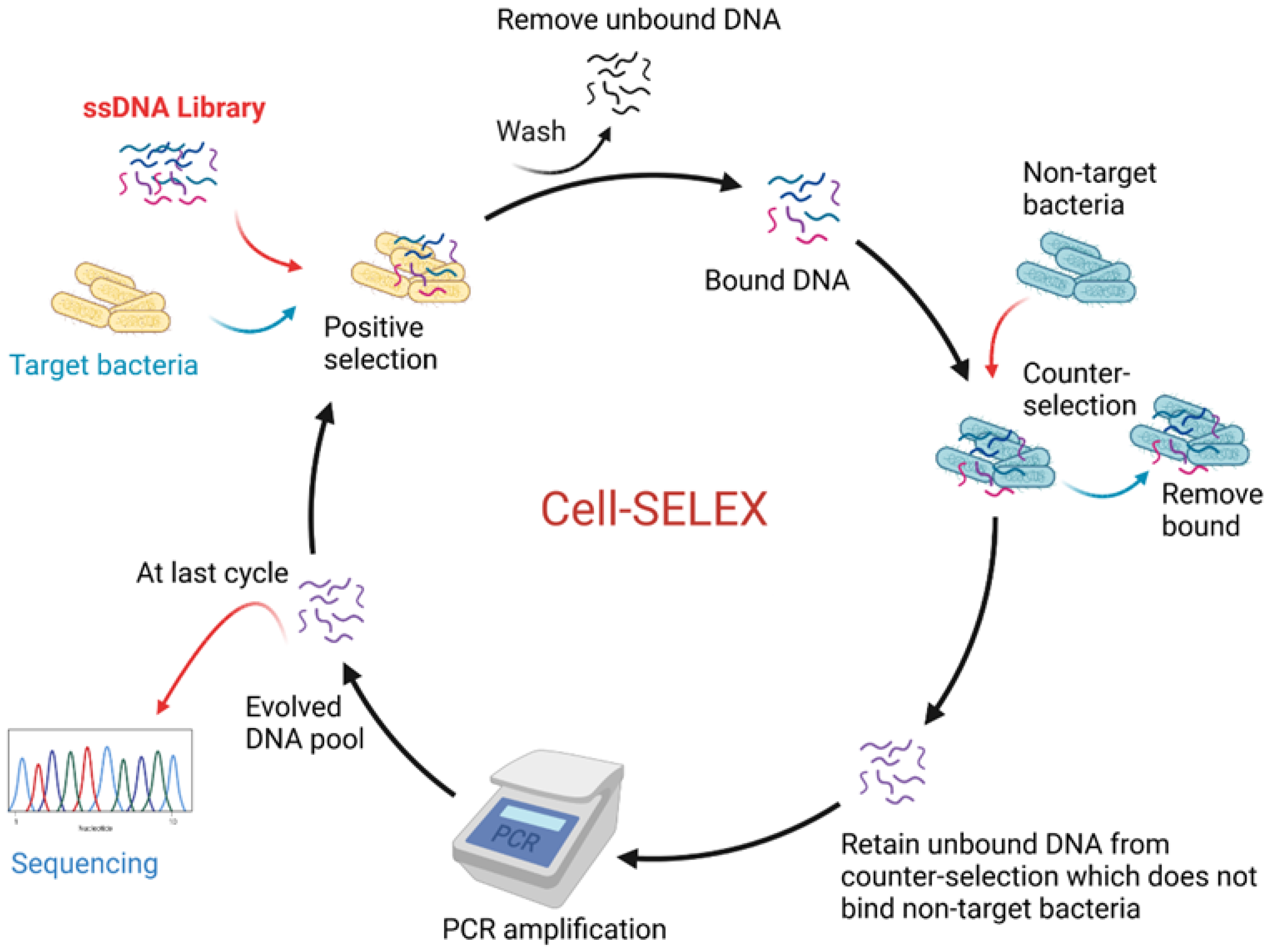
3.4.1. Aptamers
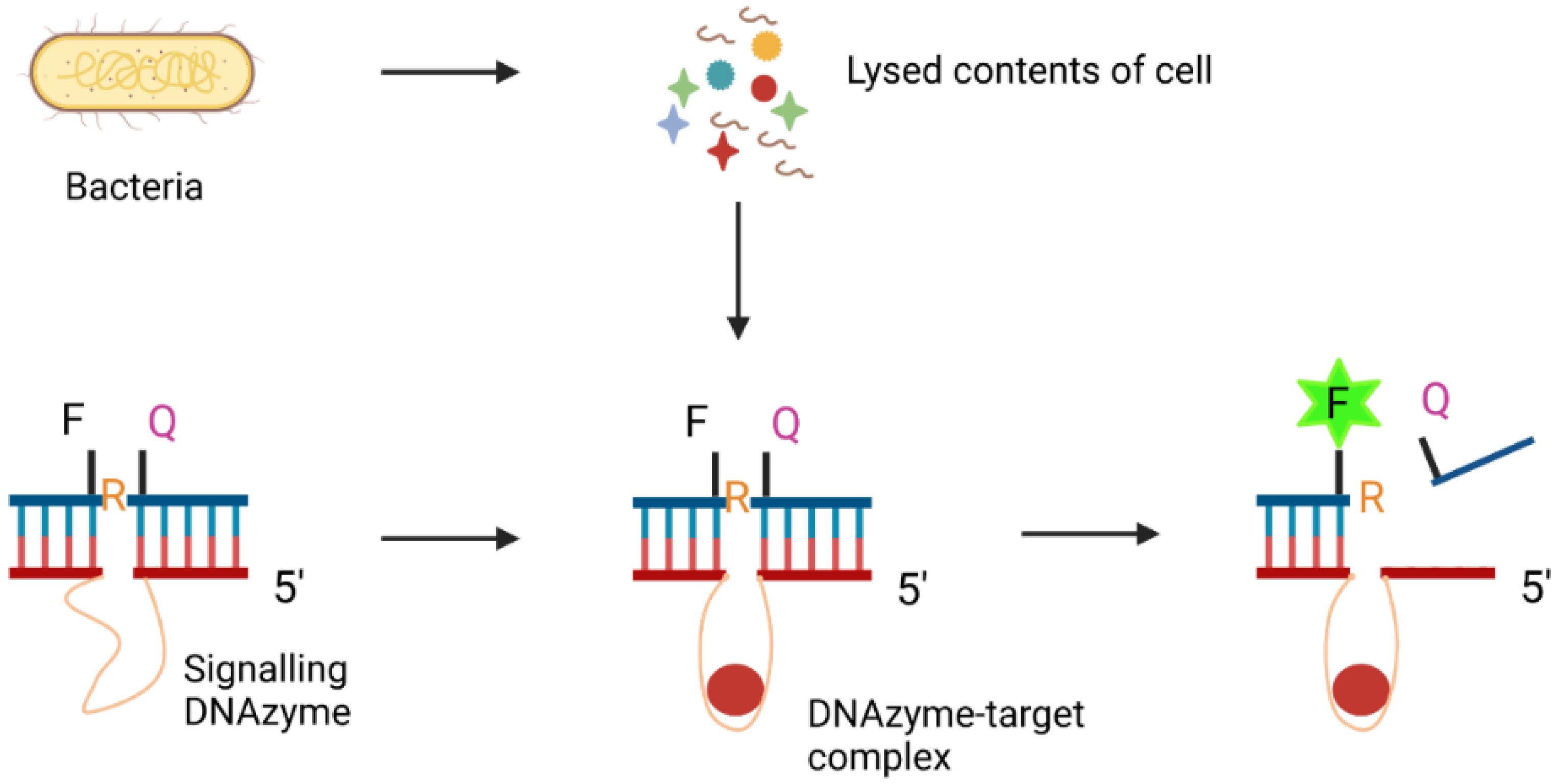
3.4.2. DNAzymes
3.4.3. Peptide Nucleic Acids (PNAs)
3.5. CRISPR-Cas
3.6. Bacteriophages
3.7. Molecularly Imprinted Polymers (MIPs)
3.8. Antibiotics
3.8.1. Vancomycin
3.8.2. Amoxicillin
3.8.3. Ampicillin
3.8.4. Neomycin
3.9. Chemical Compounds
4. Future Perspectives
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. The Top 10 Causes of Death. Available online: http://www.who.int/en/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 16 March 2021).
- World Health Organisation. New Report Calls for Urgent Action to Avert Antimicrobial Resistance Crisis. Available online: https://www.who.int/news/item/29-04-2019-new-report-calls-for-urgent-action-to-avert-antimicrobial-resistance-crisis (accessed on 16 March 2021).
- Gu, W.; Miller, S.; Chiu, C.Y. Clinical Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 319–338. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.B.; Petrich, A.; Smieja, M. Molecular diagnosis of respiratory virus infections. Crit. Rev. Clin. Lab. Sci. 2011, 48, 217–249. [Google Scholar] [CrossRef]
- Sin, M.L.; Mach, K.E.; Wong, P.K.; Liao, J.C. Advances and challenges in biosensor-based diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 2014, 14, 225–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudd, K.E.; Johnson, S.C.; Agesa, K.M.; Shackelford, K.A.; Tsoi, D.; Kievlan, D.R.; Colombara, D.V.; Ikuta, K.S.; Kissoon, N.; Finfer, S.; et al. Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020, 395, 200–211. [Google Scholar] [CrossRef] [Green Version]
- Urdea, M.; Penny, L.A.; Olmsted, S.S.; Giovanni, M.Y.; Kaspar, P.; Shepherd, A.; Wilson, P.; Dahl, C.A.; Buchsbaum, S.; Moeller, G.; et al. Requirements for high impact diagnostics in the developing world. Nature 2006, 444, 73–79. [Google Scholar] [CrossRef]
- Dhiman, A.; Kalra, P.; Bansal, V.; Bruno, J.G.; Sharma, T.K. Aptamer-based point-of-care diagnostic platforms. Sens. Actuators B Chem. 2017, 246, 535–553. [Google Scholar] [CrossRef]
- Morales, M.A.; Halpern, J.M. Guide to Selecting a Biorecognition Element for Biosensors. Bioconjug. Chem. 2018, 29, 3231–3239. [Google Scholar] [CrossRef]
- Lim, E.K.; Kim, T.; Paik, S.; Haam, S.; Huh, Y.M.; Lee, K. Nanomaterials for theranostics: Recent advances and future challenges. Chem. Rev. 2015, 115, 327–394. [Google Scholar] [CrossRef] [PubMed]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef]
- Bahadir, E.B.; Sezgintürk, M.K. Applications of commercial biosensors in clinical, food, environmental, and biothreat/biowarfare analyses. Anal. Biochem. 2015, 478, 107–120. [Google Scholar] [CrossRef]
- Sharma, A.; Sharma, N.; Kumari, A.; Lee, H.J.; Kim, T.Y.; Tripathi, K.M. Nano-carbon based sensors for bacterial detection and discrimination in clinical diagnosis: A junction between material science and biology. Appl. Mater. Today 2020, 18, 100467. [Google Scholar] [CrossRef]
- Coris Bioconcept Clostridium K-SeT. Available online: https://www.corisbio.com/products/clostridium-k-set (accessed on 1 June 2021).
- Thermofisher Scientific OxoidTM Clostridium difficile Test Kit. Available online: https://www.thermofisher.com/order/catalog/product/DR1107A#/DR1107A (accessed on 1 June 2021).
- Meridian Bioscience ImmunoCard STAT! E. coli O157 Plus. Available online: https://www.meridianbioscience.com/human-condition/gastrointestinal/e-coli/immunocard-stat-e-coli-o157-plus/ (accessed on 1 June 2021).
- Thermofisher Scientific Escherichia coli O157 Latex Test. Available online: https://www.thermofisher.com/order/catalog/product/DR0620M#/DR0620M (accessed on 1 June 2021).
- Thermofisher Scientific WellcogenTM Haemophilus influenzae b Rapid Latex Agglutination. Test. Available online: https://www.thermofisher.com/order/catalog/product/R30858801#/R30858801 (accessed on 1 June 2021).
- Quidel QuickVue, H. pylori Test. Available online: https://www.quidel.com/immunoassays/rapid-h-pylori-tests (accessed on 1 June 2021).
- Wilburn Medical USA Beckman Coulter 395160A Icon HP (H.Pylori) Test Kit. Available online: https://wilburnmedicalusa.com/beckman-coulter-395160a-icon-hp-h-pylori-test-kit/ (accessed on 1 June 2021).
- Coris Bioconcept Pylori-Strip. Available online: https://www.corisbio.com/products/pylori-strip (accessed on 1 June 2021).
- Coris Bioconcept Pylori K-SeT. Available online: https://www.corisbio.com/products/pylori-k-set (accessed on 1 June 2021).
- Thermofisher Scientific RemelTM Catarrhalis Test Disc. Available online: https://www.thermofisher.com/order/catalog/product/R21121#/R21121 (accessed on 1 June 2021).
- Thermofisher Scientific BactiStaphTM Latex Agglutination Test Kit. Available online: https://www.thermofisher.com/order/catalog/product/R21143#/R21143 (accessed on 1 June 2021).
- Thermofisher Scientific DrySpotTM Pneumo Latex Agglutination Test. Available online: https://www.thermofisher.com/order/catalog/product/DR0420M#/DR0420M (accessed on 1 June 2021).
- Abbott BinaxNOWTM Streptococcus Pneumoniae Antigen Card. Available online: https://www.globalpointofcare.abbott/en/product-details/binaxnow-streptococcus-pneumoniae-ww.html (accessed on 1 June 2021).
- Thermofisher Scientific Infectious Mononucleosis Test Using Latex. Agglutination. Available online: https://www.thermofisher.com/order/catalog/product/DR0780M#/DR0780M (accessed on 1 June 2021).
- Quidel QuickVue Influenza A + B Test. Available online: https://www.quidel.com/immunoassays/rapid-influenza-tests/quickvue-influenza-test (accessed on 1 June 2021).
- Abbott Alere BinaxNOW® Influenza A & B Card. Available online: https://www.globalpointofcare.abbott/en/product-details/binaxnow-influenza-a-and-b.html (accessed on 1 June 2021).
- Lucira Health COVID-19 All-In-One Test Kit. Available online: https://www.lucirahealth.com/ (accessed on 1 June 2021).
- Abbott BinaxNOW COVID-19 Ag Card Home Test. Available online: https://www.globalpointofcare.abbott/en/product-details/binaxnow-covid-19-home-test-us.html (accessed on 1 June 2021).
- Ellume Health Ellume COVID-19 Home Test. Available online: https://www.ellumehealth.com/products/consumer-products/covid-home-test (accessed on 1 June 2021).
- Abbott BinaxNOW® Malaria. Available online: https://www.globalpointofcare.abbott/en/product-details/binaxnow-malaria.html (accessed on 1 June 2021).
- Prada, Y.A.; Soler, M.; Guzmán, F.; Castillo, J.J.; Lechuga, L.M.; Mejía-Ospino, E. Design and characterization of high-affinity synthetic peptides as bioreceptors for diagnosis of cutaneous leishmaniasis. Anal. Bioanal. Chem. 2021, 413, 4545–4555. [Google Scholar] [CrossRef]
- Curk, T.; Brackley, C.A.; Farrell, J.D.; Xing, Z.; Joshi, D.; Direito, S.; Bren, U.; Angioletti-Uberti, S.; Dobnikar, J.; Eiser, E.; et al. Computational design of probes to detect bacterial genomes by multivalent binding. Proc. Natl. Acad. Sci. USA 2020, 117, 8719–8726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, A.D.; Dekker, J.P. From the pipeline to the bedside: Advances and challenges in clinical metagenomics. J. Infect. Dis. 2020, 221, S331–S340. [Google Scholar] [CrossRef] [PubMed]
- O’Flaherty, B.M.; Li, Y.; Tao, Y.; Paden, C.R.; Queen, K.; Zhang, J.; Dinwiddie, D.L.; Gross, S.M.; Schroth, G.P.; Tong, S. Comprehensive viral enrichment enables sensitive respiratory virus genomic identification and analysis by next generation sequencing. Genome Res. 2018, 28, 869–877. [Google Scholar] [CrossRef] [Green Version]
- Wylie, K.M.; Wylie, T.N.; Buller, R.; Herter, B.; Cannella, M.T.; Storch, G.A. Detection of viruses in clinical samples by use of metagenomic sequencing and targeted sequence capture. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Arutyunov, D.; Szymanski, C.M.; Evoy, S. Bacteriophage based probes for pathogen detection. Analyst 2012, 137, 3405–3421. [Google Scholar] [CrossRef]
- Crivianu-Gaita, V.; Thompson, M. Aptamers, antibody scFv, and antibody Fab’ fragments: An overview and comparison of three of the most versatile biosensor biorecognition elements. Biosens. Bioelectron. 2016, 85, 32–45. [Google Scholar] [CrossRef] [PubMed]
- Foudeh, A.M.; Fatanat Didar, T.; Veres, T.; Tabrizian, M. Microfluidic designs and techniques using lab-on-a-chip devices for pathogen detection for point-of-care diagnostics. Lab Chip 2012, 12, 3249–3266. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Lee, J.; Kim, Y.; Kim, J.; Lee, J.; Park, S. 3D-printed microfluidic magnetic preconcentrator for the detection of bacterial pathogen using an ATP luminometer and antibody-conjugated magnetic nanoparticles. J. Microbiol. Methods 2017, 132, 128–133. [Google Scholar] [CrossRef]
- Jang, H.; Hwang, E.Y.; Kim, Y.; Choo, J.; Jeong, J.; Lim, D.W. Surface-enhanced raman scattering and fluorescence-based dual nanoprobes for multiplexed detection of bacterial pathogens. J. Biomed. Nanotechnol. 2016, 12, 1938–1951. [Google Scholar] [CrossRef] [PubMed]
- Ilhan, H.; Guven, B.; Dogan, U.; Torul, H.; Evran, S.; Çetin, D.; Suludere, Z.; Saglam, N.; Boyaci, İ.H.; Tamer, U. The coupling of immunomagnetic enrichment of bacteria with paper-based platform. Talanta 2019, 201, 245–252. [Google Scholar] [CrossRef]
- Peláez, E.C.; Estevez, M.C.; Mongui, A.; Menéndez, M.C.; Toro, C.; Herrera-Sandoval, O.L.; Robledo, J.; García, M.J.; Del Portillo, P.; Lechuga, L.M. Detection and Quantification of HspX Antigen in Sputum Samples Using Plasmonic Biosensing: Toward a Real Point-of-Care (POC) for Tuberculosis Diagnosis. ACS Infect. Dis. 2020, 6, 1110–1120. [Google Scholar] [CrossRef]
- Kaushik, A.; Yndart, A.; Kumar, S.; Jayant, R.D.; Vashist, A.; Brown, A.N.; Li, C.Z.; Nair, M. A sensitive electrochemical immunosensor for label-free detection of Zika-virus protein. Sci. Rep. 2018, 8, 3–7. [Google Scholar] [CrossRef] [Green Version]
- Reid, R.; Chatterjee, B.; Das, S.J.; Ghosh, S.; Sharma, T.K. Application of aptamers as molecular recognition elements in lateral flow assays. Anal. Biochem. 2020, 593, 113574. [Google Scholar] [CrossRef] [PubMed]
- Oue, S.; Okamoto, A.; Yano, T.; Kagamiyama, H. Redesigning the substrate specificity of an enzyme by cumulative effects of the mutations of non-active site residues. J. Biol. Chem. 1999, 274, 2344–2349. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Lu, L.; Xu, S.; Wang, L. One-pot synthesis of gold nanoclusters with bright red fluorescence and good biorecognition Abilities for visualization fluorescence enhancement detection of E. coli. Talanta 2015, 134, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Vocadlo, D.J.; Davies, G.J.; Laine, R.; Withers, S.G. Catalysis by hen egg-white lysozyme proceeds via a covalent intermediate. Nature 2001, 412, 835–838. [Google Scholar] [CrossRef] [PubMed]
- Couniot, N.; Vanzieleghem, T.; Rasson, J.; Van Overstraeten-Schlögel, N.; Poncelet, O.; Mahillon, J.; Francis, L.A.; Flandre, D. Lytic enzymes as selectivity means for label-free, microfluidic and impedimetric detection of whole-cell bacteria using ALD-Al2O3 passivated microelectrodes. Biosens. Bioelectron. 2015, 67, 154–161. [Google Scholar] [CrossRef]
- Clemente, A.; Alba-Patiño, A.; Rojo-Molinero, E.; Russell, S.M.; Borges, M.; Oliver, A.; De La Rica, R. Rapid Detection of Pseudomonas aeruginosa Biofilms via Enzymatic Liquefaction of Respiratory Samples. ACS Sens. 2020, 5, 3956–3963. [Google Scholar] [CrossRef]
- Pavan, S.; Berti, F. Short peptides as biosensor transducers. Anal. Bioanal. Chem. 2012, 402, 3055–3070. [Google Scholar] [CrossRef]
- Rodrigues, J.L.; Ferreira, D.; Rodrigues, L.R. Synthetic biology strategies towards the development of new bioinspired technologies for medical applications. In Bioinspired Materials for Medical Applications; Rodrigues, L., Mota, M., Eds.; Woodhead Publishing: Cambridge, UK, 2017; pp. 451–497. ISBN 9780081007464. [Google Scholar]
- Xu, C.; Akakuru, O.U.; Zheng, J.; Wu, A. Applications of iron oxide-based magnetic nanoparticles in the diagnosis and treatment of bacterial infections. Front. Bioeng. Biotechnol. 2019, 7, 141. [Google Scholar] [CrossRef] [PubMed]
- Mannoor, M.S.; Zhang, S.; Link, A.J.; McAlpine, M.C. Electrical detection of pathogenic bacteria via immobilized antimicrobial peptides. Proc. Natl. Acad. Sci. USA 2010, 107, 19207–19212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoyos-Nogués, M.; Gil, F.J.; Mas-Moruno, C. Antimicrobial peptides: Powerful biorecognition elements to detect bacteria in biosensing technologies. Molecules 2018, 23, 1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Ren, C.; Hu, F.; Gao, Y.; Wang, Z.; Li, H.; Liu, J.; Liu, B.; Yang, C. Detection of Bacterial Alkaline Phosphatase Activity by Enzymatic in Situ Self-Assembly of the AIEgen-Peptide Conjugate. Anal. Chem. 2020, 92, 5185–5190. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Mei, Q.; Guo, X.; Xu, Y.; Yang, D.; Sánchez, B.J.; Sheng, B.; Liu, C.; Hu, Z.; Yu, G.; et al. Antimicrobial peptide based magnetic recognition elements and Au@Ag-GO SERS tags with stable internal standards: A three in one biosensor for isolation, discrimination and killing of multiple bacteria in whole blood. Chem. Sci. 2018, 9, 8781–8795. [Google Scholar] [CrossRef] [Green Version]
- Dao, T.N.T.; Lee, E.Y.; Koo, B.; Jin, C.E.; Lee, T.Y.; Shin, Y. A microfluidic enrichment platform with a recombinase polymerase amplification sensor for pathogen diagnosis. Anal. Biochem. 2018, 544, 87–92. [Google Scholar] [CrossRef]
- Azmi, S.; Jiang, K.; Stiles, M.; Thundat, T.; Kaur, K. Detection of Listeria monocytogenes with short peptide fragments from class IIa bacteriocins as recognition elements. ACS Comb. Sci. 2015, 17, 156–163. [Google Scholar] [CrossRef]
- Etayash, H.; Jiang, K.; Thundat, T.; Kaur, K. Impedimetric detection of pathogenic gram-positive bacteria using an antimicrobial peptide from class IIa bacteriocins. Anal. Chem. 2014, 86, 1693–1700. [Google Scholar] [CrossRef]
- Arcidiacono, S.; Pivarnik, P.; Mello, C.M.; Senecal, A. Cy5 labeled antimicrobial peptides for enhanced detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2008, 23, 1721–1727. [Google Scholar] [CrossRef]
- De Miranda, J.L.; Oliveira, M.D.L.; Oliveira, I.S.; Frias, I.A.M.; Franco, O.L.; Andrade, C.A.S. A simple nanostructured biosensor based on clavanin A antimicrobial peptide for gram-negative bacteria detection. Biochem. Eng. J. 2017, 124, 108–114. [Google Scholar] [CrossRef]
- Akram, A.R.; Avlonitis, N.; Scholefield, E.; Vendrell, M.; McDonald, N.; Aslam, T.; Craven, T.H.; Gray, C.; Collie, D.S.; Fisher, A.J.; et al. Enhanced avidity from a multivalent fluorescent antimicrobial peptide enables pathogen detection in a human lung model. Sci. Rep. 2019, 9, 8422. [Google Scholar] [CrossRef] [Green Version]
- Wright, A.T.; Anslyn, E.V. Differential receptor arrays and assays for solution-based molecular recognition. Chem. Soc. Rev. 2006, 35, 14–28. [Google Scholar] [CrossRef]
- Kulagina, N.V.; Shaffer, K.M.; Ligler, F.S.; Taitt, C.R. Antimicrobial peptides as new recognition molecules for screening challenging species. Sens. Actuators B Chem. 2007, 121, 150–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, M.Q.; Wang, X.C.; Dou, W.T.; Chen, G.R.; James, T.D.; Zhou, D.M.; He, X.P. Supramolecular fluorogenic peptide sensor array based on graphene oxide for the differential sensing of ebola virus. Chem. Commun. 2020, 56, 5735–5738. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Lai, D.; Lei, Q.; Xu, Z.; Wang, F.; Hou, H.; Chen, L.; Wu, J.; Ren, Y.; Liang, M.M.; et al. Systematic evaluation of IgG responses to SARS-CoV-2 spike protein-derived peptides for monitoring COVID-19 patients. Cell. Mol. Immunol. 2021, 18, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.F.; Chen, J.; Hu, J.l.; Long, Q.X.; Deng, H.J.; Liu, P.; Fan, K.; Liao, P.; Liu, B.Z.; Wu, G.C.; et al. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of coronavirus disease 2019. J. Infect. Dis. 2020, 222, 189–195. [Google Scholar] [CrossRef]
- Pomplun, S.; Jbara, M.; Quartararo, A.J.; Zhang, G.; Brown, J.S.; Lee, Y.C.; Ye, X.; Hanna, S.; Pentelute, B.L. De Novo Discovery of High-Affinity Peptide Binders for the SARS-CoV-2 Spike Protein. ACS Cent. Sci. 2021, 7, 156–163. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Mcconnell, E.M.; Cozma, I.; Morrison, D.; Li, Y. Biosensors made of synthetic functional nucleic acids toward better human health. Anal. Chem. 2020, 92, 327–344. [Google Scholar] [CrossRef] [PubMed]
- Saadati, A.; Hassanpour, S.; de la Guardia, M.; Mosafer, J.; Hashemzaei, M.; Mokhtarzadeh, A.; Baradaran, B. Recent advances on application of peptide nucleic acids as a bioreceptor in biosensors development. TrAC Trends Anal. Chem. 2019, 114, 56–68. [Google Scholar] [CrossRef]
- Wang, C.H.; Wu, J.J.; Lee, G. Bin Screening of highly-specific aptamers and their applications in paper-based microfluidic chips for rapid diagnosis of multiple bacteria. Sens. Actuators B Chem. 2019, 284, 395–402. [Google Scholar] [CrossRef]
- Sousa, D.; Ferreira, D.; Rodrigues, J.L.; Rodrigues, L.R. Nanotechnology in Targeted Drug Delivery and Therapeutics. In Applications of Targeted Nano Drugs and Delivery Systems; Mohapatra, S.S., Ranjan, S., Dasgupta, N., Mishra, R.K., Thomas, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 357–409. [Google Scholar]
- Dwivedi, H.P.; Smiley, R.D.; Jaykus, L.A. Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Appl. Microbiol. Biotechnol. 2010, 87, 2323–2334. [Google Scholar] [CrossRef]
- Meirinho, S.G.; Dias, L.G.; Peres, A.M.; Rodrigues, L.R. Voltammetric aptasensors for protein disease biomarkers detection: A review. Biotechnol. Adv. 2016, 34, 941–953. [Google Scholar] [CrossRef] [Green Version]
- Dias, L.G.; Meirinho, S.G.; Veloso, A.C.A.; Rodrigues, L.R.; Peres, A.M. Electronic tongues and aptasensors. In Bioinspired Materials for Medical Applications; Rodrigues, L., Mota, M., Eds.; Woodhead Publishing: Cambridge, UK, 2017; pp. 371–402. ISBN 9780081007464. [Google Scholar]
- Meirinho, S.G.; Dias, L.G.; Peres, A.M.; Rodrigues, L.R. Electrochemical aptasensor for human osteopontin detection using a DNA aptamer selected by SELEX. Anal. Chim. Acta 2017, 987, 25–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’Donoghue, M.B.; Tan, W. Development of DNA aptamers using cell-selex. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef]
- Meirinho, S.G.; Dias, L.G.; Peres, A.M.; Rodrigues, L.R. Development of an electrochemical RNA-aptasensor to detect human osteopontin. Biosens. Bioelectron. 2015, 71, 332–341. [Google Scholar] [CrossRef] [Green Version]
- Savory, N.; Lednor, D.; Tsukakoshi, K.; Abe, K.; Yoshida, W.; Ferri, S.; Jones, B.V.; Ikebukuro, K. In silico maturation of binding-specificity of DNA aptamers against Proteus mirabilis. Biotechnol. Bioeng. 2013, 110, 2573–2580. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.L.; Zhou, Y.X.; Wu, S.M.; Pan, Q.; Xia, B.; Zhang, X.L. CFP10 and ESAT6 aptamers as effective Mycobacterial antigen diagnostic reagents. J. Infect. 2014, 69, 569–580. [Google Scholar] [CrossRef]
- Weldingh, K.; Andersen, P. ESAT-6/CFP10 skin test predicts disease in M. tuberculosis-infected Guinea pigs. PLoS ONE 2008, 3, 1978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wu, H.; Yang, Y.; Yan, R.; Zhao, Y.; Wang, Y.; Chen, A.; Shao, S.; Jiang, P.; Li, Y.Q. Bacterial species-identifiable magnetic nanosystems for early sepsis diagnosis and extracorporeal photodynamic blood disinfection. Nanoscale 2018, 10, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.; Gu, H.; Duan, N.; Wu, S.; Ma, X.; Xia, Y.; Wang, H.; Wang, Z. A chemiluminescent aptasensor based on rolling circle amplification and Co2+/N-(aminobutyl)-N-(ethylisoluminol) functional flowerlike gold nanoparticles for Salmonella typhimurium detection. Talanta 2017, 164, 275–282. [Google Scholar] [CrossRef]
- Murakami, T.; Sumaoka, J.; Komiyama, M. Sensitive isothermal detection of nucleic-acid sequence by primer generation-rolling circle amplification. Nucleic Acids Res. 2009, 37, e19. [Google Scholar] [CrossRef] [Green Version]
- Trunzo, N.E.; Hong, K.L. Recent progress in the identification of aptamers against bacterial origins and their diagnostic applications. Int. J. Mol. Sci. 2020, 21, 5074. [Google Scholar] [CrossRef]
- Park, K.S. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens. Bioelectron. 2018, 102, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Wu, J.; Gu, J.; Shen, L.; Mao, L. Application of aptamers in virus detection and antiviral therapy. Front. Microbiol. 2019, 10, 1462. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Li, Q.; Liao, Z.; Sun, Y.; Cheng, Q.; Song, Y.; Song, E.; Tan, W. Magnetism-Resolved Separation and Fluorescence Quantification for Near-Simultaneous Detection of Multiple Pathogens. Anal. Chem. 2018, 90, 9621–9628. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, H.; Luan, C.; Liu, Y.; Chen, B.; Zhao, Y. Aptamer-based hydrogel barcodes for the capture and detection of multiple types of pathogenic bacteria. Biosens. Bioelectron. 2018, 100, 404–410. [Google Scholar] [CrossRef]
- Sande, M.G.; Çaykara, T.; Silva, C.J.; Rodrigues, L.R. New solutions to capture and enrich bacteria from complex samples. Med. Microbiol. Immunol. 2020, 209, 335–341. [Google Scholar] [CrossRef] [Green Version]
- Shahrokhian, S.; Ranjbar, S. Aptamer immobilization on amino-functionalized metal-organic frameworks: An ultrasensitive platform for the electrochemical diagnostic of: Escherichia coli O157:H7. Analyst 2018, 143, 3191–3201. [Google Scholar] [CrossRef]
- Abbaspour, A.; Norouz-Sarvestani, F.; Noori, A.; Soltani, N. Aptamer-conjugated silver nanoparticles for electrochemical dual-aptamer-based sandwich detection of Staphylococcus aureus. Biosens. Bioelectron. 2015, 68, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, X.; Xia, Y.; Wu, S.; Duan, N.; Ma, X.; Wang, Z. Selection, identification and application of a DNA aptamer against Staphylococcus aureus enterotoxin A. Anal. Methods 2014, 6, 690–697. [Google Scholar] [CrossRef]
- Pang, Y.; Wan, N.; Shi, L.; Wang, C.; Sun, Z.; Xiao, R.; Wang, S. Dual-recognition surface-enhanced Raman scattering(SERS)biosensor for pathogenic bacteria detection by using vancomycin-SERS tags and aptamer-Fe3O4@Au. Anal. Chim. Acta 2019, 1077, 288–296. [Google Scholar] [CrossRef]
- Hamula, C.L.A.; Peng, H.; Wang, Z.; Tyrrell, G.J.; Li, X.F.; Le, X.C. An improved SELEX technique for selection of DNA aptamers binding to M-type 11 of Streptococcus pyogenes. Methods 2016, 97, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Sypabekova, M.; Bekmurzayeva, A.; Wang, R.; Li, Y.; Nogues, C.; Kanayeva, D. Selection, characterization, and application of DNA aptamers for detection of Mycobacterium tuberculosis secreted protein MPT64. Tuberculosis 2017, 104, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Meng, X.; Sun, Y.; Li, Q.; Zhao, R.; Zhang, Y.; Wang, J.; Yi, Z. Aptamer-based fluorometric assay for direct identification of methicillin-resistant Staphylococcus aureus from clinical samples. J. Microbiol. Methods 2018, 153, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Gao, X.; Gao, R.; Jia, L. Selective capture and sensitive fluorometric determination of Pseudomonas aeruginosa by using aptamer modified magnetic nanoparticles. Microchim. Acta 2018, 185. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.T.; Seo Bin, H.; Kim, B.C.; Kim, S.K.; Song, C.S.; Gu, M.B. Highly sensitive sandwich-type SPR based detection of whole H5Nx viruses using a pair of aptamers. Biosens. Bioelectron. 2016, 86, 293–300. [Google Scholar] [CrossRef]
- Chen, C.; Zou, Z.; Chen, L.; Ji, X.; He, Z. Functionalized magnetic microparticle-based colorimetric platform for influenza A virus detection. Nanotechnology 2016, 27, 435102. [Google Scholar] [CrossRef]
- Sung, H.J.; Kayhan, B.; Ben-Yedidia, T.; Arnon, R. A DNA aptamer prevents influenza infection by blocking the receptor binding region of the viral hemagglutinin. J. Biol. Chem. 2004, 279, 48410–48419. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.H.; Zeng, H. Aptamer-Based ELISA Assay for Highly Specific and Sensitive Detection of Zika NS1 Protein. Anal. Chem. 2017, 89, 12743–12748. [Google Scholar] [CrossRef]
- Giamberardino, A.; Labib, M.; Hassan, E.M.; Tetro, J.A.; Springthorpe, S.; Sattar, S.A.; Berezovski, M.V.; DeRosa, M.C. Ultrasensitive norovirus detection using DNA aptasensor technology. PLoS ONE 2013, 8, 79087. [Google Scholar] [CrossRef] [Green Version]
- Bai, C.; Lu, Z.; Jiang, H.; Yang, Z.; Liu, X.; Ding, H.; Li, H.; Dong, J.; Huang, A.; Fang, T.; et al. Aptamer selection and application in multivalent binding-based electrical impedance detection of inactivated H1N1 virus. Biosens. Bioelectron. 2018, 110, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.K.; Griffiths, C.; Lea, S.M.; James, W. Structural characterization of an anti-gp120 RNA aptamer that neutralizes R5 strains of HIV-1. RNA 2005, 11, 873–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbasi, A.D.; Hussain, Z.; Yang, K.L. Aptamer laden liquid crystals biosensing platform for the detection of HIV-1 glycoprotein-120. Molecules 2021, 26, 2893. [Google Scholar] [CrossRef]
- Chekin, F.; Bagga, K.; Subramanian, P.; Jijie, R.; Singh, S.K.; Kurungot, S.; Boukherroub, R.; Szunerits, S. Nucleic aptamer modified porous reduced graphene oxide/MoS2 based electrodes for viral detection: Application to human papillomavirus (HPV). Sens. Actuators B Chem. 2018, 262, 991–1000. [Google Scholar] [CrossRef]
- Leija-Montoya, A.G.; Benítez-Hess, M.L.; Toscano-Garibay, J.D.; Alvarez-Salas, L.M. Characterization of an RNA aptamer against HPV-16 L1 virus-like particles. Nucleic Acid Ther. 2014, 24, 344–355. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Song, J.; Wei, X.; Huang, M.; Sun, M.; Zhu, L.; Lin, B.; Shen, H.; Zhu, Z.; Yang, C. Discovery of Aptamers Targeting the Receptor-Binding Domain of the SARS-CoV-2 Spike Glycoprotein. Anal. Chem. 2020, 92, 9895–9900. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Fang, X.; Liu, X.; Ou, H.; Zhang, H.; Wang, J.; Li, Q.; Cheng, H.; Zhang, W.; Luo, Z. Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers. Chem. Commun. 2020, 56, 10235–10238. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, Q.; Chen, J.; Ni, X.; Dai, J. A DNA Aptamer Based Method for Detection of SARS-CoV-2 Nucleocapsid Protein. Virol. Sin. 2020, 35, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, A.; Weber, A.; Bayin, M.; Breuers, S.; Fieberg, V.; Famulok, M.; Mayer, G. A SARS-CoV-2 Spike Binding DNA Aptamer that Inhibits Pseudovirus Infection by an RBD-Independent Mechanism. Angew. Chem. Int. Ed. 2021, 60, 10279–10285. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Cai, H.; Park, M.; Wall, T.A.; Stott, M.A.; Alfson, K.J.; Griffiths, A.; Carrion, R.; Patterson, J.L.; Hawkins, A.R.; et al. Multiplexed efficient on-chip sample preparation and sensitive amplification-free detection of Ebola virus. Biosens. Bioelectron. 2017, 91, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G. Predicting the uncertain future of aptamer-based diagnostics and therapeutics. Molecules 2015, 20, 6866–6887. [Google Scholar] [CrossRef] [PubMed]
- Lakhin, A.V.; Tarantul, V.Z.; Gening, L.V. Aptamers: Problems, solutions and prospects. Acta Nat. 2013, 5, 34–43. [Google Scholar] [CrossRef] [Green Version]
- Bauer, M.; Strom, M.; Hammond, D.S.; Shigdar, S. Anything you can do, I can do better: Can aptamers replace antibodies in clinical diagnostic applications? Molecules 2019, 24, 4377. [Google Scholar] [CrossRef] [Green Version]
- Silverman, S.K. DNA as a versatile chemical component for catalysis, encoding, and stereocontrol. Angew. Chem. Int. Ed. 2010, 49, 7180–7201. [Google Scholar] [CrossRef]
- Breaker, R.R.; Joyce, G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1, 223–229. [Google Scholar] [CrossRef]
- Swearingen, C.B.; Wernette, D.P.; Cropek, D.M.; Lu, Y.; Sweedler, J.V.; Bohn, P.W. Immobilization of a catalytic DNA molecular beacon on Au for Pb(II) detection. Anal. Chem. 2005, 77, 442–448. [Google Scholar] [CrossRef] [Green Version]
- Geng, X.; Zhang, M.; Wang, X.; Sun, J.; Zhao, X.; Zhang, L.; Wang, X.; Shen, Z. Selective and sensitive detection of chronic myeloid leukemia using fluorogenic DNAzyme probes. Anal. Chim. Acta 2020, 1123, 28–35. [Google Scholar] [CrossRef]
- Zheng, L.; Qi, P.; Zhang, D. DNA-templated fluorescent silver nanoclusters for sensitive detection of pathogenic bacteria based on MNP-DNAzyme-AChE complex. Sens. Actuators B Chem. 2018, 276, 42–47. [Google Scholar] [CrossRef]
- Kang, D.K.; Ali, M.M.; Zhang, K.; Huang, S.S.; Peterson, E.; Digman, M.A.; Gratton, E.; Zhao, W. Rapid detection of single bacteria in unprocessed blood using Integrated Comprehensive Droplet Digital Detection. Nat. Commun. 2014, 5, 5427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ali, M.M.; Slepenkin, A.; Peterson, E.; Zhao, W. A Simple DNAzyme-Based Fluorescent Assay for Klebsiella pneumoniae. ChemBioChem 2019, 20, 906–910. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Wolfe, M.; Tram, K.; Gu, J.; Filipe, C.D.M.; Li, Y.; Brennan, J.D. A DNAzyme-Based Colorimetric Paper Sensor for Helicobacter pylori. Angew. Chem. 2019, 131, 10012–10016. [Google Scholar] [CrossRef]
- Kim, S.U.; Batule, B.S.; Mun, H.; Byun, J.Y.; Shim, W.B.; Kim, M.G. Colorimetric molecular diagnosis of the HIV gag gene using DNAzyme and a complementary DNA-extended primer. Analyst 2018, 143, 695–699. [Google Scholar] [CrossRef]
- Anantharaj, A.; Das, S.J.; Sharanabasava, P.; Lodha, R.; Kabra, S.K.; Sharma, T.K.; Medigeshi, G.R. Visual Detection of SARS-CoV-2 RNA by Conventional PCR-Induced Generation of DNAzyme Sensor. Front. Mol. Biosci. 2020, 7, 444. [Google Scholar] [CrossRef] [PubMed]
- Xi, H.; Juhas, M.; Zhang, Y. G-quadruplex based biosensor: A potential tool for SARS-CoV-2 detection. Biosens. Bioelectron. 2020, 167, 112494. [Google Scholar] [CrossRef]
- Lee, H.T.; Kim, S.K.; Yoon, J.W. Antisense peptide nucleic acids as a potential anti-infective agent. J. Microbiol. 2019, 57, 423–430. [Google Scholar] [CrossRef]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Vilaivan, T.; Chailapakul, O.; Henry, C.S. Multiplex Paper-Based Colorimetric DNA Sensor Using Pyrrolidinyl Peptide Nucleic Acid-Induced AgNPs Aggregation for Detecting MERS-CoV, MTB, and HPV Oligonucleotides. Anal. Chem. 2017, 89, 5428–5435. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.S.; Lytton-Jean, A.K.R.; Hurst, S.J.; Mirkin, C.A. Silver nanoparticle—Oligonucleotide conjugates based on DNA with triple cyclic disulfide moieties. Nano Lett. 2007, 7, 2112–2115. [Google Scholar] [CrossRef] [Green Version]
- Machado, A.; Almeida, C.; Salgueiro, D.; Henriques, A.; Vaneechoutte, M.; Haesebrouck, F.; Vieira, M.J.; Rodrigues, L.; Azevedo, N.F.; Cerca, N. Fluorescence in situ Hybridization method using Peptide Nucleic Acid probes for rapid detection of Lactobacillus and Gardnerella spp. BMC Microbiol. 2013, 13, 82. [Google Scholar] [CrossRef] [Green Version]
- Rocha, R.; Sousa, J.M.; Cerqueira, L.; Vieira, M.J.; Almeida, C.; Azevedo, N.F. Development and application of Peptide Nucleic Acid Fluorescence in situ Hybridization for the specific detection of Listeria monocytogenes. Food Microbiol. 2019, 80, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Almeida, C.; Azevedo, N.F.; Santos, S.; Keevil, C.W.; Vieira, M.J. Discriminating multi-species populations in biofilms with peptide nucleic acid fluorescence in situ hybridization (PNA FISH). PLoS ONE 2011, 6, 14786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahour, F.; Pournaghi-Azar, M.H.; Alipour, E.; Hejazi, M.S. Detection and discrimination of recombinant plasmid encoding hepatitis C virus core/E1 gene based on PNA and double-stranded DNA hybridization. Biosens. Bioelectron. 2013, 45, 287–291. [Google Scholar] [CrossRef]
- Oliveira, R.; Almeida, C.; Azevedo, N.F. Detection of Microorganisms by Fluorescence In Situ Hybridization Using Peptide Nucleic Acid. In Methods in Molecular Biology; Nielsen, P., Ed.; Humana Press Inc.: New York, NY, USA, 2020; Volume 2105, pp. 217–230. [Google Scholar]
- Talebian, S.; Wallace, G.G.; Schroeder, A.; Stellacci, F.; Conde, J. Nanotechnology-based disinfectants and sensors for SARS-CoV-2. Nat. Nanotechnol. 2020, 15, 618–621. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Shang, X.; Huang, X. Next-generation pathogen diagnosis with CRISPR/Cas-based detection methods. Emerg. Microbes Infect. 2020, 9, 1682–1691. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [Green Version]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-based diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef]
- Khambhati, K.; Bhattacharjee, G.; Singh, V. Current progress in CRISPR-based diagnostic platforms. J. Cell. Biochem. 2019, 120, 2721–2725. [Google Scholar] [CrossRef] [PubMed]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef] [Green Version]
- Ai, J.W.; Zhou, X.; Xu, T.; Yang, M.; Chen, Y.; He, G.Q.; Pan, N.; Cai, Y.; Li, Y.; Wang, X.; et al. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg. Microbes Infect. 2019, 8, 1361–1369. [Google Scholar] [CrossRef] [Green Version]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. SHERLOCK: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020, 16, e1008705. [Google Scholar] [CrossRef]
- Eisenstein, M. My enemy’s enemy is my friend. Nat. Methods 2006, 3, 338. [Google Scholar] [CrossRef]
- Santos, S.B.; Cunha, A.P.; Macedo, M.; Nogueira, C.L.; Brandão, A.; Costa, S.P.; Melo, L.D.R.; Azeredo, J.; Carvalho, C.M. Bacteriophage-receptor binding proteins for multiplex detection of Staphylococcus and Enterococcus in blood. Biotechnol. Bioeng. 2020, 117, 3286–3298. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, M.; Fan, E.; Ouyang, H.; Yue, H.; Su, X.; Liao, G.; Wang, L.; Lu, S.; Fu, Z. Highly Specific Bacteriophage-Affinity Strategy for Rapid Separation and Sensitive Detection of Viable Pseudomonas aeruginosa. Anal. Chem. 2017, 89, 1916–1921. [Google Scholar] [CrossRef]
- Ackermann, H.W. Tailed bacteriophages: The order caudovirales. Adv. Virus Res. 1998, 51, 135–201. [Google Scholar] [CrossRef] [PubMed]
- Liana, A.E.; Marquis, C.P.; Gunawan, C.; Gooding, J.J.; Amal, R. T4 bacteriophage conjugated magnetic particles for E. coli capturing: Influence of bacteriophage loading, temperature and tryptone. Colloids Surf. B Biointerfaces 2017, 151, 47–57. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Chen, W.; Ma, Y.; Pan, G. Molecularly imprinted polymers as receptor mimics for selective cell recognition. Chem. Soc. Rev. 2018, 47, 5574–5587. [Google Scholar] [CrossRef]
- Khan, M.A.R.; Aires Cardoso, A.R.; Sales, M.G.F.; Merino, S.; Tomás, J.M.; Rius, F.X.; Riu, J. Artificial receptors for the electrochemical detection of bacterial flagellar filaments from Proteus mirabilis. Sens. Actuators B Chem. 2017, 244, 732–741. [Google Scholar] [CrossRef]
- Golabi, M.; Kuralay, F.; Jager, E.W.H.; Beni, V.; Turner, A.P.F. Electrochemical bacterial detection using poly(3-aminophenylboronic acid)-based imprinted polymer. Biosens. Bioelectron. 2017, 93, 87–93. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Chen, X.; Zhang, L.; Gao, J.; Ma, Q. Electrochemiluminescence Detection of Escherichia coli O157:H7 Based on a Novel Polydopamine Surface Imprinted Polymer Biosensor. ACS Appl. Mater. Interfaces 2017, 9, 5430–5436. [Google Scholar] [CrossRef]
- Idil, N.; Hedström, M.; Denizli, A.; Mattiasson, B. Whole cell based microcontact imprinted capacitive biosensor for the detection of Escherichia coli. Biosens. Bioelectron. 2017, 87, 807–815. [Google Scholar] [CrossRef]
- Shan, X.; Yamauchi, T.; Yamamoto, Y.; Shiigi, H.; Nagaoka, T. A rapid and specific bacterial detection method based on cell-imprinted microplates. Analyst 2018, 143, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Piletsky, S.; Canfarotta, F.; Poma, A.; Bossi, A.M.; Piletsky, S. Molecularly Imprinted Polymers for Cell Recognition. Trends Biotechnol. 2020, 38, 368–387. [Google Scholar] [CrossRef] [PubMed]
- Cai, D.; Ren, L.; Zhao, H.; Xu, C.; Zhang, L.; Yu, Y.; Wang, H.; Lan, Y.; Roberts, M.F.; Chuang, J.H.; et al. A molecular-imprint nanosensor for ultrasensitive detection of proteins. Nat. Nanotechnol. 2010, 5, 597–601. [Google Scholar] [CrossRef] [Green Version]
- Cui, F.; Zhou, Z.; Zhou, H.S. Molecularly imprinted polymers and surface imprinted polymers based electrochemical biosensor for infectious diseases. Sensors 2020, 20, 996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Shen, X.L.; Zeng, Q.; Wang, H.S.; Wang, L.S. A multi-walled carbon nanotubes based molecularly imprinted polymers electrochemical sensor for the sensitive determination of HIV-p24. Talanta 2017, 164, 121–127. [Google Scholar] [CrossRef]
- Yang, S.; Ouyang, H.; Su, X.; Gao, H.; Kong, W.; Wang, M.; Shu, Q.; Fu, Z. Dual-recognition detection of Staphylococcus aureus using vancomycin-functionalized magnetic beads as concentration carriers. Biosens. Bioelectron. 2016, 78, 174–180. [Google Scholar] [CrossRef]
- Bu, T.; Yao, X.; Huang, L.; Dou, L.; Zhao, B.; Yang, B.; Li, T.; Wang, J.; Zhang, D. Dual recognition strategy and magnetic enrichment based lateral flow assay toward Salmonella enteritidis detection. Talanta 2020, 206, 120204. [Google Scholar] [CrossRef]
- Kell, A.J.; Stewart, G.; Ryan, S.; Peytavi, R.; Boissinot, M.; Huletsky, A.; Bergeron, M.G.; Simard, B. Vancomycin-modified nanoparticles for efficient targeting and preconcentration of gram-positive and gram-negative bacteria. ACS Nano 2008, 2, 1777–1788. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhou, X.; Zhu, M.; Xing, D. Sensitive detection of Listeria monocytogenes based on highly efficient enrichment with vancomycin-conjugated brush-like magnetic nano-platforms. Biosens. Bioelectron. 2017, 91, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Li, F.; Li, F.; Xiong, Y.; Xu, H. Vancomycin modified PEGylated-magnetic nanoparticles combined with PCR for efficient enrichment and detection of Listeria monocytogenes. Sens. Actuators B Chem. 2017, 247, 546–555. [Google Scholar] [CrossRef]
- Wang, C.W.; Gu, B.; Liu, Q.Q.; Pang, Y.F.; Xiao, R.; Wang, S.Q. Combined use of vancomycin-modified Ag-coated magnetic nanoparticles and secondary enhanced nanoparticles for rapid surface-enhanced Raman scattering detection of bacteria. Int. J. Nanomed. 2018, 13, 1159–1178. [Google Scholar] [CrossRef] [Green Version]
- Kong, K.F.; Schneper, L.; Mathee, K. Beta-lactam antibiotics: From antibiosis to resistance and bacteriology. Apmis 2010, 118, 1–36. [Google Scholar] [CrossRef] [Green Version]
- Hasan, N.; Guo, Z.; Wu, H.F. Large protein analysis of Staphylococcus aureus and Escherichia coli by MALDI TOF mass spectrometry using amoxicillin functionalized magnetic nanoparticles. Anal. Bioanal. Chem. 2016, 408, 6269–6281. [Google Scholar] [CrossRef]
- Carter, A.P.; Clemons, W.M.; Brodersen, D.E.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 2000, 407, 340–348. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, Q.; Xu, S.; Zuo, L.; You, X.; Hu, H.Y. Aminoglycoside-based novel probes for bacterial diagnostic and therapeutic applications. Chem. Commun. 2017, 53, 1366–1369. [Google Scholar] [CrossRef]
- Pang, X.; Xiao, Q.; Cheng, Y.; Ren, E.; Lian, L.; Zhang, Y.; Gao, H.; Wang, X.; Leung, W.; Chen, X.; et al. Bacteria-responsive nanoliposomes as smart sonotheranostics for multidrug resistant bacterial infections. ACS Nano 2019, 13, 2427–2438. [Google Scholar] [CrossRef]
- Fu, X.; Wang, Z.; Li, L.; Dong, S.; Li, Z.; Jiang, Z.; Wang, Y.; Shui, W. Novel Chemical Ligands to Ebola Virus and Marburg Virus Nucleoproteins Identified by Combining Affinity Mass Spectrometry and Metabolomics Approaches. Sci. Rep. 2016, 6, 29680. [Google Scholar] [CrossRef]
- Vaca, D.J.; Thibau, A.; Schütz, M.; Kraiczy, P.; Happonen, L.; Malmström, J.; Kempf, V.A.J. Interaction with the host: The role of fibronectin and extracellular matrix proteins in the adhesion of Gram-negative bacteria. Med. Microbiol. Immunol. 2020, 209, 277–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Pathogen Class | Biorecognition Element | Company | Product | Type of Test | Refs. |
|---|---|---|---|---|---|
| Clostridium difficile | Antibodies specific to C. difficile antigen glutamate dehydrogenase | Corisbio | Clostridium K-SeT | Immunochromatographic assay | [14] |
| N/A | Thermo Scientific | Oxoid™ Clostridium difficile Test Kit | Rapid latex agglutination | [15] | |
| Escherichia coli serogroup O157 | N/A | Meridian Bioscience, USA | ImmunoCard STAT! E. coli O157 Plus | Lateral flow immunoassay | [16] |
| Antibodies specific to O157 serogroup antigen | Thermo Scientific | Escherichia coli O157 Latex Test | Colorimetric latex agglutination | [17] | |
| Haemophilus influenzae b | Antibodies specific to H. influenzae type b antigen | Thermo Scientific | Wellcogen™ Haemophilus influenzae b | Rapid Latex Agglutination | [18] |
| Helicobacter pylori | Antibodies specific to H. pylori | Quidel, USA | QuickVue H. pylori Test | Solid phase immunoassay | [19] |
| Anti-H. pylori lgG antibody | Beckman Coulter, USA | Icon HP | Immunoassay | [20] | |
| Antibodies specific to H. pylori antigens | Corisbio | Pylori-Strip/Pylori K-SeT | Immunochromatographic assay | [21,22] | |
| Moraxella catarrhalis | Indoxyl butyrate detects the enzyme butyrate esterase of M. catarrhalis | Thermo Scientific | Remel™ Catarrhalis Test Disc | Colorimetric chemical identification | [23] |
| Staphylococcus aureus | A protein detects clumping factor and protein A of S. aureus | Thermo Scientific | BactiStaph™ Latex Agglutination Test Kit | Agglutination of protein-coated latex particles | [24] |
| Streptococcus pneumoniae | Specific rabbit antibody | Thermo Scientific | DrySpot™ Pneumo Latex Agglutination Test | Agglutination of antibody-coated latex particles | [25] |
| N/A | Abbott | BinaxNOW™ Streptococcus pneumoniae Antigen Card | Lateral flow immunoassay | [26] | |
| Epstein-Barr virus | Heterophile antibody detects bovine red cell mononucleosis antigen | Thermo Scientific | Infectious Mononucleosis Test using Latex Agglutination | Latex Agglutination | [27] |
| Influenza A and B virus | Antibodies specific to Influenza type A and type B antigens | Quidel, USA | QuickVue Influenza A + B test | Immunoassay | [28] |
| Antibody detects nucleoprotein antigens | Abbott | Alere BinaxNOW® Influenza A & B Card | Immunochromatographic assay | [29] | |
| Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) | N/A | Lucira Health | COVID-19 All-In-One Test Kit (FDA approved) | Amplification of viral genetic material | [30] |
| Antibody | Abbott | BinaxNOW COVID-19 Ag Card Home Test (FDA Emergency Use Authorization) | Rapid antigen | [31] | |
| Antibody | Ellume Health | Ellume COVID-19 Home Test (FDA Emergency Use Authorization) | Rapid antigen | [32] | |
| Plasmodium falciparum | Antibodies detect histidine-rich protein II antigen specific to P. falciparum and a pan-malarial antigen | Abbott | BinaxNOW® Malaria | Immunochromatographic assay | [33] |
| Bacteria | Target (Analyte) | Detection System | Refs. |
|---|---|---|---|
| Escherichia coli O157:H7 and Salmonella typhimurium | Whole bacteria | Aptamer-modified fluorescent magnetic multifunctional nanoprobes | [92] |
| Staphylococcus aureus and Escherichia coli | Whole bacteria | Multiplex aptamer-based hydrogel barcodes | [93,94] |
| Escherichia coli O157:H7 | Whole bacteria | Electrochemical biosensor with amino-functionalised metal–organic frame integrated with aptamers | [95] |
| Staphylococcus aureus | Whole bacteria | Electrochemical detection by dual-aptamer-based sandwich with silver nanoparticles | [96] |
| Staphylococcus aureus | Enterotoxin A protein | Graphene oxide-based fluorescent bioassay | [97] |
| Staphylococcus aureus | Whole bacteria | Surface-enhanced Raman spectroscopy (SERS) biosensor | [98] |
| Campylobacter jenuni | Whole bacteria | N/A | [77] |
| Streptococcus pyogenes | M11 M-type serotype whole bacteria | N/A | [99] |
| Mycobacterium tuberculosis | MPT64 secreted protein | Enzyme linked oligonucleotide assay | [100] |
| Methicillin-resistant Staphylococcus aureus (MRSA) | Penicillin binding protein 2a (PBP2a) | Fluorometric assay | [101] |
| Pseudomonas aeruginosa | Whole bacteria | Fluorometric assay | [102] |
| Virus | Target (Analyte) | Detection System | Refs. |
|---|---|---|---|
| Avian influenza strain H5Nx | Whole virus | Sandwich-type surface plasmon resonance (SPR)-based biosensor assay | [103] |
| Influenza A strain H3N2 | Globular region of hemagglutinin | Aptamer-functionalised magnetic microparticle-based colorimetric method | [104,105] |
| Zika | Zika NS1 Protein | Aptamer-Based enzyme-linked immunosorbent assay (ELISA) | [106] |
| Norovirus | Murine norovirus and capsids of a human norovirus strain GII.3 | Electrochemical sensor | [107] |
| H1N1 | Inactivated H1N1 virus particles | Electrochemical impedance sensor | [108] |
| Human immuno-deficiency virus Type 1 (HIV-1) | Glycoprotein-120 (gp-120) | Liquid crystal optical sensor | [109,110] |
| Human papillomavirus (HPV) | L1-major capsid protein of HPV | Electrochemical impedance sensor | [111,112] |
| SARS-CoV-2 | Receptor-binding domain (RBD) of the spike glycoprotein | N/A | [113] |
| SARS-CoV-2 | Nucleocapsid protein | ELISA and a gold nanoparticle immunochromatographic strip | [114] |
| SARS-CoV-2 | Nucleocapsid protein | N/A | [115] |
| SARS-CoV-2 | Spike glycoprotein | N/A | [116] |
| Ebola | Viral RNA | Antiresonant reflecting optical waveguide | [117] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sande, M.G.; Rodrigues, J.L.; Ferreira, D.; Silva, C.J.; Rodrigues, L.R. Novel Biorecognition Elements against Pathogens in the Design of State-of-the-Art Diagnostics. Biosensors 2021, 11, 418. https://doi.org/10.3390/bios11110418
Sande MG, Rodrigues JL, Ferreira D, Silva CJ, Rodrigues LR. Novel Biorecognition Elements against Pathogens in the Design of State-of-the-Art Diagnostics. Biosensors. 2021; 11(11):418. https://doi.org/10.3390/bios11110418
Chicago/Turabian StyleSande, Maria G., Joana L. Rodrigues, Débora Ferreira, Carla J. Silva, and Ligia R. Rodrigues. 2021. "Novel Biorecognition Elements against Pathogens in the Design of State-of-the-Art Diagnostics" Biosensors 11, no. 11: 418. https://doi.org/10.3390/bios11110418
APA StyleSande, M. G., Rodrigues, J. L., Ferreira, D., Silva, C. J., & Rodrigues, L. R. (2021). Novel Biorecognition Elements against Pathogens in the Design of State-of-the-Art Diagnostics. Biosensors, 11(11), 418. https://doi.org/10.3390/bios11110418








