Highly Sensitive Luminescent Bioassay Using Recombinant Escherichia coli Biosensor for Rapid Detection of Low Cr(VI) Concentration in Environmental Water
Abstract
:1. Introduction
2. Materials and Methods
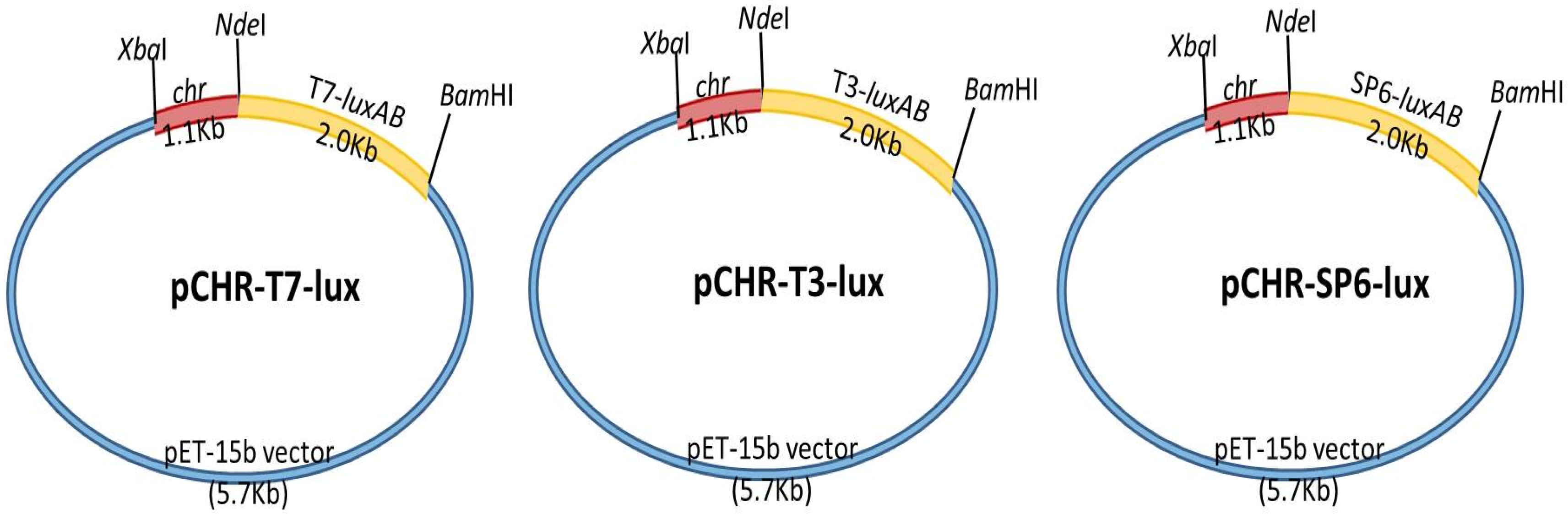
2.1. Bacterial Strains, Gene Cloning, and Biosensor Construction
2.2. Bacterial Growth
2.3. Determination of Optimal Conditions
2.4. Establishment of Calibration Curve and Measurement of Real Water Samples
3. Results and Discussion
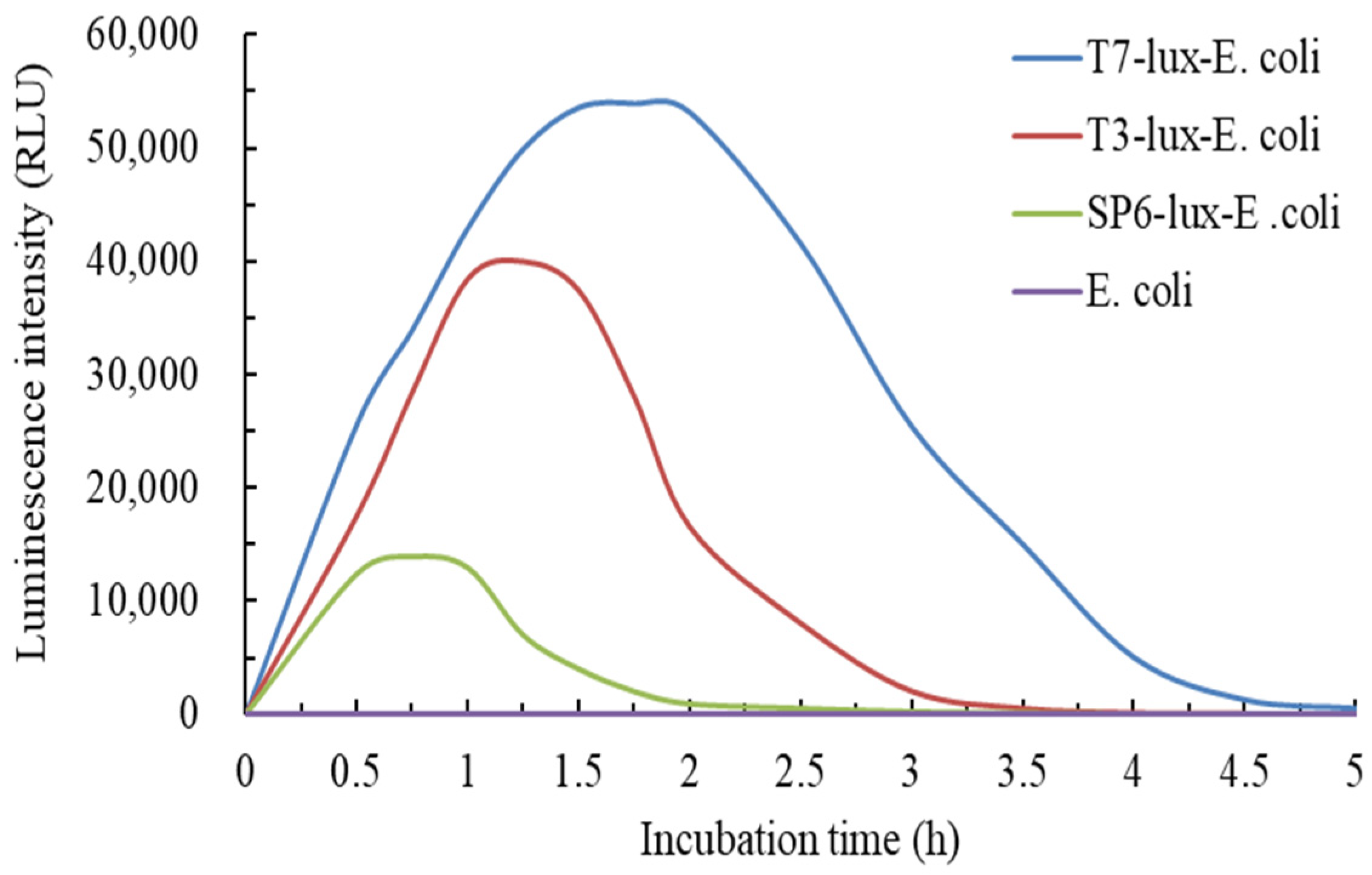
3.1. Time-Dependent Induction of Three Recombinant E. coli Biosensors with Cr(VI)
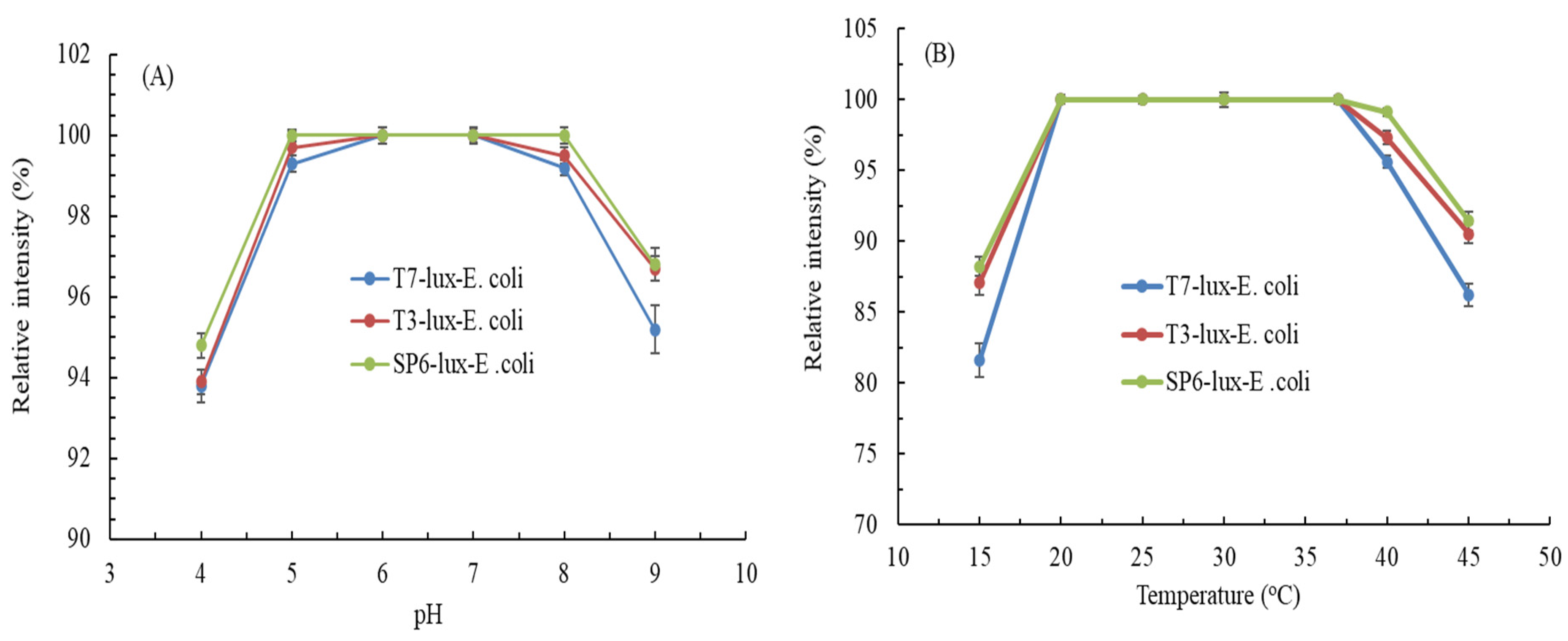
3.2. Effects of Culture Conditions on Luminescence Intensity
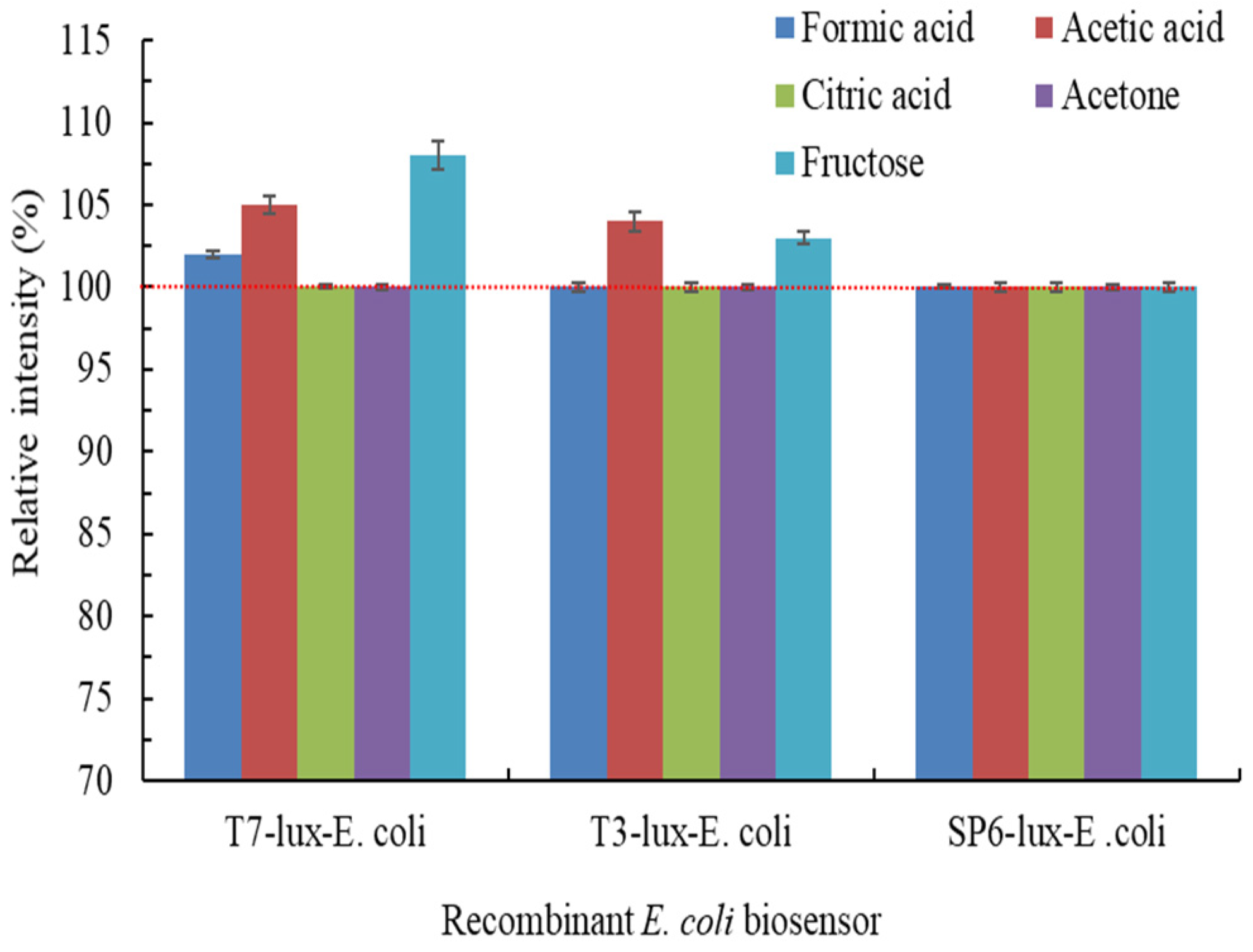
3.3. Effects of Coexisting Carbon Source, Cr(VI) Oxyanion Form, and Coexisting Ion on Luminescence Intensity
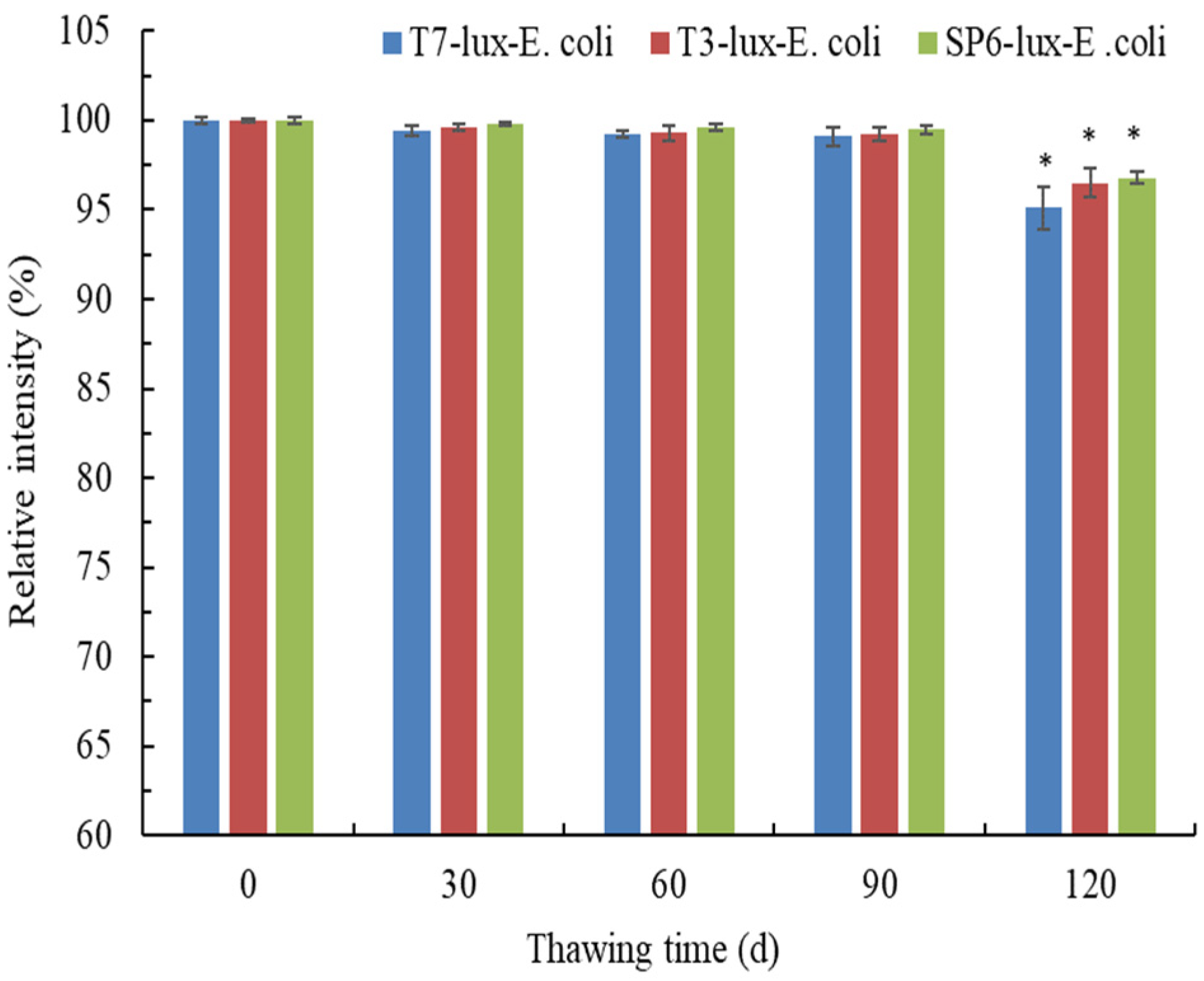
3.4. Effects of Thawing Time on Luminescence Intensity
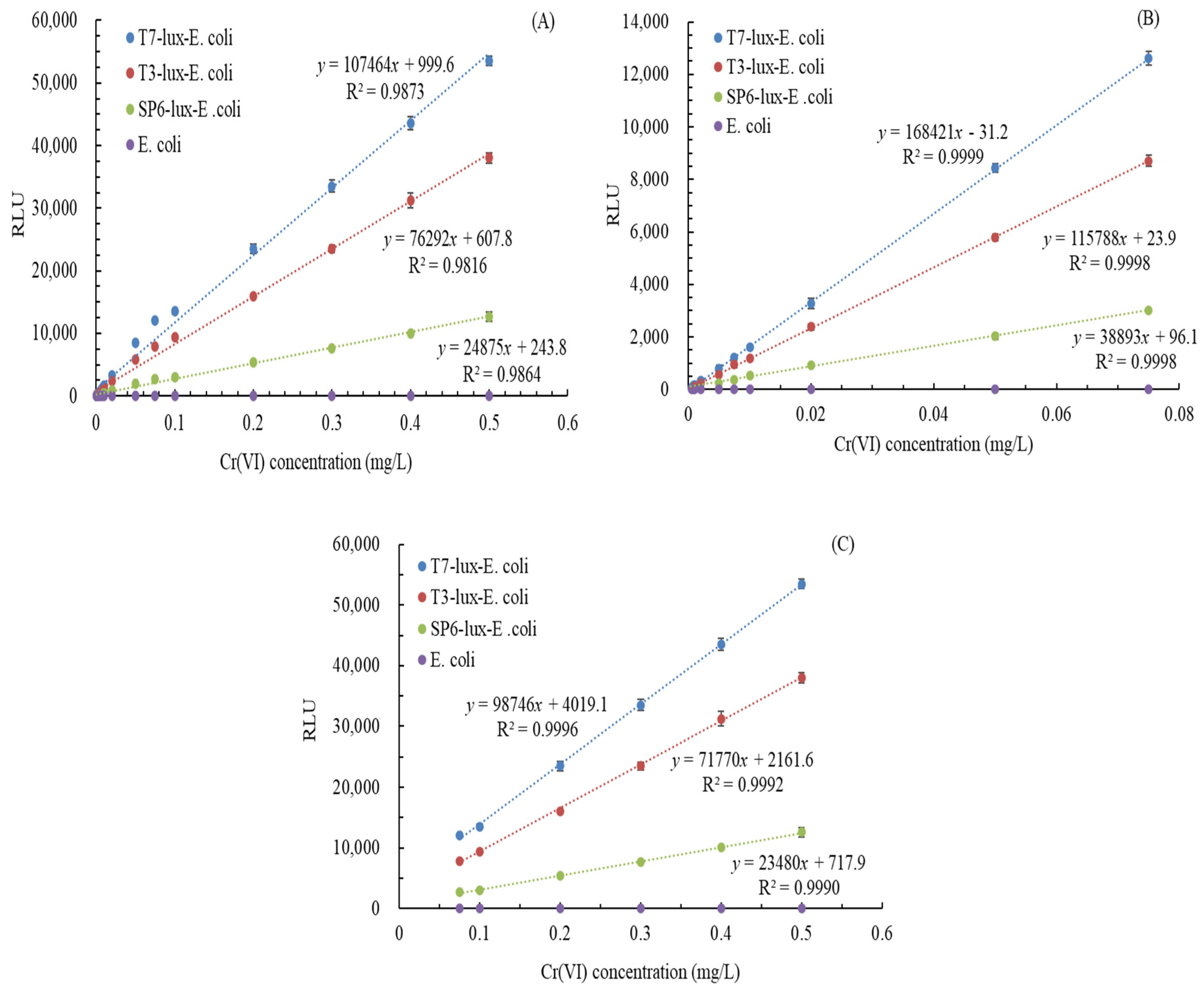
3.5. Relationship of Cr(VI) Concentration with Luminescence Intensity
3.6. Cr(VI) Detection in Real Water Samples by Using Our Three Recombinant Luminescent E. coli Biosensors
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pellerin, C.; Booker, S.M. Reflections on hexavalent chromium: Health hazards of an industrial heavyweight. Environ. Health Perspect. 2000, 108, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Rahman, A.; Nahar, N.; Nawani, N.N.; Jass, J.; Hossain, K.; Saud, Z.A.; Saha, A.K.; Ghosh, S.; Olsson, B.; Mandal, A. Bioremediation of hexavalent chromium (VI) by a soil-borne bacterium, Enterobacter cloacae B2-DHA. J. Environ. Sci. Health Part A 2015, 50, 1136–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, M.; Peterson, E.; Quievryn, G.; Zhitkovich, A. Human nucleotide excision repair efficiently removes chromium-DNA phosphate adducts and protects cells against chromate toxicity. J. Biol. Chem. 2004, 279, 30419–30424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhitkovich, A. Importance of chromium-DNA adducts in mutagenicity and toxicity of chromium(VI). Chem. Res. Toxicol. 2005, 18, 3–11. [Google Scholar] [CrossRef]
- Coelho, C.; Branco, R.; Natal-da-Luz, T.; Sousa, J.P.; Morais, P.V. Evaluation of bacterial biosensors to determine chromate bioavailability and to assess ecotoxicity of soils. Chemosphere 2015, 128, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.H.; Cheng, C.Y.; Liu, M.H.; Chen, T.Y.; Hsieh, M.C.; Chung, Y.C. Utility of Ochrobactrum anthropi YC152 in a microbial fuel cell as an early warning device for hexavalent chromium determination. Sensors 2016, 16, 1272. [Google Scholar] [CrossRef] [Green Version]
- Tauriainen, S.M.; Virta, M.P.J.; Karp, M.K. Detecting bioavailable toxic metals and metalloids from natural water samples using luminescent sensor bacteria. Wat. Res. 2000, 34, 2661–2666. [Google Scholar] [CrossRef]
- Duffy, G.; Maguire, I.; Heery, B.; Gers, P.; Ducrée, J.; Regan, F. ChromiSense: A colourimetric lab-on-a-disc sensor for chromium speciation in water. Talanta 2018, 178, 392–399. [Google Scholar] [CrossRef]
- Wang, G.H.; Tsai, T.H.; Kui, C.C.; Cheng, C.Y.; Huang, T.L.; Chung, Y.C. Analysis of bioavailable toluene by using recombinant luminescent bacterial biosensors with different promoters. J. Biol. Eng. 2021, 15, 2. [Google Scholar] [CrossRef]
- Branco, R.; Cristóvão, A.; Morais, P.V. Highly Sensitive, highly specific whole-cell bioreporters for the detection of chromate in environmental samples. PLoS ONE 2013, 8, e54005. [Google Scholar]
- Michel, C.; Ouerd, A.; Battaglia-Brunet, F.; Ignatiadis, I. Cr(VI) quantification using an amperometric enzyme-based sensor: Interference and physical and chemical factors controlling the biosensor response in ground waters. Biosens. Bioelectron. 2006, 22, 285–290. [Google Scholar] [CrossRef]
- Nepomuscene, N.J.; Daniel, D.; Krastanov, A. Biosensor to detect chromium in wastewater. Biotechnol. Biotechnol. Equip. 2007, 21, 377–381. [Google Scholar] [CrossRef] [Green Version]
- Bohrn, U.; Muchaa, A.; Werner, C.F.; Trattner, B.; Bäcker, M.; Krumbe, C.; Schienlea, M.; Stütz, E.; Schmitt-Landsiedel, D.; Fleischer, M.; et al. A critical comparison of cell-based sensor systems for the detection of Cr(VI) in aquatic environment. Sens. Actuators B Chem. 2013, 182, 58–65. [Google Scholar] [CrossRef]
- Gurung, A.; Oh, S.; Kim, K.D.; Shin, B. Semi-continuous detection of toxic hexavalent chromium using a sulfur-oxidizing bacteria biosensor. J. Environ. Manag. 2012, 106, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Panda, J.; Sarkar, P. Biosensing and bioremediation of Cr(VI) by cell free extract of Enterobacter aerogenes T2. J. Environ. Sci. Health Part A 2014, 49, 600–608. [Google Scholar] [CrossRef]
- Wu, L.C.; Tsai, T.H.; Liu, M.H.; Kuo, J.L.; Chang, Y.C.; Chung, Y.C. A green microbial fuel cell-based biosensor for in situ chromium (VI) measurement in electroplating wastewater. Sensors 2017, 17, 2461. [Google Scholar] [CrossRef]
- Wu, L.C.; Wang, G.H.; Tsai, T.H.; Lo, S.Y.; Cheng, C.Y.; Chung, Y.C. Three-stage single-chambered microbial fuel cell biosensor inoculated with Exiguobacterium aestuarii YC211 for continuous chromium (VI) measurement. Sensors 2019, 19, 1418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branco, R.; Chung, A.P.; Johnston, T.; Gure, V.; Morais, P.; Zhitkovich, A. The chromate-inducible chrBACF operon from the transposable element TnOtChr confers resistance to chromium (VI) and superoxide. J. Bacteriol. 2008, 190, 6996–7003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.Y.; Cheng, C.Y.; Chen, C.K.; Hsieh, M.C.; Lin, S.T.; Ho, K.Y.; Li, J.W.; Lin, C.P.; Chung, Y.C. Hexavalent chromium removal and bioelectricity generation by Ochrobactrum sp. YC211 under different oxygen conditions. J. Environ. Sci. Health Part A 2016, 51, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Baldiris, R.; Acosta-Tapia, N.; Montes, A.; Hernández, J.; Vivas-Reyes, R. Reduction of hexavalent chromium and detection of chromate reductase (ChrR) in Stenotrophomonas maltophilia. Molecules 2018, 23, 406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chai, L.; Ding, C.; Tang, C.; Yang, W.; Yang, Z.; Wang, Y.; Liao, Q.; Li, J. Discerning three novel chromate reduce and transport genes of highly efficient Pannonibacter phragmitetus BB: From genome to gene and protein. Ecotoxicol. Environ. Saf. 2018, 162, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Hora, A.; Shetty, V.K. Partial purification and characterization of chromate reductase of a novel Ochrobactrum sp. strain Cr-B4. Prep. Biochem. Biotechnol. 2015, 45, 769–784. [Google Scholar] [CrossRef]
- Magrisso, S.; Erel, Y.; Belkin, S. Microbial reporters of metal bioavailability. Microb. Biotechnol. 2008, 1, 320–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, C.; Choi, S. Technology and applications of microbial biosensor. Open J. Appl. Biosens. 2013, 2, 83–93. [Google Scholar] [CrossRef] [Green Version]
- Eun, H.M. RNA polymerases. In Enzymology Primer for Recombinant DNA Technology; Elsevier Science Publishing Co Inc: San Diego, CA, USA; Amsterdam, The Netherlands, 1996; pp. 491–566. [Google Scholar]
- Martin, C.T.; Esposito, E.A.; Theis, K.; Gong, P. Structure and function in promoter escape by T7 RNA polymerase. Prog. Nucleic Acid Res. Mol. Biol. 2005, 80, 323–347. [Google Scholar]
- Zhu, L.P.; Li, Z.F.; Sun, X.; Li, S.G.; Li, Y.Z. Characteristics and activity analysis of epothilone operon promoters from Sorangium cellulosum strains in Escherichia coli. Appl. Microbiol. Biotechnol. 2013, 97, 6857–6866. [Google Scholar] [CrossRef]
- Zhang, N.; Darbari, V.C.; Glyde, R.; Zhang, X.; Buck, M. The bacterial enhancer-dependent RNA polymerase. Biochem. J. 2016, 473, 3741–3753. [Google Scholar] [CrossRef] [Green Version]
- Duval, J.F.L.; Pagnout, C. Decoding the time-dependent response of bioluminescent metal-detecting whole-cell bacterial sensors. ACS Sens. 2019, 4, 1373–1383. [Google Scholar] [CrossRef] [Green Version]
- Brutesco, C.; Prévéral, S.; Escoffier, C.; Descamps, E.C.; Prudent, E.; Cayron, J.; Dumas, L.; Ricquebourg, M.; Adryanczyk-Perrier, G.; de Groot, A.; et al. Bacterial host and reporter gene optimization for genetically encoded whole cell biosensors. Environ. Sci. Pollut. Res. Int. 2017, 24, 52–65. [Google Scholar] [CrossRef]
- Smutok, O.; Broda, D.; Smutok, H.; Dmytruk, K.; Gonchar, M. Chromate-reducing activity of Hansenula polymorpha recombinant cells over-producing flavocytochrome b2. Chemosphere 2011, 83, 449–454. [Google Scholar] [CrossRef]
- Tehrani, G.A.; Mirzaahmadi, S.; Bandehpour, M.; Laloei, F.; Eidi, A.; Valinasab, T.; Kazemi, B. Molecular cloning and expression of the luciferase coding genes of Vibrio fischeri. Afr. J. Biotechnol. 2011, 10, 4018–4023. [Google Scholar]
- Mergeay, M.; Nies, D.; Schlrgrl, H.G.; Gerits, J.; Charles, P. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 1985, 162, 328–334. [Google Scholar] [CrossRef] [Green Version]
- Lace, A.; Ryan, D.; Bowkett, M.; Cleary, L. Chromium monitoring in water by colorimetry using optimised 1,5-diphenylcarbazide method. Int. J. Environ. Res. Public Health. 2019, 16, 1803. [Google Scholar] [CrossRef] [Green Version]
- Dollard, M.A.; Billard, P. Whole-cell bacterial sensors for the monitoring of phosphate bioavailability. J. Microbiol. Methods 2003, 55, 221–229. [Google Scholar] [CrossRef]
- Zhang, Q.; Song, Y.; Amor, K.; Huang, W.E.; Porcelli, D.; Thompson, I. Monitoring Cr toxicity and remediation processes-Combining a whole-cell bioreporter and Cr isotope techniques. Water Res. 2019, 153, 295–303. [Google Scholar] [CrossRef] [PubMed]
- De Las Heras, A.; de Lorenzo, V. Engineering whole-cell biosensors with no antibiotic markers for monitoring aromatic compounds in the environment. Methods Mol. Biol. 2012, 834, 261–281. [Google Scholar] [PubMed]
- Blazeck, J.; Alper, H.S. Promoter engineering: Recent advances in controlling transcription at the most fundamental level. Biotechnol. J. 2013, 8, 46–58. [Google Scholar] [CrossRef] [PubMed]
- Pagga, U. Testing biodegradability with standardized methods. Chemosphere 1997, 35, 2953–2972. [Google Scholar] [CrossRef]
- Espinosa-Urgel, M.; Serrano, L.; Ramos, J.L.; Ferna´ndez-Escamilla, A.M. Engineering biological approaches for detection of toxic compounds: A new microbial biosensor based on the Pseudomonas putida TtgR Repressor. Mol. Biotechnol. 2015, 57, 558–564. [Google Scholar] [CrossRef]
| Primer | Primer Sequence |
|---|---|
| Lux-Forward primer | |
| NdeI-T7-luxABf | CGCA↓TATGTAATACGACTCACTATAGGGATGAAGTTTGGAAATATTTG |
| NdeI -T3-luxABf | CGCA↓TATGGCAATTAACCCTCACTAAAGGATGAAGTTTGGAAATATTTG |
| NdeI -SP6-luxABf | CGCA↓TATGATTTAGGTGACACTATAGATGAAGTTTGGAAATATTTG |
| Lux-Reverse primer | |
| BamHI-luxABr | CGG↓GATCCTTAAGGCAGATTCTTTTC |
| Chr-Forward primer | |
| XbaI-chrf | CGT↓CTAGAGATTGCTTATTCCTATTGCCA |
| Chr-Reverse primer | |
| NdeI-chrr | CGCA↓TATGTCATACGCTGAGGGTCCCTTT |
| Sensor | LOD (mg/L) | Operating Conditions | References |
|---|---|---|---|
| gfp-based recombinant O. tritici biosensor | 0.388 | 37 °C, pH 7, detection time: 5 h, batch | [5] |
| O. anthropi YC152 MFC-based biosensor | 0.0125 | 35 °C, pH 7, detection time: 15 min, batch | [6] |
| gfp-based recombinant E. coli biosensor | 0.0194 | 37 °C, pH 7, detection time: 5 h, batch | [10] |
| V79 cell biosensor | 0.97 | 37 °C, pH 7, detection time: 3 h, batch | [13] |
| E. aestuarii YC211 MFC-based biosensor | 2.5 | 30 °C, pH 7, detection time: 15 min, batch | [16] |
| three-stage single-chambered MFC biosensor | 5 | 30 °C, pH 7, detection time: 6.6 min, continuous flow | [17] |
| flavocytochrome b2–based H. polymorpha recombinant biosensor | 0.52 | 24 °C, pH 6.3, detection time: 20 min, batch | [31] |
| T7-biosensor | 0.0005 | 37 °C, pH 7, detection time: 1.5 h, batch | This study |
| T3-biosensor | 0.001 | 37 °C, pH 7, detection time: 1.0 h, batch | This study |
| SP6-biosensor | 0.005 | 37 °C, pH 7, detection time: 0.5 h, batch | This study |
| Industrial Effluents | Domestic Effluents | Surface Water | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A | B | C | A | B | C | A | B | C | |
| DPC method | 0.482 * | 6.72 | 2.31 | 0.08 | 1.21 | 0.63 | 0.0212 | ND ** | 0.0154 |
| T7-biosensor | 0.461 (−4.4%) *** | 6.83 (1.6%) | 2.42 (4.8%) | 0.076 (−5.0%) | 1.23 (1.7%) | 0.68 (7.9%) | 0.0221 (4.2%) | 0.0061 (–) | 0.0158 (2.6%) |
| T3-biosensor | 0.491 (1.9%) | 6.43 (−4.3%) | 2.21 (−4.3%) | 0.083 (3.8%) | 1.18 (−2.5%) | 0.59 (−6.3%) | 0.0243 (14.6%) | 0.0065 (–) | 0.0137 (−11.0%) |
| SP6-biosensor | 0.478 (−0.8%) | 6.62 (−1.5%) | 2.25 (−2.6%) | 0.081 (1.3%) | 1.19 (−1.7%) | 0.62 (−1.6%) | 0.0251 (18.4%) | 0.0083 (–) | 0.0171 (11.0%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.-H.; Cheng, C.-Y.; Tsai, T.-H.; Chiang, P.-K.; Chung, Y.-C. Highly Sensitive Luminescent Bioassay Using Recombinant Escherichia coli Biosensor for Rapid Detection of Low Cr(VI) Concentration in Environmental Water. Biosensors 2021, 11, 357. https://doi.org/10.3390/bios11100357
Wang G-H, Cheng C-Y, Tsai T-H, Chiang P-K, Chung Y-C. Highly Sensitive Luminescent Bioassay Using Recombinant Escherichia coli Biosensor for Rapid Detection of Low Cr(VI) Concentration in Environmental Water. Biosensors. 2021; 11(10):357. https://doi.org/10.3390/bios11100357
Chicago/Turabian StyleWang, Guey-Horng, Chiu-Yu Cheng, Teh-Hua Tsai, Pin-Kuan Chiang, and Ying-Chien Chung. 2021. "Highly Sensitive Luminescent Bioassay Using Recombinant Escherichia coli Biosensor for Rapid Detection of Low Cr(VI) Concentration in Environmental Water" Biosensors 11, no. 10: 357. https://doi.org/10.3390/bios11100357
APA StyleWang, G.-H., Cheng, C.-Y., Tsai, T.-H., Chiang, P.-K., & Chung, Y.-C. (2021). Highly Sensitive Luminescent Bioassay Using Recombinant Escherichia coli Biosensor for Rapid Detection of Low Cr(VI) Concentration in Environmental Water. Biosensors, 11(10), 357. https://doi.org/10.3390/bios11100357






