Aptamer Embedded Arch-Cruciform DNA Assemblies on 2-D VS2 Scaffolds for Sensitive Detection of Breast Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Instruments
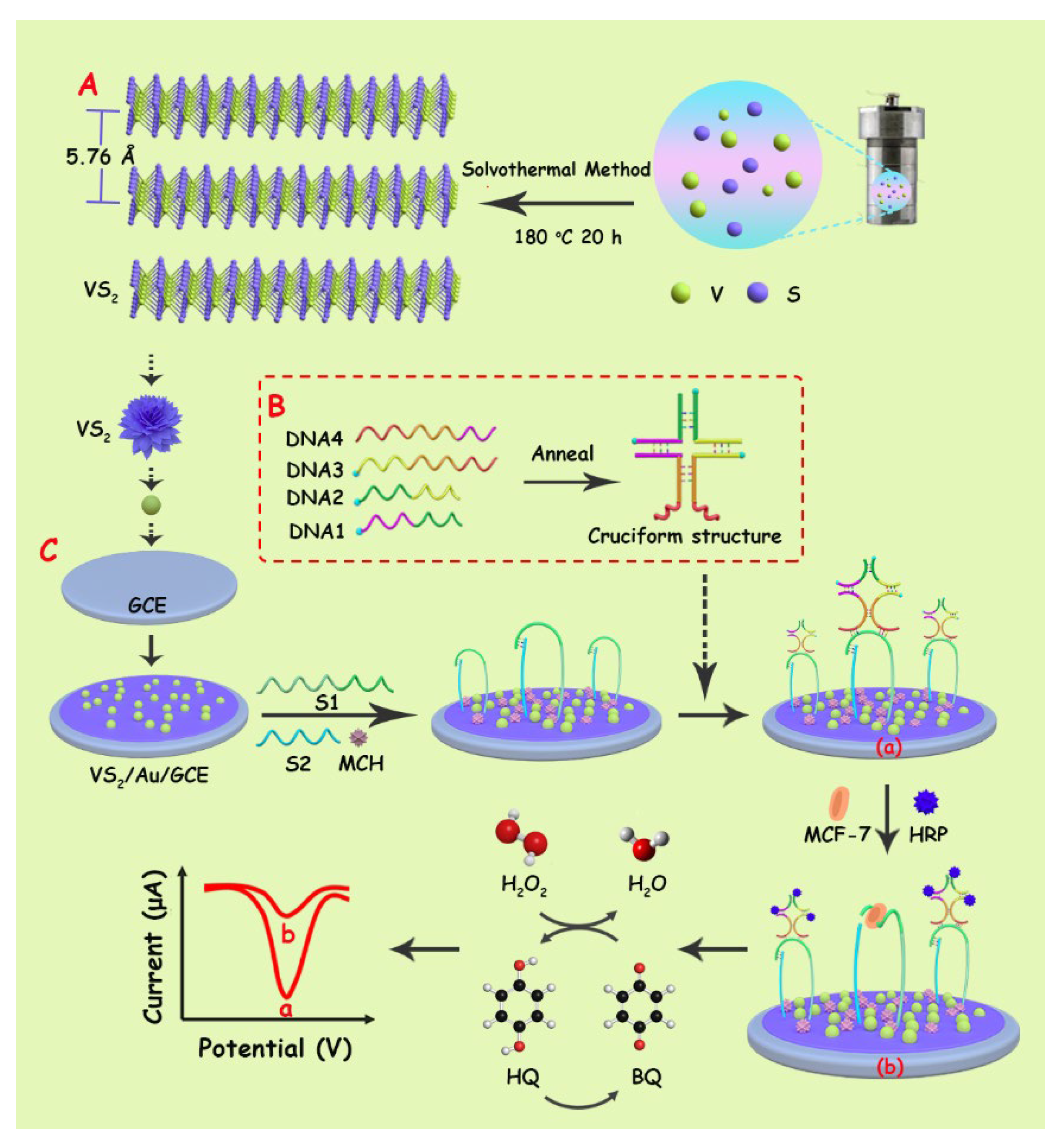
2.2. Preparation of VS2 Nanosheets
2.3. Preparation of Cruciform DNA
2.4. Cytotoxicity Test of Arch and Cruciform DNA
2.5. Preparation of Biosensor
3. Results and Discussion
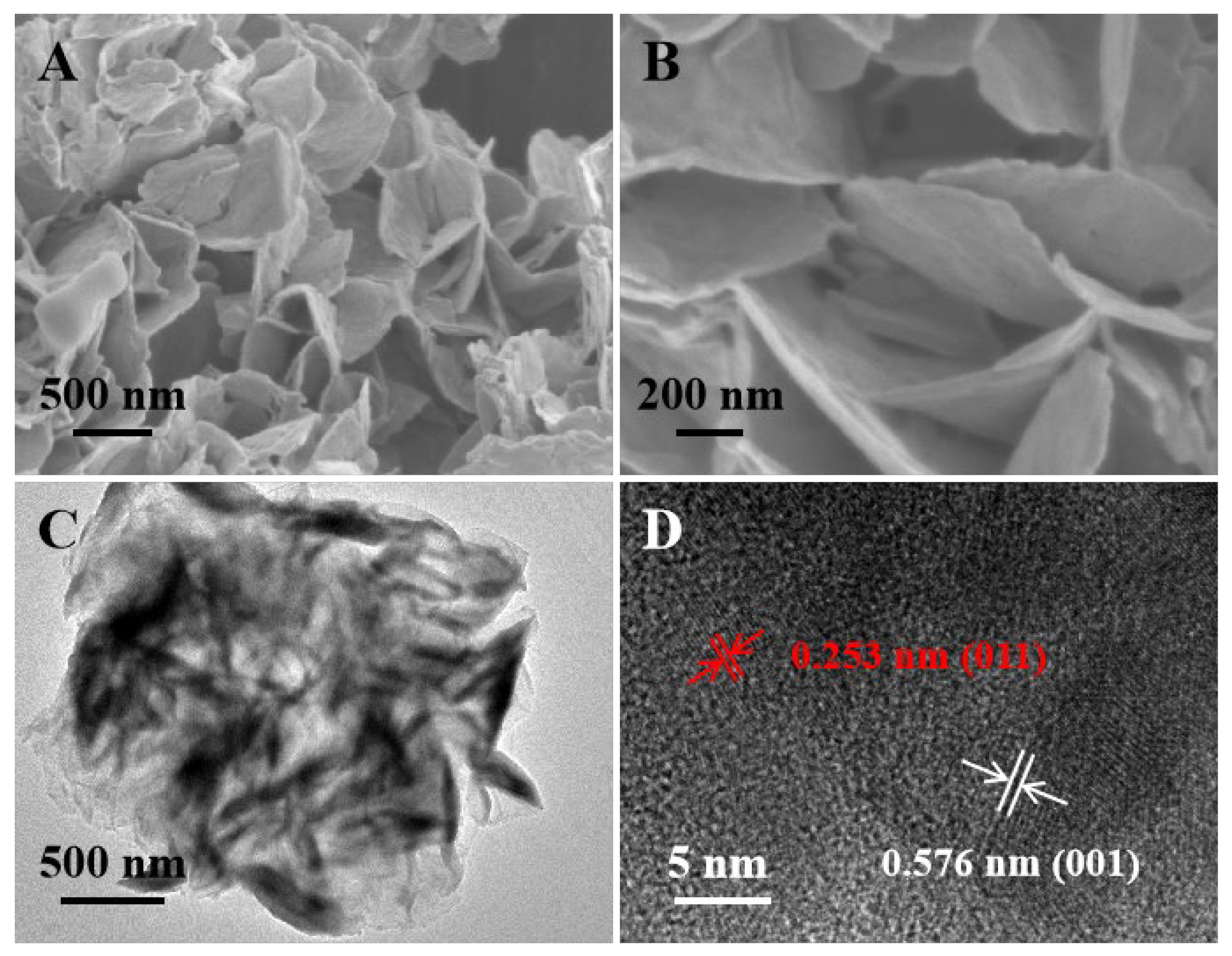
3.1. Characterization of VS2
3.2. Electrochemical Characterization
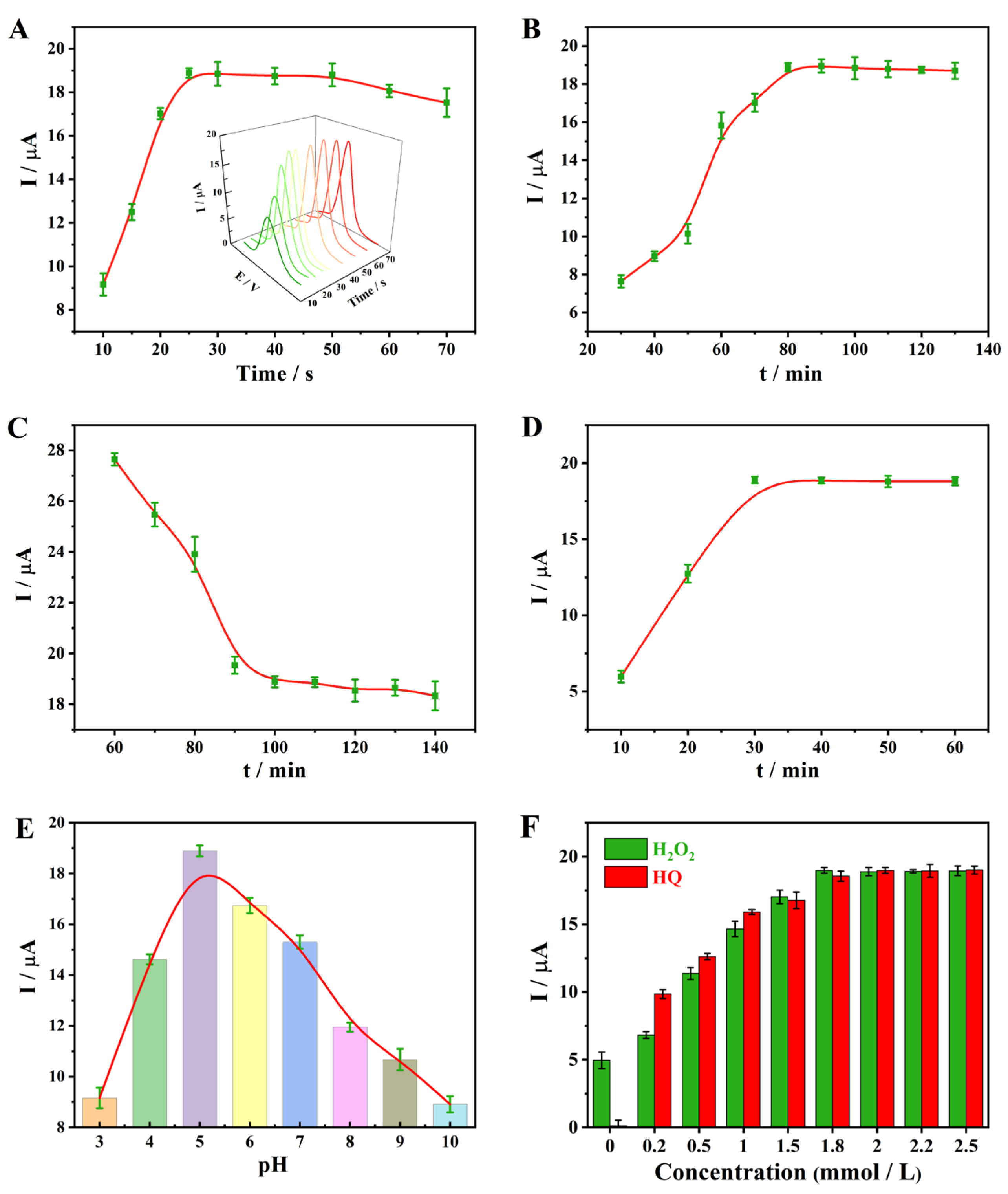
3.3. Optimization of Experimental Conditions
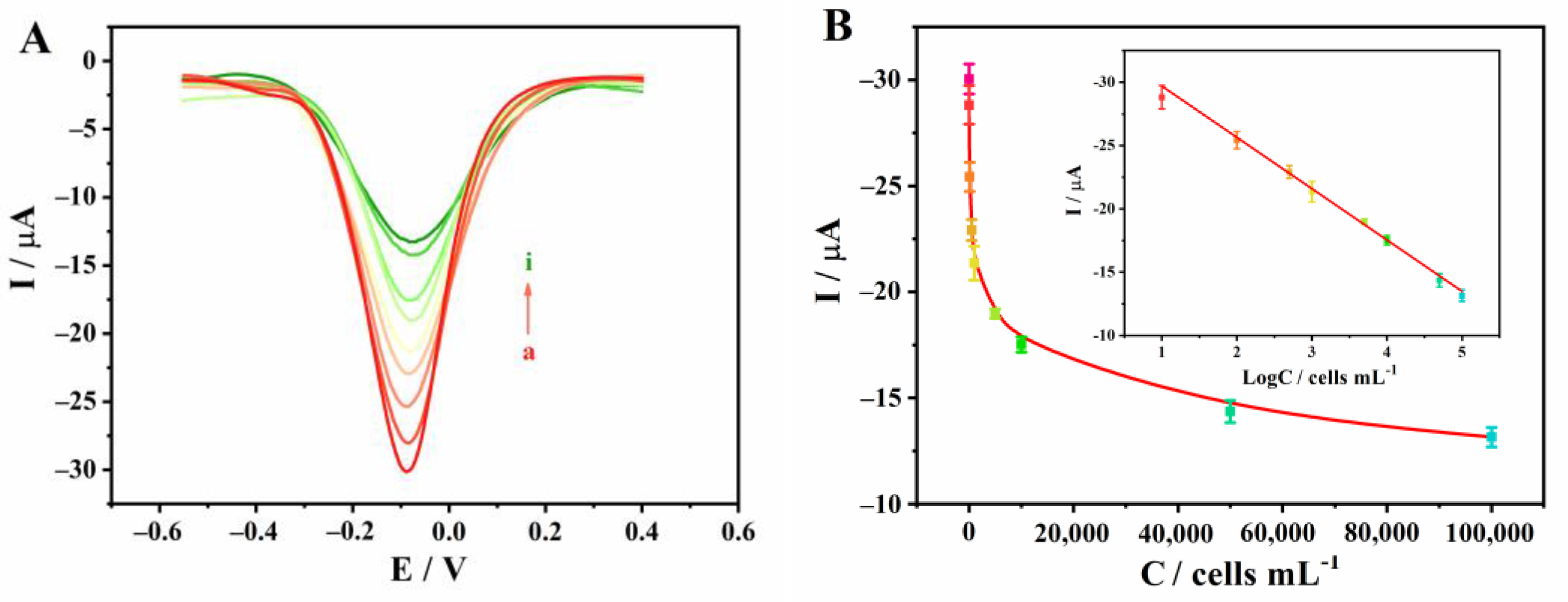
3.4. Analysis Performance of Sensor
3.5. Specificity, Reproducibility, Stability and Real Sample Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, K.; He, M.-Q.; Zhai, F.-H.; He, R.-H.; Yu, Y.-L. A novel electrochemical biosensor based on polyadenine modified aptamer for label-free and ultrasensitive detection of human breast cancer cells. Talanta 2017, 166, 87–92. [Google Scholar] [CrossRef]
- Li, T.; Fan, Q.; Liu, T.; Zhu, X.; Zhao, J.; Li, G. Detection of breast cancer cells specially and accurately by an electrochemical method. Biosens. Bioelectron. 2010, 25, 2686–2689. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef] [Green Version]
- Sheng, Q.; Cheng, N.; Bai, W.; Zheng, J. Ultrasensitive electrochemical detection of breast cancer cells based on DNA-rolling-circle-amplification-directed enzyme-catalyzed polymerization. Chem. Commun. 2015, 51, 2114–2117. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wang, Q.; Li, Q.; Yang, X.; Wang, K.; Nie, W. Surface plasmon resonance biosensor for sensitive detection of microRNA and cancer cell using multiple signal amplification strategy. Biosens. Bioelectron. 2017, 87, 433–438. [Google Scholar] [CrossRef]
- Li, Y.; Huan, K.; Deng, D.; Tang, L.; Wang, J.; Luo, L. Facile Synthesis of ZnMn2O4@rGO Microspheres for Ultrasensitive Electrochemical Detection of Hydrogen Peroxide from Human Breast Cancer Cells. ACS Appl. Mater. Interfaces 2019, 12, 3430–3437. [Google Scholar] [CrossRef] [PubMed]
- Bernsdorf, M.; Berthelsen, A.K.; Wielenga, V.T.; Kroman, N.; Teilum, D.; Binderup, T.; Tange, U.B.; Andersson, M.; Kjær, A.; Loft, A.; et al. Preoperative PET/CT in early-stage breast cancer. Ann. Oncol. 2012, 23, 2277–2282. [Google Scholar] [CrossRef]
- Luo, Y.; Pan, Q.; Yang, H.; Peng, L.; Zhang, W.; Li, F. Fibroblast Activation Protein–Targeted PET/CT with 68Ga-FAPI for Imaging IgG4-Related Disease: Comparison to 18F-FDG PET/CT. J. Nucl. Med. 2021, 62, 266–271. [Google Scholar] [CrossRef]
- Warner, E.; Plewes, D.B.; Hill, K.A.; Causer, P.A.; Zubovits, J.T.; Jong, R.A.; Cutrara, M.R.; DeBoer, G.; Yaffe, M.J.; Messner, S.J.; et al. Surveillance of BRCA1 and BRCA2 Mutation Carriers With Magnetic Resonance Imaging, Ultrasound, Mammography, and Clinical Breast Examination. JAMA 2004, 292, 1317–1325. [Google Scholar] [CrossRef] [Green Version]
- Hananouchi, T.; Chen, Y.; Jerban, S.; Teramoto, M.; Ma, Y.; Dorthe, E.; Chang, E.; Du, J.; D’Lima, D. A Useful Combination of Quantitative Ultrashort Echo Time MR Imaging and a Probing Device for Biomechanical Evaluation of Articular Cartilage. Biosensors 2021, 11, 52. [Google Scholar] [CrossRef]
- Galgano, S.; Viets, Z.; Fowler, K.; Gore, L.; Thomas, J.V.; McNamara, M.; McConathy, J. Practical Considerations for Clinical PET/MR Imaging. Magn. Reson. Imaging Clin. N. Am. 2017, 25, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Giesel, F.L.; Adeberg, S.; Syed, M.; Lindner, T.; Jiménez-Franco, L.D.; Mavriopoulou, E.; Staudinger, F.; Tonndorf-Martini, E.; Regnery, S.; Rieken, S.; et al. FAPI-74 PET/CT Using Either 18F-AlF or Cold-Kit 68Ga Labeling: Biodistribution, Radiation Dosimetry, and Tumor Delineation in Lung Cancer Patients. J. Nucl. Med. 2021, 62, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Galanzha, E.I.; Kim, J.-W.; Zharov, V.P. Nanotechnology-based molecular photoacoustic and photothermal flow cytometry platform forin-vivodetection and killing of circulating cancer stem cells. J. Biophotonics 2009, 2, 725–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittal, S.; Kaur, H.; Gautam, N.; Mantha, A.K. Biosensors for breast cancer diagnosis: A review of bioreceptors, biotransducers and signal amplification strategies. Biosens. Bioelectron. 2017, 88, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zou, L.; Yang, X.; Liu, X.; Nie, W.; Zheng, Y.; Cheng, Q.; Wang, K. Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens. Bioelectron. 2019, 135, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Liu, H.; Ge, L.; Wang, Y.; Ge, S.; Yu, J.; Yan, M. Aptamer-Based electrochemiluminescent detection of MCF-7 cancer cells based on carbon quantum dots coated mesoporous silica nanoparticles. Electrochim. Acta 2014, 146, 262–269. [Google Scholar] [CrossRef]
- Wang, X.; Cheng, S.; Wang, X.; Wei, L.; Kong, Q.; Ye, M.; Luo, X.; Xu, J.; Zhang, C.; Xian, Y. pH-Sensitive Dye-Based Nanobioplatform for Colorimetric Detection of Heterogeneous Circulating Tumor Cells. ACS Sens. 2021, 6, 1925–1932. [Google Scholar] [CrossRef]
- Luo, J.; Liang, D.; Li, X.; Deng, L.; Wang, Z.; Yang, M. Aptamer-based photoelectrochemical assay for the determination of MCF-7. Microchim. Acta 2020, 187, 1–7. [Google Scholar] [CrossRef]
- Huang, R.; Xi, Z.; He, N. Applications of aptamers for chemistry analysis, medicine and food security. Sci. China Ser. B Chem. 2015, 58, 1122–1130. [Google Scholar] [CrossRef]
- Vajhadin, F.; Ahadian, S.; Travas-Sejdic, J.; Lee, J.; Mazloum-Ardakani, M.; Salvador, J.; Aninwene, G.E.; Bandaru, P.; Sun, W.; Khademhossieni, A. Electrochemical cytosensors for detection of breast cancer cells. Biosens. Bioelectron. 2020, 151, 111984. [Google Scholar] [CrossRef]
- Lu, F.; Yang, L.; Hou, T.; Li, F. Label-free and “signal-on” homogeneous photoelectrochemical cytosensing strategy for ultrasensitive cancer cell detection. Chem. Commun. 2020, 56, 11126–11129. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Liu, B.; Wei, G. Two-Dimensional Material-Based Colorimetric Biosensors: A Review. Biosensors 2021, 11, 259. [Google Scholar] [CrossRef] [PubMed]
- Kasani, S.P.K.; Curtin, K.; Wu, N. A review of 2D and 3D plasmonic nanostructure array patterns: Fabrication, light management and sensing applications. Nanophotonics 2019, 8, 2065–2089. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Huang, K.-J.; Wu, X. Recent advances in transition-metal dichalcogenides based electrochemical biosensors: A review. Biosens. Bioelectron. 2017, 97, 305–316. [Google Scholar] [CrossRef]
- Yu, D.; Pang, Q.; Gao, Y.; Wei, Y.; Wang, C.; Chen, G.; Du, F. Hierarchical flower-like VS2 nanosheets — A high rate-capacity and stable anode material for sodium-ion battery. Energy Storage Mater. 2018, 11, 1–7. [Google Scholar] [CrossRef]
- Li, W.; Sari, H.M.K.; Li, X. Emerging Layered Metallic Vanadium Disulfide for Rechargeable Metal-Ion Batteries: Progress and Opportunities. ChemSusChem 2020, 13, 1172–1202. [Google Scholar] [CrossRef]
- Xua, D.; Wanga, H.; Qiua, R.; Wangb, Q.; Maoa, Z.; Jiangc, Y.; Wanga, R.; Hea, B.; Gonga, Y.; Lib, D.; et al. Coupling of bowl-like VS2 nanosheet arrays and carbon nanofiber enables ultrafast Na+-Storage and robust flexibility for sodium-ion hybrid capacitors. Energy Storage Mater. 2020, 28, 91–100. [Google Scholar] [CrossRef]
- Mikhaleva, N.S.; Visotin, M.A.; Kuzubov, A.A.; Popov, Z.I. VS2/Graphene Heterostructures as Promising Anode Material for Li-Ion Batteries. J. Phys. Chem. C 2017, 121, 24179–24184. [Google Scholar] [CrossRef]
- Liu, Y.-Y.; Xu, L.; Guo, X.-T.; Lv, T.-T.; Pang, H. Vanadium sulfide based materials: Synthesis, energy storage and conversion. J. Mater. Chem. A 2020, 8, 20781–20802. [Google Scholar] [CrossRef]
- Xie, X.-C.; Shuai, H.-L.; Wu, X.; Huang, K.-J.; Wang, L.-N.; Wang, R.-M.; Chen, Y. Engineering ultra-enlarged interlayer carbon-containing vanadium disulfide composite for high-performance sodium and potassium ion storage. J. Alloy. Compd. 2020, 847, 156288. [Google Scholar] [CrossRef]
- Tian, L.; Zhang, Y.; Wang, L.; Geng, Q.; Liu, D.; Duan, L.; Wang, Y.; Cui, J. Ratiometric Dual Signal-Enhancing-Based Electrochemical Biosensor for Ultrasensitive Kanamycin Detection. ACS Appl. Mater. Interfaces 2020, 12, 52713–52720. [Google Scholar] [CrossRef]
- Wang, M.; Chen, Y.; Cai, W.; Feng, H.; Du, T.; Liu, W.; Jiang, H.; Pasquarelli, A.; Weizmann, Y.; Wang, X. In situ self-assembling Au-DNA complexes for targeted cancer bioimaging and inhibition. Proc. Natl. Acad. Sci. USA 2020, 117, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-X.; Huang, K.-J.; Lin, F.; Fang, L.-X. Ultrasensitive electrochemical sensing platform based on graphene wrapping SnO2 nanocorals and autonomous cascade DNA duplication strategy. Talanta 2017, 175, 168–176. [Google Scholar] [CrossRef]
- He, P.; Yan, M.; Zhang, G.; Sun, R.; Chen, L.; An, Q.; Mai, L. Layered VS2 Nanosheet-Based Aqueous Zn Ion Battery Cathode. Adv. Energy Mater. 2017, 7, 1601920. [Google Scholar] [CrossRef]
- Huang, L.; Deng, H.; Zhong, X.; Zhu, M.; Chai, Y.; Yuan, R.; Yuan, Y. Wavelength distinguishable signal quenching and enhancing toward photoactive material 3,4,9,10-perylenetetracarboxylic dianhydride for simultaneous assay of dual metal ions. Biosens. Bioelectron. 2019, 145, 111702. [Google Scholar] [CrossRef]
- Li, Y.; Hu, M.; Huang, X.; Wang, M.; He, L.; Song, Y.; Jia, Q.; Zhou, N.; Zhang, Z.; Du, M. Multicomponent zirconium-based metal-organic frameworks for impedimetric aptasensing of living cancer cells. Sens. Actuators B Chem. 2020, 306, 127608. [Google Scholar] [CrossRef]
- Yang, B.; Chen, B.; He, M.; Yin, X.; Xu, C.; Hu, B. Aptamer-Based Dual-Functional Probe for Rapid and Specific Counting and Imaging of MCF-7 Cells. Anal. Chem. 2018, 90, 2355–2361. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhou, C.; Sun, X.; Jian, Y.; Kong, Q.; Cui, K.; Ge, S.; Yu, J. Polyhedral-AuPd nanoparticles-based dual-mode cytosensor with turn on enable signal for highly sensitive cell evalution on lab-on-paper device. Biosens. Bioelectron. 2018, 117, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Fu, Y.; Su, H.; Mao, L.; Chen, M. Sensitive detection of MCF-7 human breast cancer cells by using a novel DNA-labeled sandwich electrochemical biosensor. Biosens. Bioelectron. 2018, 122, 175–182. [Google Scholar] [CrossRef]
- Cai, S.; Chen, M.; Liu, M.; He, W.; Liu, Z.; Wu, D.; Xia, Y.; Yang, H.; Chen, J. A signal amplification electrochemical aptasensor for the detection of breast cancer cell via free-running DNA walker. Biosens. Bioelectron. 2016, 85, 184–189. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Chen, B.; He, M.; Wang, H.; Xiao, G.; Yang, B.; Hu, B. Simultaneous detection of MCF-7 and HepG2 cells in blood by ICP-MS with gold nanoparticles and quantum dots as elemental tags. Biosens. Bioelectron. 2017, 90, 343–348. [Google Scholar] [CrossRef]
- Yaman, Y.T.; Akbal, Ö.; Abaci, S. Development of clay-protein based composite nanoparticles modified single-used sensor platform for electrochemical cytosensing application. Biosens. Bioelectron. 2019, 132, 230–237. [Google Scholar] [CrossRef]
- Liu, N.; Song, J.; Lu, Y.; Davis, J.J.; Gao, F.; Luo, X. Electrochemical Aptasensor for Ultralow Fouling Cancer Cell Quantification in Complex Biological Media Based on Designed Branched Peptides. Anal. Chem. 2019, 91, 8334–8340. [Google Scholar] [CrossRef] [PubMed]
- Shenab, C.; Zhongb, L.; Xiongab, L.; Liub, C.; Yuc, L.; Chuab, X.; Luoab, X.; Zhaob, M.; Liuab, B. A novel sandwich-like cytosensor based on aptamers-modified magnetic beads and carbon dots/cobalt oxyhydroxide nanosheets for circulating tumor cells detection. Sens. Actuators B Chem. 2021, 331, 129399. [Google Scholar] [CrossRef]
- Shen, H.; Liu, L.; Yuan, Z.; Liu, Q.; Li, B.; Zhang, M.; Tang, H.; Zhang, J.; Zhao, S. Novel cytosensor for accurate detection of circulating tumor cells based on a dual-recognition strategy and BSA@Ag@Ir metallic-organic nanoclusters. Biosens. Bioelectron. 2021, 179, 113102. [Google Scholar] [CrossRef] [PubMed]



| Detection Techniques | Experimental Methods | Linear Range (cell/mL) | LOD (cell/mL) | Ref. |
|---|---|---|---|---|
| EIS | Zr-MOF compound material | 1 × 102 ~ 1 × 105 | 31 | [36] |
| ICP-MS | Aptamer based bifunctional probe | 2 × 102 ~ 1.2 × 104 | 81 | [37] |
| Colorimetry | PH-AuPd-NPs combined 3D-rGO | 50 ~ 1 × 107 | 32 | [38] |
| DPV | AuNGs/MWCNT-NH2 | 1 × 102 ~ 1 × 106 | 80 | [39] |
| CC | DNA walker | 0 ~ 5 × 102 | 47 | [40] |
| ICP-MS | Magnetic bead binding anti-EpCAM | 2 × 102 ~ 4 × 103 | 50 | [41] |
| EIS | Clay-protein based nanocomposites | 1.5 × 102 ~ 7.5 × 106 | 148 | [42] |
| DPV | Branched chain peptide modified electrode interface | 50 ~ 1 × 106 | 20 | [43] |
| Fluorescence | aptamer-modified magnetic beads | 10 ~ 1 × 105 | 5 | [44] |
| DPV | BSA@Ag@Ir metallic-organic nanoclusters | 3 ~ 3 × 106 | 1 | [45] |
| DPV | Arch DNA, cruciform DNA, material signal amplification, enzyme amplification | 10 ~ 1 × 105 | 5 | This work |
| Sample | Concentration (cells/mL) | Recovery Rate (%) | |
|---|---|---|---|
| Amount Added | Measured Value | ||
| PBS | 10 | 9 | 90.0 |
| 100 | 92 | 92.0 | |
| 500 | 479 | 95.8 | |
| 1000 | 983 | 98.3 | |
| 2000 | 2063 | 103.2 | |
| 5000 | 4760 | 95.2 | |
| Blood sample | 10 | 8 | 80.0 |
| 100 | 83 | 83.0 | |
| 500 | 389 | 77.8 | |
| 1000 | 841 | 84.1 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quan, J.; Wang, Y.; Zhang, J.; Huang, K.; Wang, X.; Jiang, H. Aptamer Embedded Arch-Cruciform DNA Assemblies on 2-D VS2 Scaffolds for Sensitive Detection of Breast Cancer Cells. Biosensors 2021, 11, 378. https://doi.org/10.3390/bios11100378
Quan J, Wang Y, Zhang J, Huang K, Wang X, Jiang H. Aptamer Embedded Arch-Cruciform DNA Assemblies on 2-D VS2 Scaffolds for Sensitive Detection of Breast Cancer Cells. Biosensors. 2021; 11(10):378. https://doi.org/10.3390/bios11100378
Chicago/Turabian StyleQuan, Jinfeng, Yihan Wang, Jialei Zhang, Kejing Huang, Xuemei Wang, and Hui Jiang. 2021. "Aptamer Embedded Arch-Cruciform DNA Assemblies on 2-D VS2 Scaffolds for Sensitive Detection of Breast Cancer Cells" Biosensors 11, no. 10: 378. https://doi.org/10.3390/bios11100378
APA StyleQuan, J., Wang, Y., Zhang, J., Huang, K., Wang, X., & Jiang, H. (2021). Aptamer Embedded Arch-Cruciform DNA Assemblies on 2-D VS2 Scaffolds for Sensitive Detection of Breast Cancer Cells. Biosensors, 11(10), 378. https://doi.org/10.3390/bios11100378







