Intelligent Bio-Responsive Fluorescent Au–shRNA Complexes for Regulated Autophagy and Effective Cancer Bioimaging and Therapeutics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Patients and Specimens
2.3. qRT-PCR
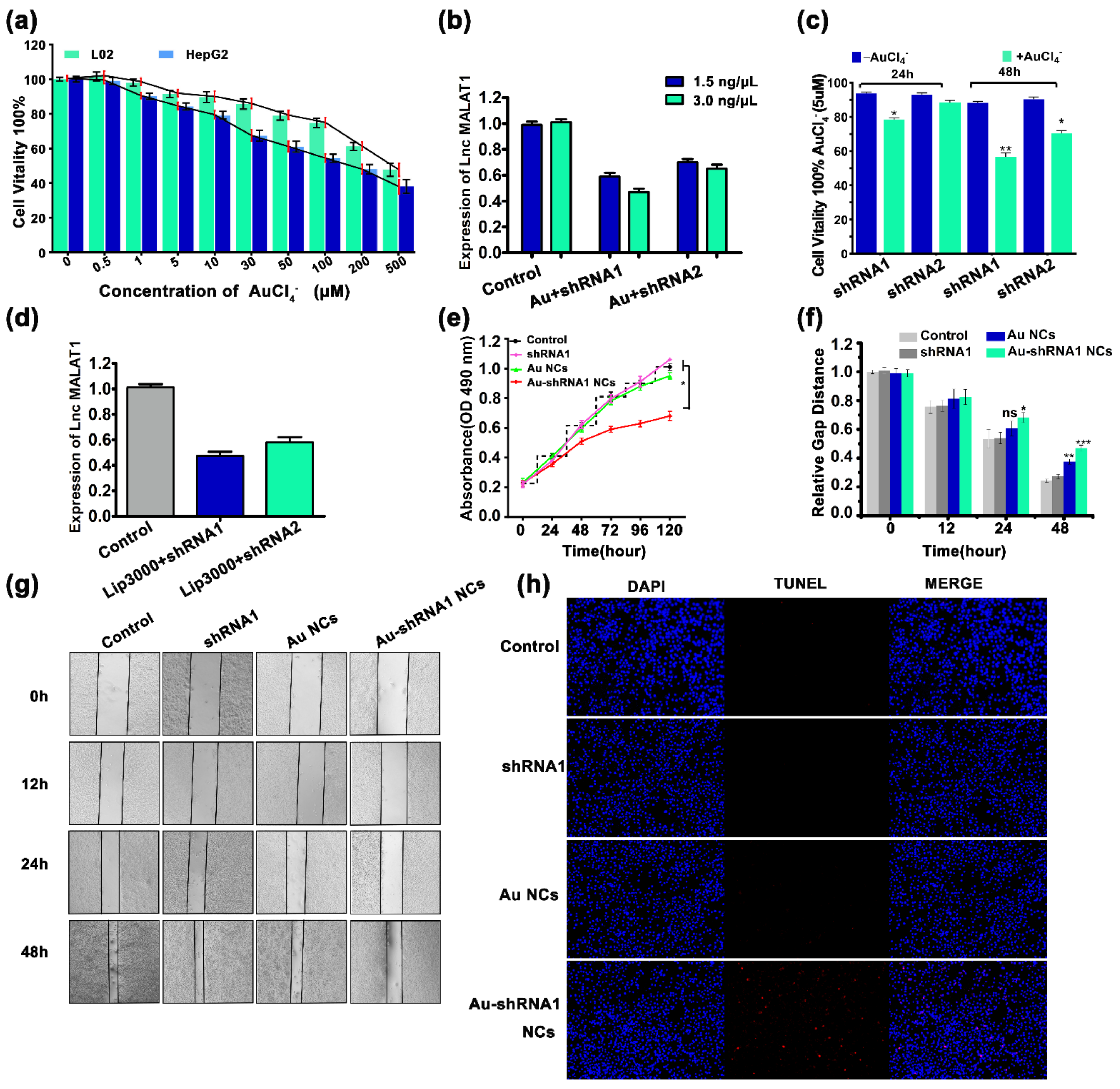
2.4. MTT Cytotoxicity Assessment and Cytostatic Test
2.5. Wound Healing Assay
2.6. In Situ Biosynthetic Au–shRNA1 NCs
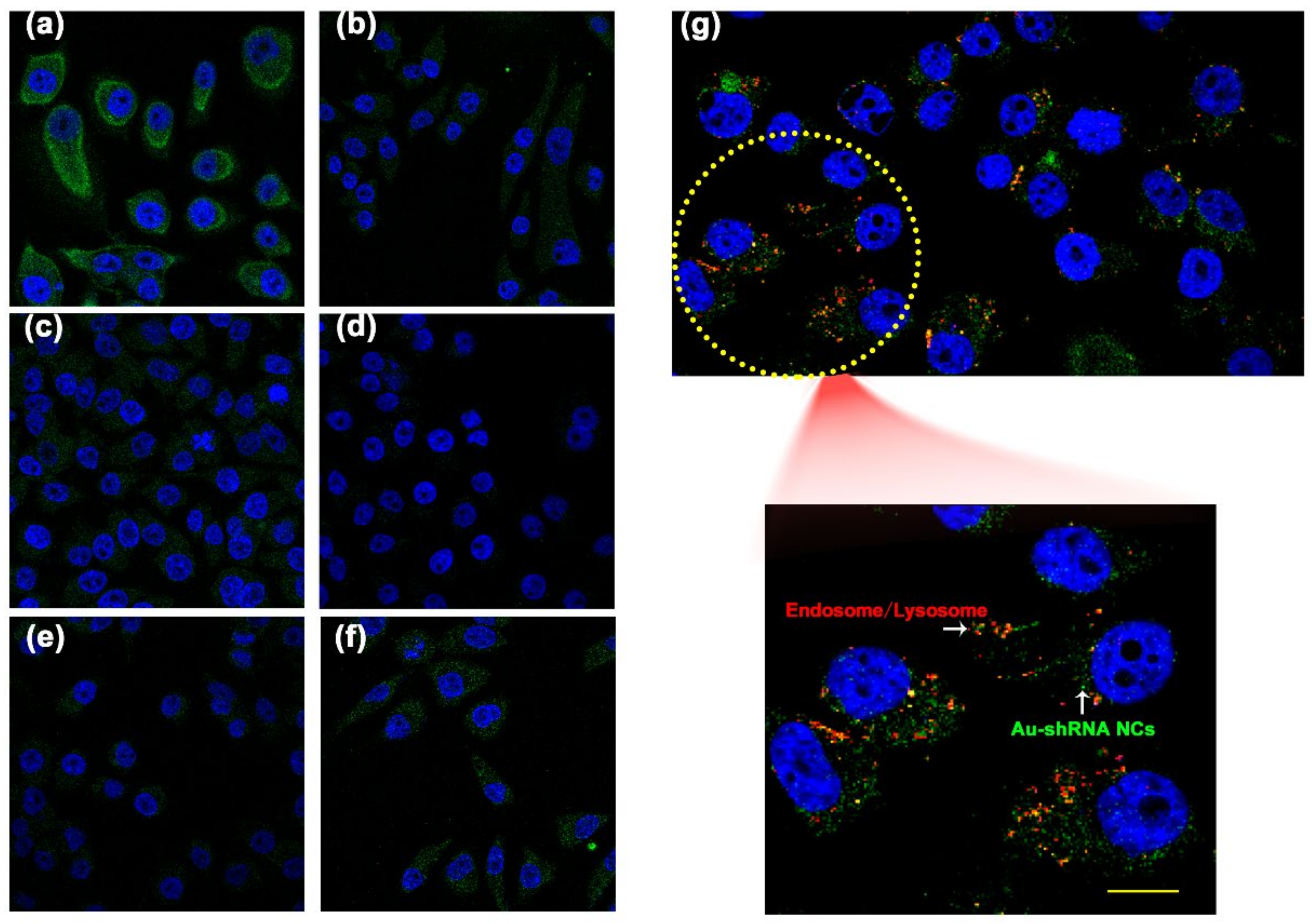
2.7. Cellular Uptake and Colocalization Studies
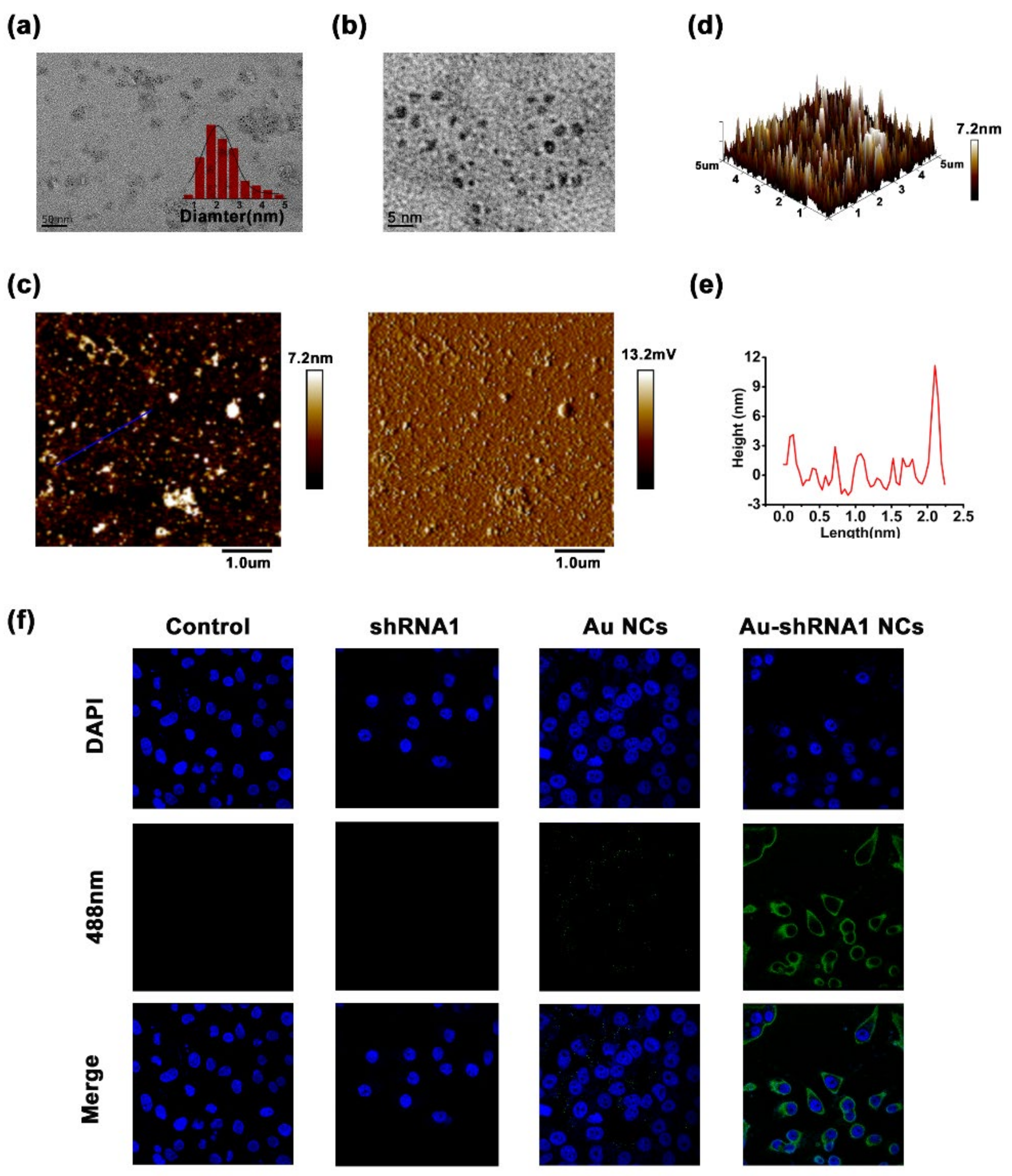
2.8. Transmission Electron Microscopy (TEM)
2.9. Atomic Force Microscopy (AFM)
2.10. Fluorescence Confocal Microscopy
2.11. Western Blot Analysis
2.12. GFP-(Microtubule-Associated Protein 1 Light Chain 3 (LC3)/Lysosomal-Associated Membrane Protein2 (LAMP2)/p62 Staining
2.13. Orthotopic Tumor Model
2.14. Statistical Analysis
3. Results and Discussion
3.1. Elevated Expression of MALAT1 Identified in HCC Patients and HCC Cell Lines
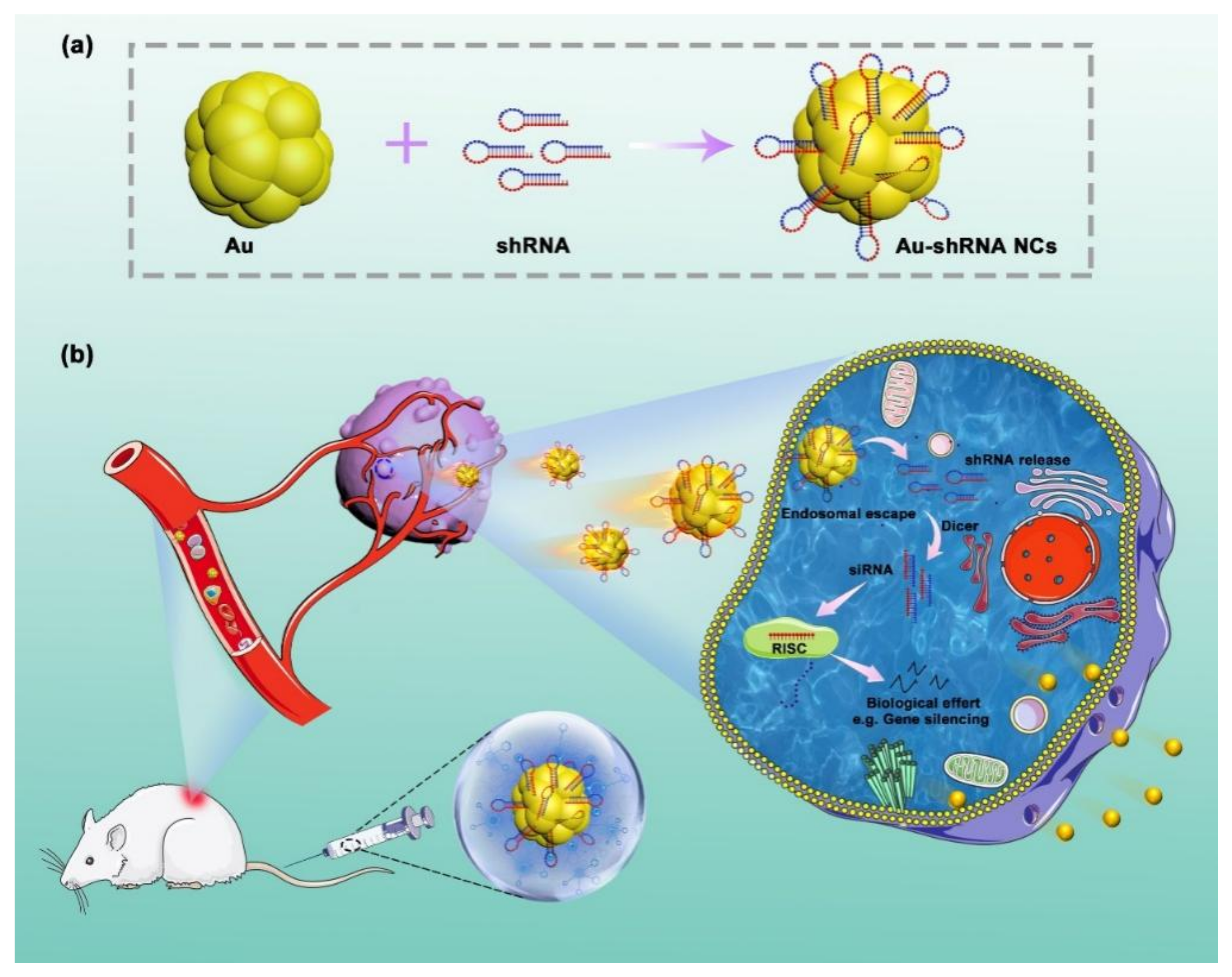
3.2. In Situ Self-Assembly of Au–shRNA NCs
3.3. Characterization of Au–shRNA NC Uptake and Escape
3.4. Effective Silencing of Target Gene MALAT1 via Au–shRNA NCs
3.5. Cell Proliferation Inhibition and Apoptosis via Au–shRNA1 NCs
3.6. Physicochemical Characteristics of Bio-Self-Assembled Au–shRNA1 NCs
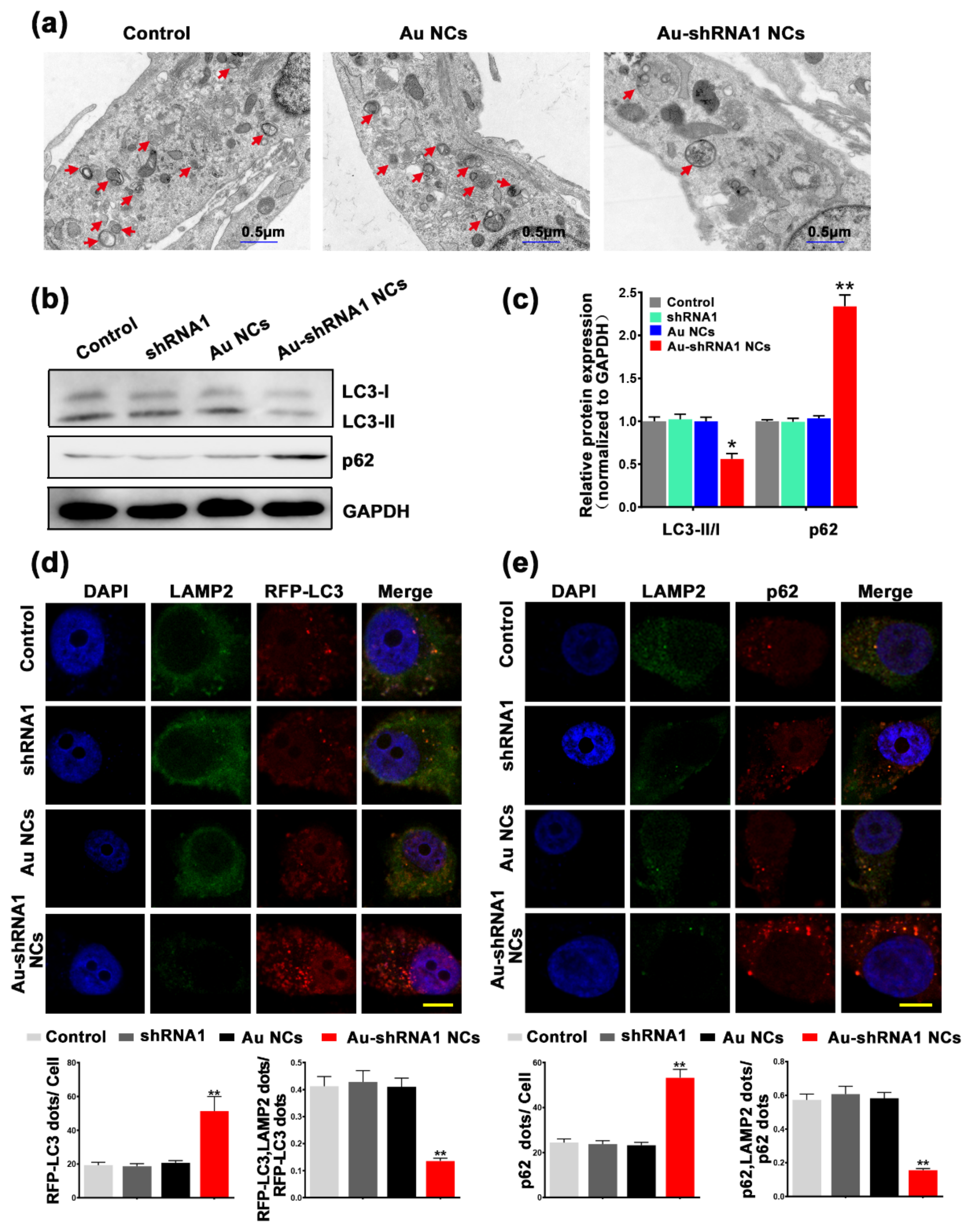
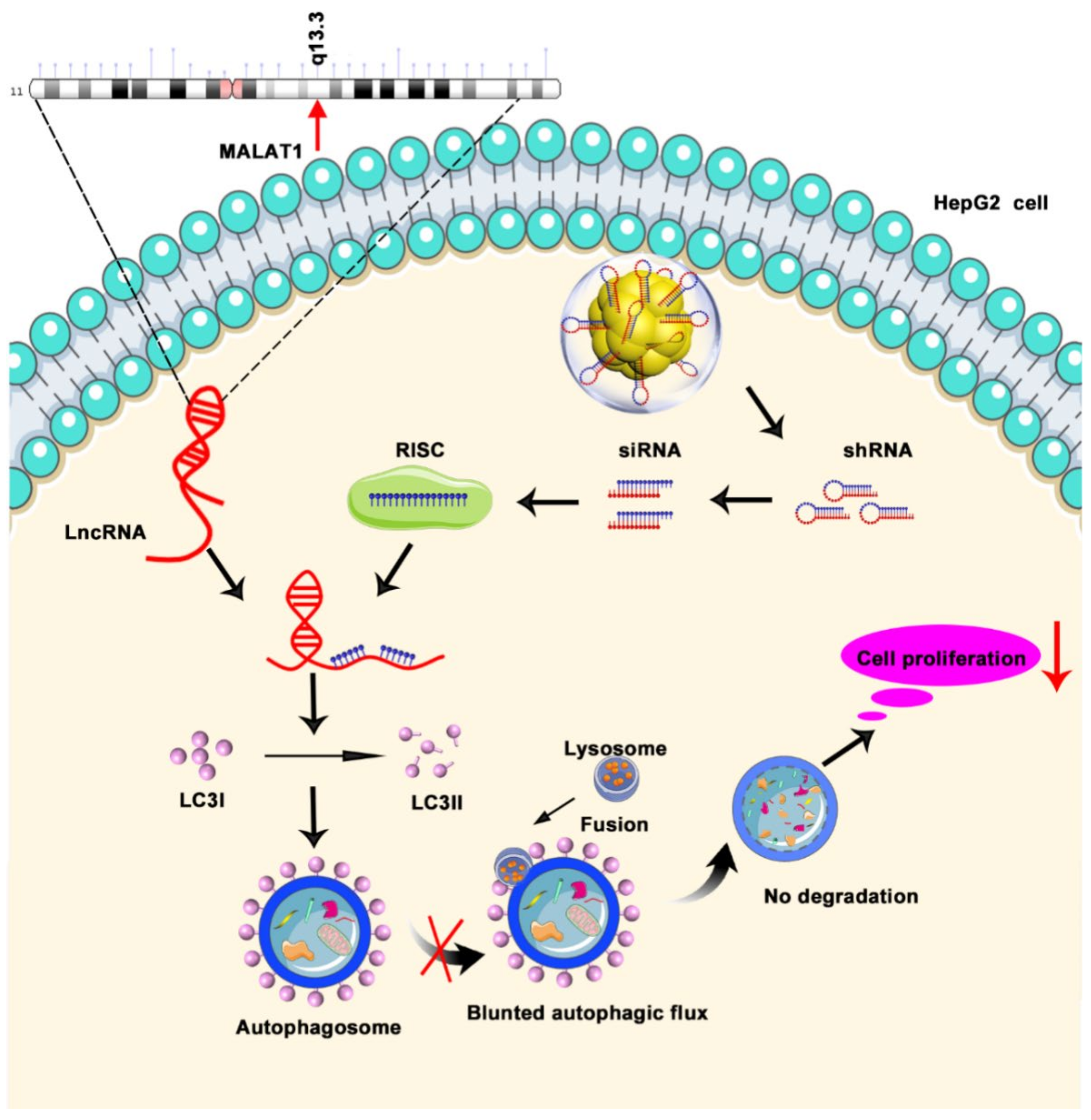
3.7. Inhibition of Autophagic Flux through Silencing of MALAT1 by Au–shRNA1 NCs
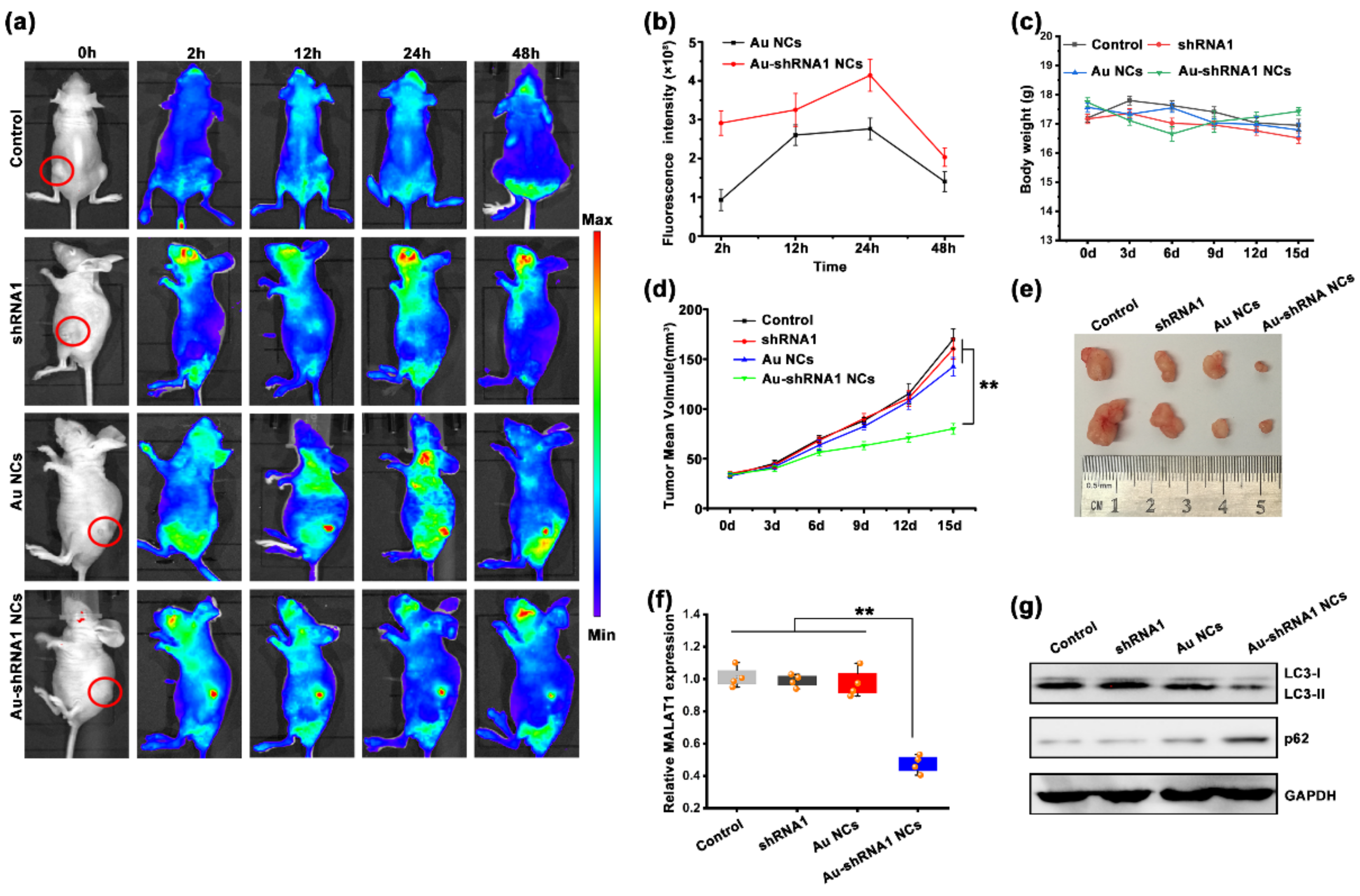
3.8. Au–shRNA1 NCs for Effective Bioimaging and Theranostics in an Orthotopic Tumor Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2015, 17, 47–62. [Google Scholar] [CrossRef]
- Geisler, S.; Coller, J. RNA in unexpected places: Long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol. 2013, 14, 699–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- E Kornienko, A.; Guenzl, P.M.; Barlow, D.P.; Pauler, F.M. Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 2013, 11, 59. [Google Scholar] [CrossRef] [Green Version]
- Wong, C.-M.; Tsang, F.H.-C.; Ng, I.O.-L. Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 137–151. [Google Scholar] [CrossRef]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Fullwood, M.J. Roles, Functions, and Mechanisms of Long Non-coding RNAs in Cancer. Genom. Proteom. Bioinform. 2016, 14, 42–54. [Google Scholar] [CrossRef] [Green Version]
- Mercer, T.; Dinger, M.; Mattick, J. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular Mechanisms of Long Noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, M.-C.; Ni, J.-J.; Cui, W.-Y.; Wang, B.-Y.; Zhuo, W. Emerging roles of lncRNA in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366. [Google Scholar] [PubMed]
- Yiren, H.; Yingcong, Y.; Sunwu, Y.; Keqin, L.; XiaoChun, T.; Senrui, C.; Ende, C.; Xizhou, L.; Yanfan, C. Long noncoding RNA MALAT1 regulates autophagy associated chemoresistance via miR-23b-3p sequestration in gastric cancer. Mol. Cancer 2017, 16, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, W.; Zhang, R.J.; Zou, S.B. Long noncoding RNA MALAT1 as a potential novel biomarker in digestive system cancers: A meta-analysis. Minerva Med. 2016, 107, 245–250. [Google Scholar] [PubMed]
- Shi, X.S.; Li, J.; Yang, R.H.; Zhao, G.R.; Zhou, H.P.; Zeng, W.X.; Zhou, M. Correlation of increased MALAT1 expression with pathological fea-tures and prognosis in cancer patients: A meta-analysis. Genet. Mol. Res. 2015, 14, 18808–18819. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xu, A.; Zhang, J.; He, X.; Pan, Y.; Cheng, G.; Qin, C.; Hua, L.; Wang, Z. Prognostic significance of long non-coding RNA MALAT-1 in various human carcinomas: A meta-analysis. Genet. Mol. Res. 2016, 15, gmr-15017433. [Google Scholar] [CrossRef]
- Fujimoto, A.; Furuta, M.; Totoki, Y.; Tsunoda, T.; Kato, M.; Shiraishi, Y.; Tanaka, H.; Taniguchi, H.; Kawakami, Y.; Ueno, M.; et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nat. Genet. 2016, 48, 500–509. [Google Scholar] [CrossRef]
- Kim, B.; Park, J.; Sailor, M.J. Rekindling RNAi Therapy: Materials Design Requirements for In Vivo siRNA Delivery. Adv. Mater. 2019, 31, e1903637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alterman, J.F.; Godinho, B.M.D.C.; Hassler, M.R.; Ferguson, C.M.; Echeverria, D.; Sapp, E.; Haraszti, R.A.; Coles, A.H.; Conroy, F.; Miller, R.; et al. A divalent siRNA chemical scaffold for potent and sustained modulation of gene expression throughout the central nervous system. Nat. Biotechnol. 2019, 37, 884–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lambeth, L.S.; Smith, C.A. Short Hairpin RNA-Mediated Gene Silencing. Methods Mol. Biol. 2012, 942, 205–232. [Google Scholar] [CrossRef]
- Patra, J.K.; Das, G.; Fraceto, L.F.; Campos, E.V.R.; Rodriguez-Torres, M.D.P.; Acosta-Torres, L.S.; Diaz-Torres, L.A.; Grillo, R.; Swamy, M.K.; Sharma, S.; et al. Nano based drug delivery systems: Recent developments and future prospects. J. Nanobiotechnol. 2018, 16, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, A.; Jha, N.K.; Dahiya, K.; Singh, V.K.; Chaurasiya, K.; Jha, A.N.; Jha, S.K.; Mishra, P.C.; Dholpuria, S.; Astya, R.; et al. Nanoparticulate RNA delivery systems in cancer. Cancer Rep. 2020, 3, e1271. [Google Scholar] [CrossRef]
- Liu, C.-J.; Chen, P.-J. Elimination of Hepatitis B in Highly Endemic Settings: Lessons Learned in Taiwan and Challenges Ahead. Viruses 2020, 12, 815. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Qin, S.-K. Features and treatment options of Chinese hepatocellular carcinoma. Chin. Clin. Oncol. 2013, 2, 38. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wang, Y.; Li, H.; Chen, L.; Liu, Q. Regulatory Networks of LncRNA MALAT-1 in Cancer. Cancer Manag. Res. 2020, 12, 10181–10198. [Google Scholar] [CrossRef]
- Guerrieri, F. Long non-coding RNAs era in liver cancer. World J. Hepatol. 2015, 7, 1971–1973. [Google Scholar] [CrossRef] [PubMed]
- LaiZhe, M.-C.; Yang, Z.; Zhou, L.; Zhu, Q.-Q.; Xie, H.-Y.; Zhang, F.; Wu, L.-M.; Chen, L.-M.; Zheng, S.-S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol. 2011, 29, 1810–1816. [Google Scholar] [CrossRef]
- Galluzzi, L.; Pietrocola, F.; Levine, B.; Kroemer, G. Metabolic Control of Autophagy. Cell 2014, 159, 1263–1276. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Liu, C.-Y.; Zhou, L.-Y.; Wang, J.; Wang, M.; Zhao, B.; Zhao, W.-K.; Jian-Xun, W.; Yan-Fang, Z.; Zhang, X.-J.; et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat. Commun. 2015, 6, 6779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Chen, H.; Gao, Y.; Wang, Y.-W.; Zhang, G.-Q.; Pan, S.-H.; Ji, L.; Kong, R.; Wang, G.; Jia, Y.-H.; et al. Long Noncoding RNA MALAT1 Promotes Aggressive Pancreatic Cancer Proliferation and Metastasis via the Stimulation of Autophagy. Mol. Cancer Ther. 2016, 15, 2232–2243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yip, K.M.; Fischer, N.; Paknia, E.; Chari, A.; Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 2020, 587, 157–161. [Google Scholar] [CrossRef] [PubMed]
- Prabhakar, N.; Peurla, M.; Shenderova, O.; Rosenholm, J.M. Fluorescent and Electron-Dense Green Color Emitting Nanodiamonds for Single-Cell Correlative Microscopy. Molecules 2020, 25, 5897. [Google Scholar] [CrossRef]
- Cai, W.; Feng, H.; Yin, L.; Wang, M.; Jiang, X.; Qin, Z.; Liu, W.; Li, C.; Jiang, H.; Weizmann, Y.; et al. Bio responsive self-assembly of Au-miRNAs for targeted cancer theranostics. EBioMedicine 2020, 54, 102740. [Google Scholar] [CrossRef]
- Wang, M.; Chen, Y.; Cai, W.; Feng, H.; Du, T.; Liu, W.; Jiang, H.; Pasquarelli, A.; Weizmann, Y.; Wang, X. In situ self-assembling Au-DNA com-plexes for targeted cancer bioimaging and inhibition. Proc. Natl. Acad. Sci. USA 2020, 117, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.R.; Kalyana-Sundaram, S.; Mahavisno, V.; Varambally, R.; Yu, J.; Briggs, B.B.; Barrette, T.R.; Anstet, M.J.; Kincead-Beal, C.; Kulkarni, P.; et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expres-sion profiles. Neoplasia 2007, 9, 166–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.-C.; Dang, Y.-W.; Wei, D.-M.; Chen, P.; Tang, R.-X.; Huang, Q.; Liu, J.-H.; Luo, D.-Z. Clinical significance and prospective molecular mechanism of MALAT1 in pancreatic cancer exploration: A comprehensive study based on the GeneChip, GEO, Oncomine, and TCGA databases. OncoTargets Ther. 2017, 10, 3991–4005. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Cheung, S.T.; So, S.; Fan, S.T.; Barry, C.; Higgins, J.; Lai, K.-M.; Ji, J.; Dudoit, S.; Ng, I.O.-L.; et al. Gene Expression Patterns in Human Liver Cancers. Mol. Biol. Cell 2002, 13, 1929–1939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wurmbach, E.; Chen, Y.-B.; Khitrov, G.; Zhang, W.; Roayaie, S.; Schwartz, M.; Fiel, I.; Thung, S.; Mazzaferro, V.M.; Bruix, J.; et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology 2007, 45, 938–947. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Shang, S.; Guo, S.; Li, X.; Zhou, H.; Liu, H.; Sun, Y.; Wang, J.; Wang, P.; Zhi, H.; et al. Lnc2Cancer 3.0: An updated re-source for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2021, 49, D1251–D1258. [Google Scholar] [CrossRef]
- Dou, J.; Gu, N. Emerging strategies for the identification and targeting of cancer stem cells. Tumor Biol. 2010, 31, 243–253. [Google Scholar] [CrossRef]
- Rehman, F.U.; Du, T.; Shaikh, S.; Jiang, X.; Chen, Y.; Li, X.; Yi, H.; Hui, J.; Chen, B.; Selke, M.; et al. Nano in nano: Biosynthesized gold and iron nanoclusters cargo neoplastic exosomes for cancer status biomarking. Nanomed. Nanotechnol. Biol. Med. 2018, 14, 2619–2631. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, G.; Li, Q.; Jiang, H.; Liu, C.; Amatore, C.; Wang, X. In vivo self-bio-imaging of tumors through in situ biosynthesized fluo-rescent gold nanoclusters. Sci. Rep. 2013, 3, 1157. [Google Scholar] [CrossRef]
- Chang, H.-C.; Wang, X.; Shiu, K.-K.; Zhu, Y.; Wang, J.; Li, Q.; Chen, B.; Jiang, H. Layer-by-layer assembly of graphene, Au and poly(toluidine blue O) films sensor for evaluation of oxidative stress of tumor cells elicited by hydrogen peroxide. Biosens. Bioelectron. 2013, 41, 789–794. [Google Scholar] [CrossRef]
- Szatrowski, T.P.; Nathan, C.F. Production of large amounts of hydrogen peroxide by human tumor cells. Cancer Res. 1991, 51, 794–798. [Google Scholar]
- Sajid, M.I.; Moazzam, M.; Kato, S.; Cho, K.Y.; Tiwari, R.K. Overcoming Barriers for siRNA Therapeutics: From Bench to Bedside. Pharmaceuticals 2020, 13, 294. [Google Scholar] [CrossRef] [PubMed]
- Pei, D.; Buyanova, M. Overcoming Endosomal Entrapment in Drug Delivery. Bioconjugate Chem. 2018, 30, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Si, Y.; Yang, Z.; Ge, Q.; Yu, L.; Yao, M.; Sun, X.; Ren, Z.; Ding, C. Long non-coding RNA Malat1 activated autophagy, hence promoting cell pro-liferation and inhibiting apoptosis by sponging miR-101 in colorectal cancer. Cell Mol. Biol. Lett. 2019, 24, 50. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Green, D.R. Autophagy-Independent Functions of the Autophagy Machinery. Cell 2019, 177, 1682–1699. [Google Scholar] [CrossRef]
- Johansen, T.; Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011, 7, 279–296. [Google Scholar] [CrossRef]
- Sancey, L.; Kotb, S.; Truillet, C.; Appaix, F.; Marais, A.; Thomas, E.; van der Sanden, B.; Klein, J.-P.; Laurent, B.; Cottier, M.; et al. Long-Term in Vivo Clearance of Gadolinium-Based AGuIX Nanoparticles and Their Biocompatibility after Systemic Injection. ACS Nano 2015, 9, 2477–2488. [Google Scholar] [CrossRef]
- Yu, M.; Xu, J.; Zheng, J. Renal Clearable Luminescent Gold Nanoparticles: From the Bench to the Clinic. Angew. Chem. Int. Ed. 2018, 58, 4112–4128. [Google Scholar] [CrossRef]
- Kapara, A.; Brunton, V.; Graham, D.; Faulds, K. Investigation of cellular uptake mechanism of functionalised gold nanoparticles into breast cancer using SERS. Chem. Sci. 2020, 11, 5819–5829. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.E.; Kintzing, J.R.; Hanna, A.; Shannon, J.M.; Gupta, M.K.; Duvall, C.L. Balancing Cationic and Hydrophobic Content of PEGylated siRNA Polyplexes Enhances Endosome Escape, Stability, Blood Circulation Time, and Bioactivity in Vivo. ACS Nano 2013, 7, 8870–8880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, M.; Librizzi, D.; Kılıç, A.; Liu, Y.; Renz, H.; Merkel, O.M.; Kissel, T. Enhancing in vivo circulation and siRNA delivery with biodegradable polyethylenimine-graft-polycaprolactone-block-poly(ethylene glycol) copolymers. Biomaterials 2012, 33, 6551–6558. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Azmi, A.; Li, Y.; Ahmad, A.; Ali, S.; Banerjee, S.; Kong, D.; Sarkar, F. Targeting CSCs in Tumor Microenvironment: The Potential Role of ROS-Associated miRNAs in Tumor Aggressiveness. Curr. Stem Cell Res. Ther. 2013, 9, 22–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Q.; Li, Y.; Yang, X.; Zhang, W.; Hao, Y.; Zhang, H.; Hou, L.; Zhang, Z. Hypoxia-specific therapeutic agents delivery nanotheranostics: A sequential strategy for ultrasound mediated on-demand tritherapies and imaging of cancer. J. Control. Release 2018, 275, 192–200. [Google Scholar] [CrossRef]
- Li, Z.; Li, J.; Tang, N. Long noncoding RNA Malat1 is a potent autophagy inducer protecting brain microvascular endothelial cells against oxygen-glucose deprivation/reoxygenation-induced injury by sponging miR-26b and upregulating ULK2 expression. Neuroscience 2017, 354, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Cao, W.; Zang, Q.; Li, G.; Guo, X.; Fan, J. The HIF-2α-MALAT1-miR-216b axis regulates multi-drug resistance of hepatocellular carcinoma cells via modulating autophagy. Biochem. Biophys. Res. Commun. 2016, 478, 1067–1073. [Google Scholar] [CrossRef]
- Huang, Z.; Zhou, L.; Chen, Z.; Nice, E.C.; Huang, C. Stress management by autophagy: Implications for chemoresistance. Int. J. Cancer 2016, 139, 23–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, W.; Yin, L.; Jiang, H.; Weizmann, Y.; Wang, X. Intelligent Bio-Responsive Fluorescent Au–shRNA Complexes for Regulated Autophagy and Effective Cancer Bioimaging and Therapeutics. Biosensors 2021, 11, 425. https://doi.org/10.3390/bios11110425
Cai W, Yin L, Jiang H, Weizmann Y, Wang X. Intelligent Bio-Responsive Fluorescent Au–shRNA Complexes for Regulated Autophagy and Effective Cancer Bioimaging and Therapeutics. Biosensors. 2021; 11(11):425. https://doi.org/10.3390/bios11110425
Chicago/Turabian StyleCai, Weijuan, Liang Yin, Hui Jiang, Yossi Weizmann, and Xuemei Wang. 2021. "Intelligent Bio-Responsive Fluorescent Au–shRNA Complexes for Regulated Autophagy and Effective Cancer Bioimaging and Therapeutics" Biosensors 11, no. 11: 425. https://doi.org/10.3390/bios11110425
APA StyleCai, W., Yin, L., Jiang, H., Weizmann, Y., & Wang, X. (2021). Intelligent Bio-Responsive Fluorescent Au–shRNA Complexes for Regulated Autophagy and Effective Cancer Bioimaging and Therapeutics. Biosensors, 11(11), 425. https://doi.org/10.3390/bios11110425





