A Label-Free Optical Detection of Pathogens in Isopropanol as a First Step towards Real-Time Infection Prevention
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation
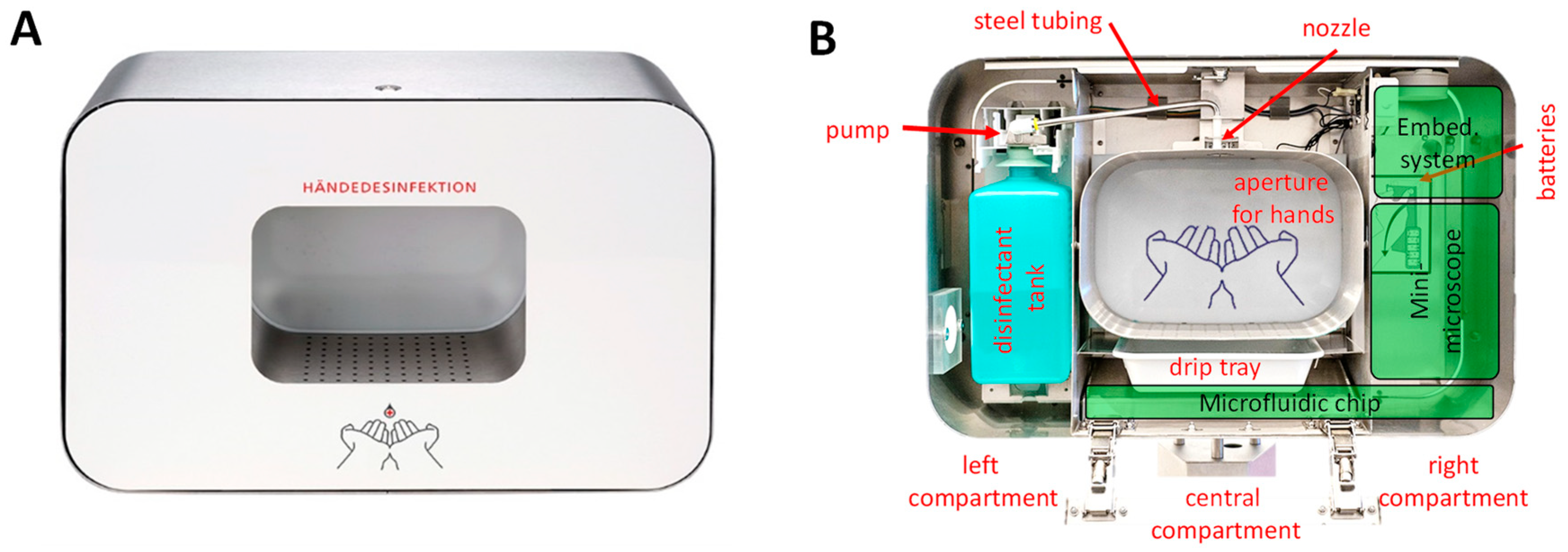
2.2. Optical Set-up
2.3. Image Analysis
2.4. Classifiers
2.4.1. Random Forest
2.4.2. Support Vector Machine
2.4.3. 1-over-4 Classification Strategy
2.4.4. Binary Classification Strategy
2.4.5. Tournament Classification Strategy
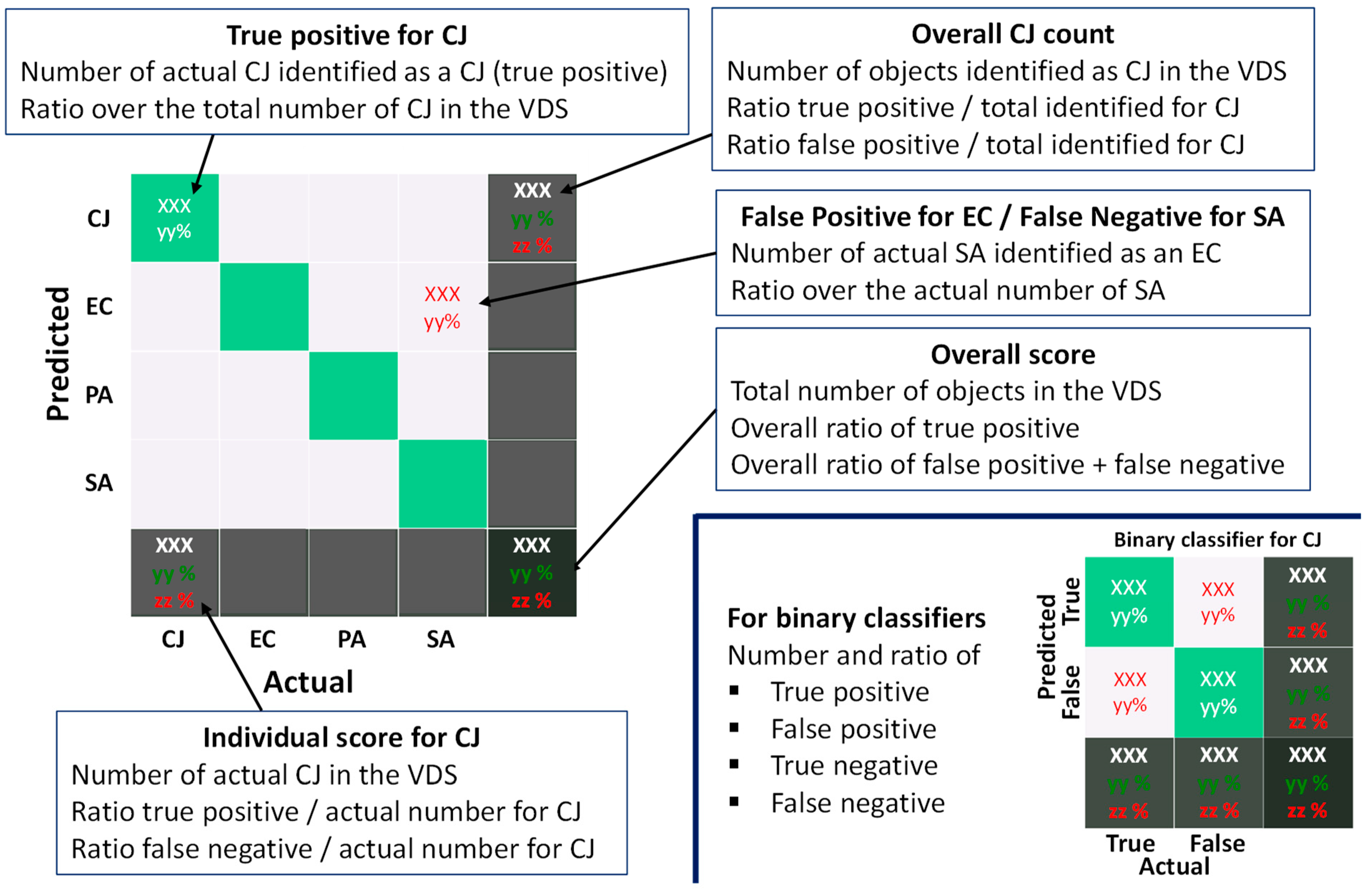
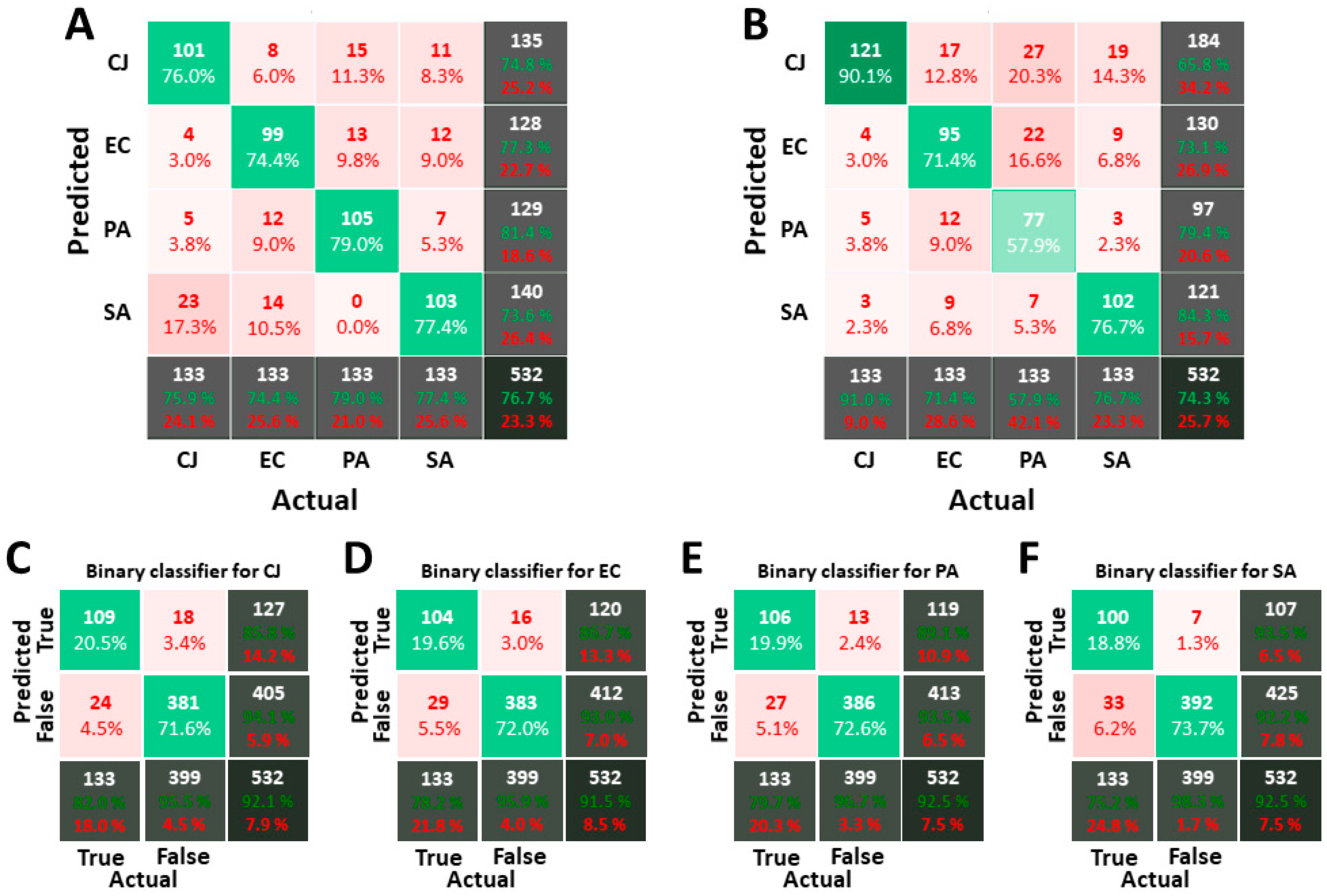
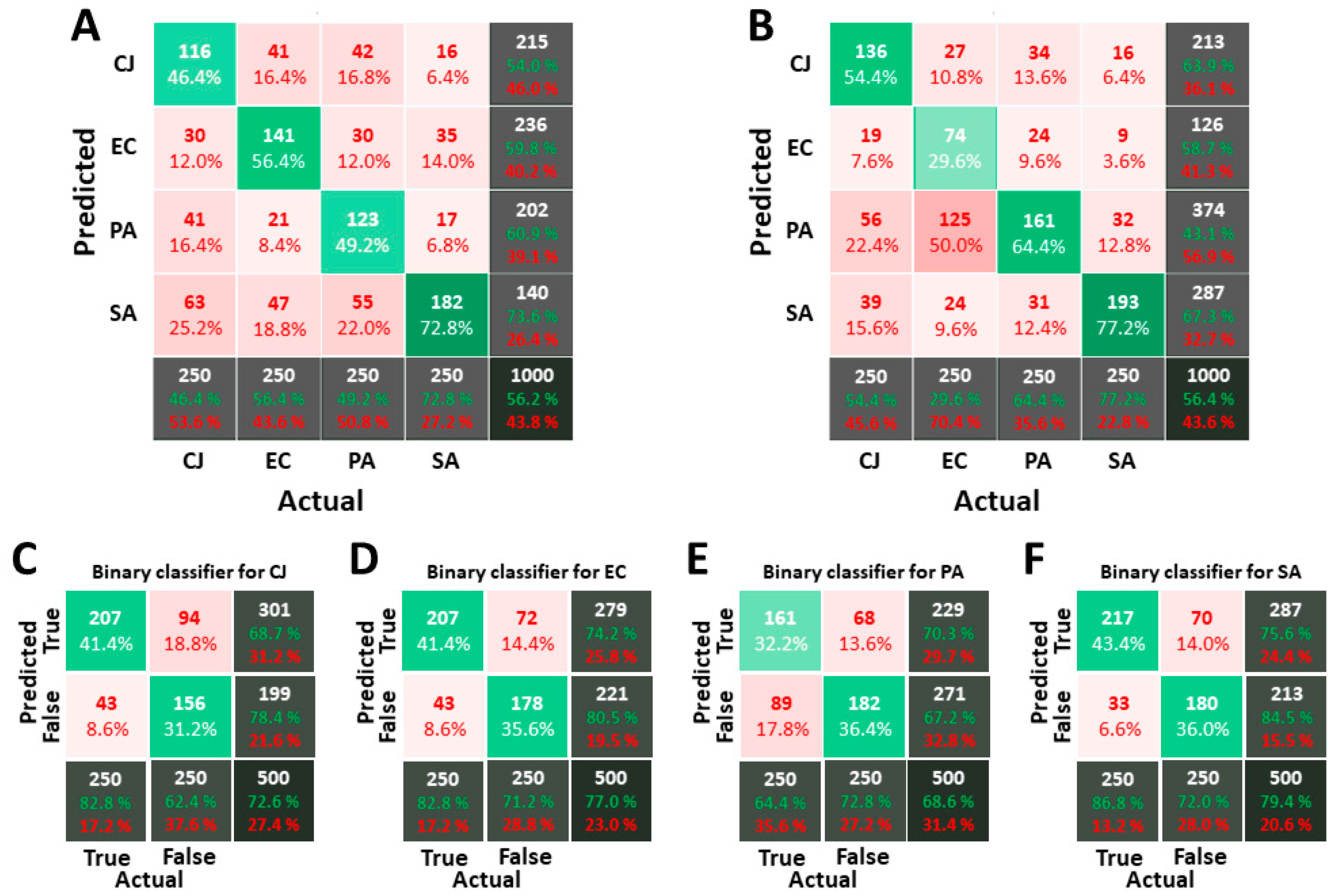
2.5. Classifier Validation and Metric
- The overall score, on the bottom-right box, that gives the ratio of objects of the VDS for which the prediction matches the actual bacterium label.
- The individual score, on the bottom row, for each bacterium, which provides the ratio of appropriate labelling for a given bacterium.
- The bacteria count which should be as homogeneous as possible, considering that the number of each bacterium in the VDS is the same. If inhomogeneity occurs, the classifier is considered to be biased.
2.6. Datasets
3. Results
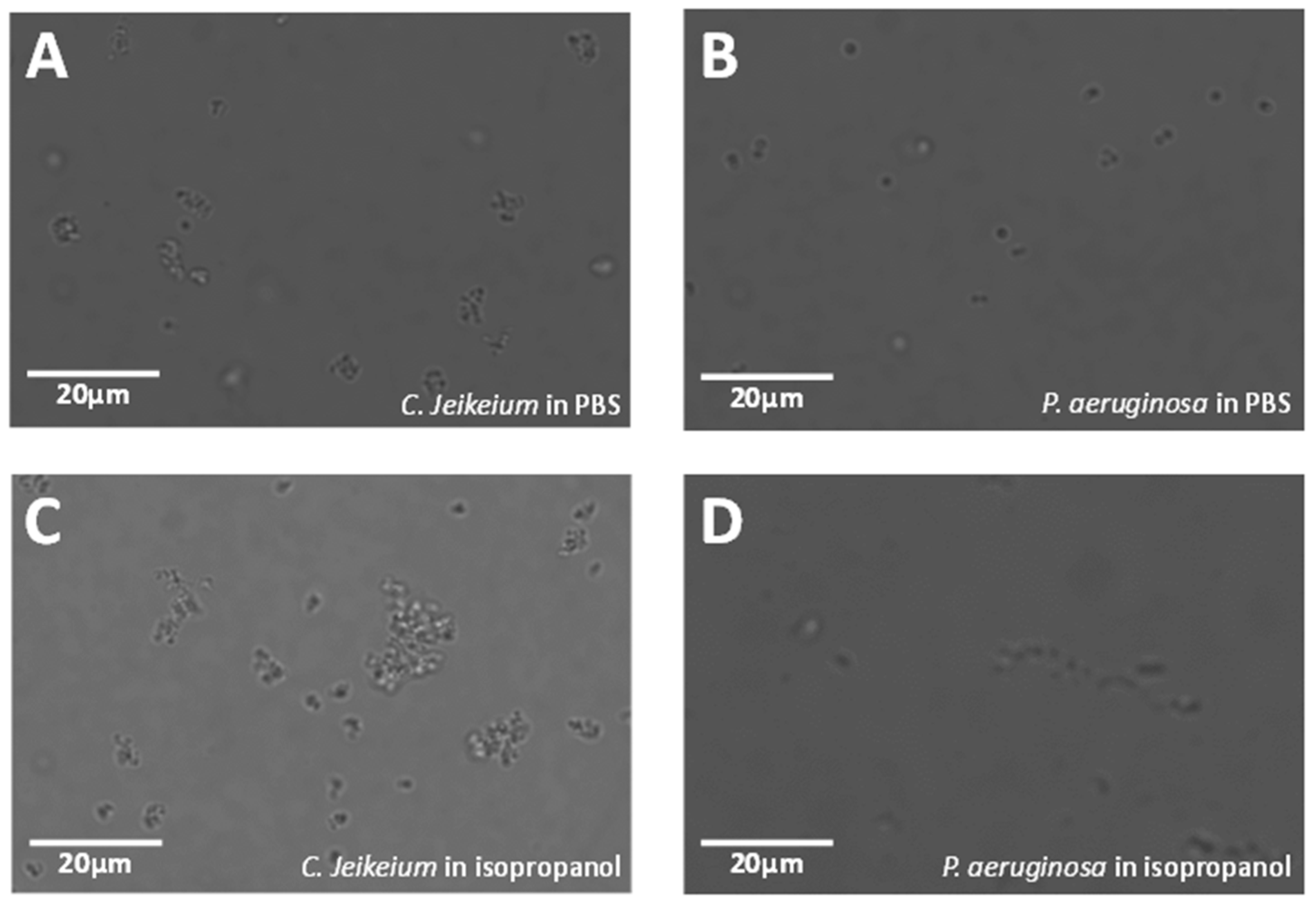
3.1. Image Processing of Images Acquired in PBS
3.2. Classification of Images Acquired in PBS
3.3. Image Processing of Images Acquired in Isopropanol
3.4. Classification of Images Acquired in Isopropanol
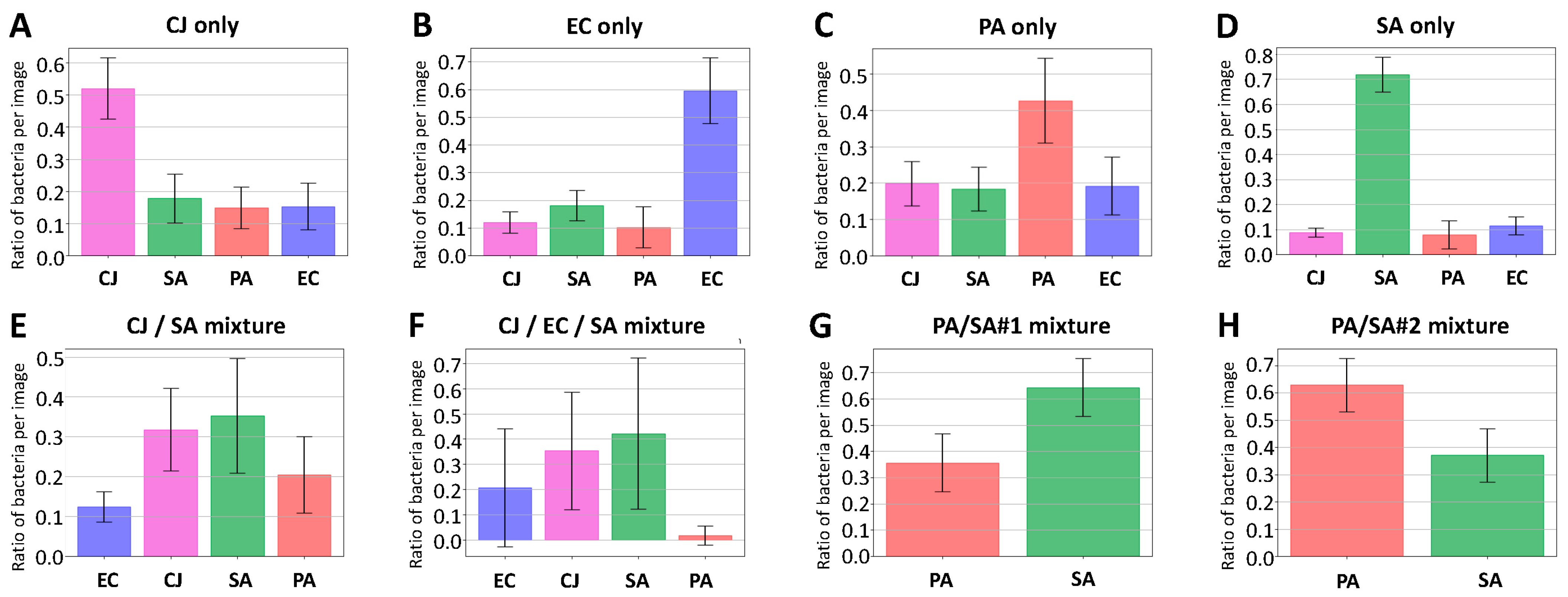
3.5. Mixture Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gastmeier, P.; Geffers, C. Nosokomiale Infektionen in Deutschland: Wie viele gibt es wirklich? DMW-Dtsch. Med. Wochenschr. 2008, 133, 1111–1115. [Google Scholar] [CrossRef] [PubMed]
- Gastmeier, P.; Brunkhorst, F.; Schrappe, M.; Kern, W.; Geffers, C. How many nosocomial infections are avoidable? DMW-Dtsch. Med. Wochenschr. 2010, 135, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Jumaa, P.A. Hand hygiene: Simple and complex. Int. J. Infect. Dis. 2005, 9, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Pittet, D. The World Health Organization (WHO) clean care is safer care promotion campaign, 2005–2018. Int. J. Infect. Dis. 2018, 73, 48. [Google Scholar] [CrossRef]
- Pittet, D.; Allegranzi, B.; Sax, H.; Dharan, S.; Pessoa-Silva, C.L.; Donaldson, L.; Boyce, J.M.; WHO Global Patient Safety Challenge. World Alliance for Patient Safety Evidence-based model for hand transmission during patient care and the role of improved practices. Lancet Infect. Dis. 2006, 6, 641–652. [Google Scholar] [CrossRef]
- Fournier, P.-E.; Drancourt, M.; Colson, P.; Rolain, J.-M.; Scola, B.L.; Raoult, D. Modern clinical microbiology: New challenges and solutions. Nat. Rev. Microbiol. 2013, 11, 574–585. [Google Scholar] [CrossRef]
- David, A.; Relman, M.D. Pathogen Discovery, Detection, and Diagnostics; National Academies Press: Washington, DC, USA, 2003. [Google Scholar]
- Peruski, A.H.; Peruski, L.F. Immunological Methods for Detection and Identification of Infectious Disease and Biological Warfare Agents. Clin. Diagn. Lab. Immunol. 2003, 10, 506–513. [Google Scholar] [CrossRef]
- Law, J.W.-F.; Ab Mutalib, N.-S.; Chan, K.-G.; Lee, L.-H. Rapid methods for the detection of foodborne bacterial pathogens: Principles, applications, advantages and limitations. Front. Microbiol. 2015, 5. [Google Scholar] [CrossRef]
- Croxatto, A.; Prod’hom, G.; Greub, G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 2012, 36, 380–407. [Google Scholar] [CrossRef]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef]
- Xu, S. Electromechanical biosensors for pathogen detection. Microchim. Acta 2012, 178, 245–260. [Google Scholar] [CrossRef]
- You, X.; Zhang, D.; Yao, K.; Huang, Y.; Liu, M.; Xie, J.; Shih, T.; Sun, H.; Chen, Z. Ultrasensitive and fast detection of pathogens using Europium-containing polystyrene nanospheres in a homemade portable NMR diagnostic system. Sens. Actuators B Chem. 2020, 320, 128370. [Google Scholar] [CrossRef]
- Yoo, S.M.; Lee, S.Y. Optical Biosensors for the Detection of Pathogenic Microorganisms. Trends Biotechnol. 2016, 34, 7–25. [Google Scholar] [CrossRef]
- Daniels, J.S.; Pourmand, N. Label-Free Impedance Biosensors: Opportunities and Challenges. Electroanalysis 2007, 19, 1239–1257. [Google Scholar] [CrossRef] [PubMed]
- Brolo, A.G. Plasmonics for future biosensors. Nat. Photonics 2012, 6, 709–713. [Google Scholar] [CrossRef]
- Bantz, K.C.; Meyer, A.F.; Wittenberg, N.J.; Im, H.; Kurtuluş, Ö.; Lee, S.H.; Lindquist, N.C.; Oh, S.-H.; Haynes, C.L. Recent progress in SERS biosensing. Phys. Chem. Chem. Phys. 2011, 13, 11551–11567. [Google Scholar] [CrossRef] [PubMed]
- Hunt, H.K.; Armani, A.M. Label-free biological and chemical sensors. Nanoscale 2010, 2, 1544–1559. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Kuwahara, M.; Hachimura, K.; Eiho, S.; Kinoshita, M. Processing of RI-Angiocardiographic Images. In Digital Processing of BioMedical Images; Preston, K., Onoe, M., Eds.; Springer: Boston, MA, USA, 1976; pp. 187–202. ISBN 978-1-4684-0769-3. [Google Scholar]
- Serra, J. Image Analysis and Mathematical Morphology; Academic Press, Inc.: Orlando, FL, USA, 1983; ISBN 978-0-12-637240-3. [Google Scholar]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning: With Applications in R.; Springer Texts in Statistics; Springer: New York, NY, USA, 2013; ISBN 978-1-4614-7137-0. [Google Scholar]
- Kievit, T.R.D. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ. Microbiol. 2009, 11, 279–288. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Park, J.-H.; Inouye, M. MqsR, a Crucial Regulator for Quorum Sensing and Biofilm Formation, Is a GCU-specific mRNA Interferase in Escherichia coli. J. Biol. Chem. 2009, 284, 28746–28753. [Google Scholar] [CrossRef]
- Yarwood, J.M.; Bartels, D.J.; Volper, E.M.; Greenberg, E.P. Quorum Sensing in Staphylococcus aureus Biofilms. J. Bacteriol. 2004, 186, 1838–1850. [Google Scholar] [CrossRef] [PubMed]
- Kwaszewska, A.; Brewczyńska, A.; Szewczyk, E. Hydrophobicity and biofilm formation of lipophilic skin corynebacteria. Pol. J. Microbiol. 2006, 55, 189–193. [Google Scholar] [PubMed]
- McDonnell, G.; Russell, A.D. Antiseptics and Disinfectants: Activity, Action, and Resistance. Clin. Microbiol. Rev. 1999, 12, 147–179. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; He, L.; Zhou, F.; Chen, S.; Ni, D.; Lei, B.; Wang, T. Segmentation, Splitting, and Classification of Overlapping Bacteria in Microscope Images for Automatic Bacterial Vaginosis Diagnosis. IEEE J. Biomed. Health Inform. 2017, 21, 1095–1104. [Google Scholar] [CrossRef]
- Wang, W.; Song, H. Cell Cluster Image Segmentation on Form Analysis. In Proceedings of the Third International Conference on Natural Computation (ICNC 2007), Haikou, China, 24–27 August 2007; Volume 4, pp. 833–836. [Google Scholar]
- Du, Y.; Duever, T.A.; Budman, H.H. Segmentation and Quantitative Analysis of Apoptosis of Chinese Hamster Ovary Cells from Fluorescence Microscopy Images. Microsc. Microanal. 2017, 23, 1–15. [Google Scholar] [CrossRef]
- Ye, J.; Chow, J.-H.; Chen, J.; Zheng, Z. Stochastic gradient boosted distributed decision trees. In Proceedings of the 18th ACM Conference on Information and Knowledge Management, Hong Kong, China, 2–6 November 2009; Association for Computing Machinery: New York, NY, USA, 2009; pp. 2061–2064. [Google Scholar]
- Zieliński, B.; Plichta, A.; Misztal, K.; Spurek, P.; Brzychczy-Włoch, M.; Ochońska, D. Deep learning approach to bacterial colony classification. PLoS ONE 2017, 12, e0184554. [Google Scholar] [CrossRef]
- Kim, G.; Ahn, D.; Kang, M.; Jo, Y.; Ryu, D.; Kim, H.; Song, J.; Ryu, J.S.; Choi, G.; Chung, H.J.; et al. Rapid and label-free identification of individual bacterial pathogens exploiting three-dimensional quantitative phase imaging and deep learning. BioRxiv 2019, 596486. [Google Scholar] [CrossRef]
- Gu, X.; Pan, L.; Liang, H.; Yang, R. Classification of Bacterial and Viral Childhood Pneumonia Using Deep Learning in Chest Radiography. In Proceedings of the 3rd International Conference on Multimedia and Image Processing, Guiyang, China, 16–19 March 2018; Association for Computing Machinery: New York, NY, USA, 2018; pp. 88–93. [Google Scholar]
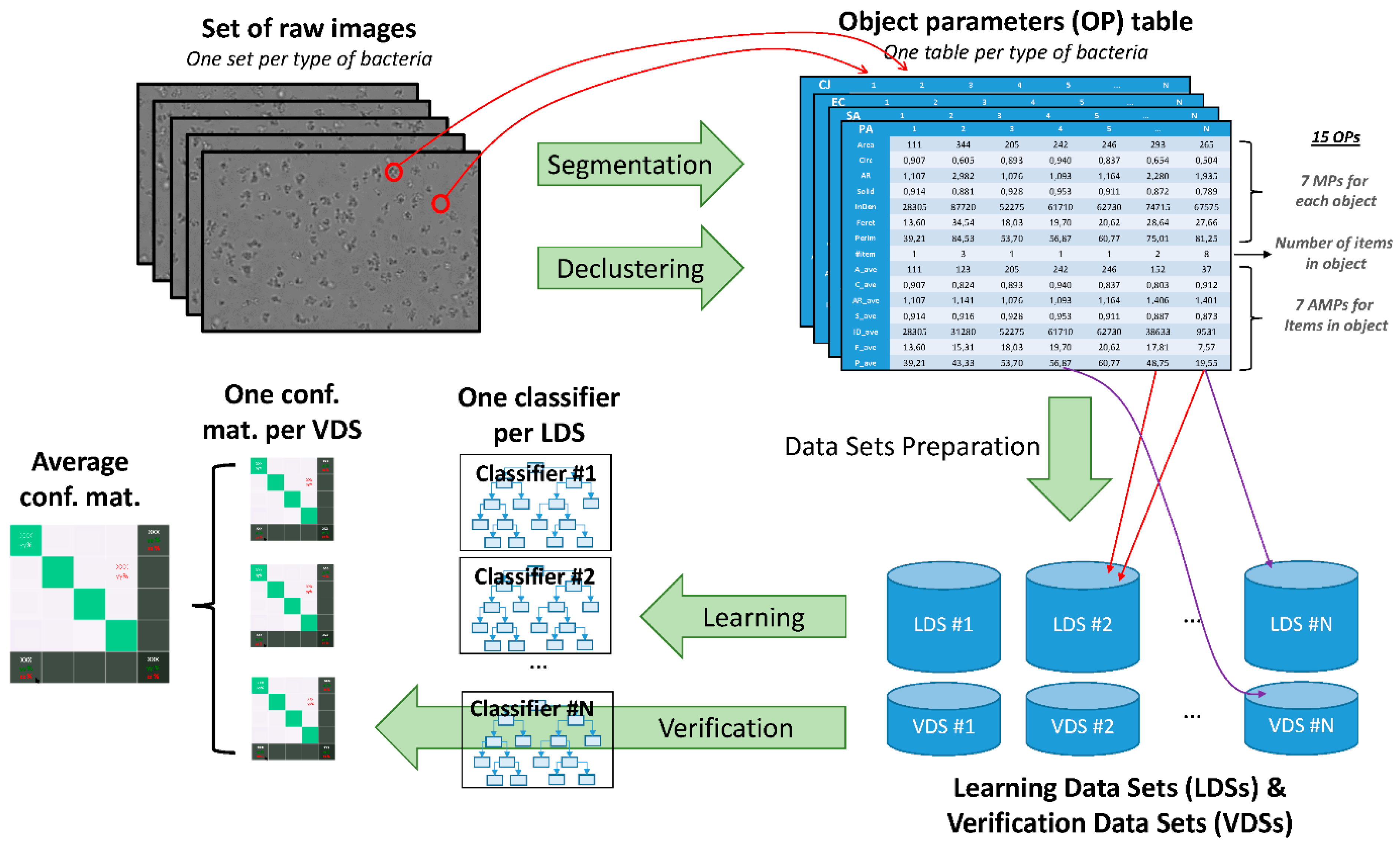
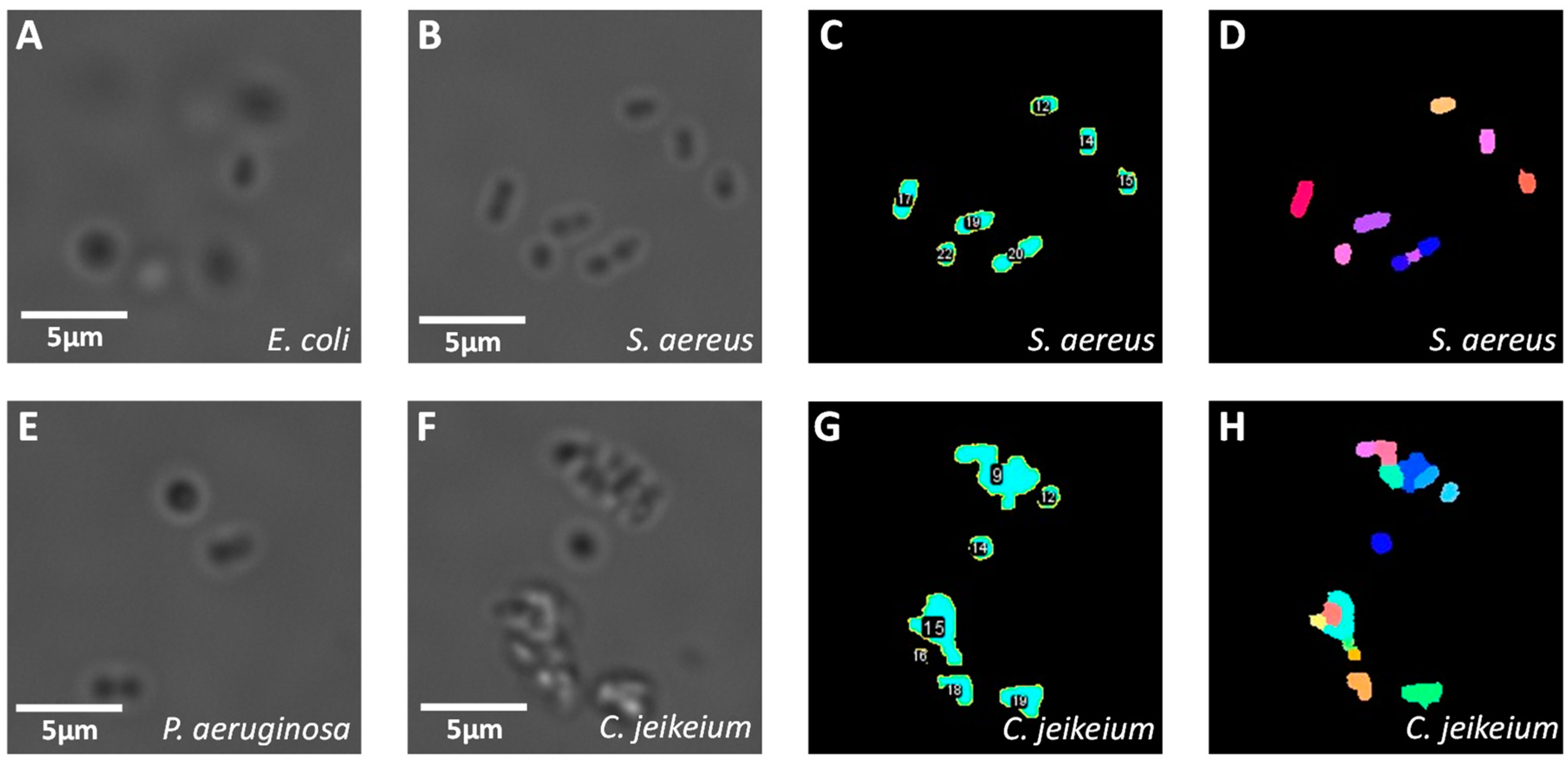
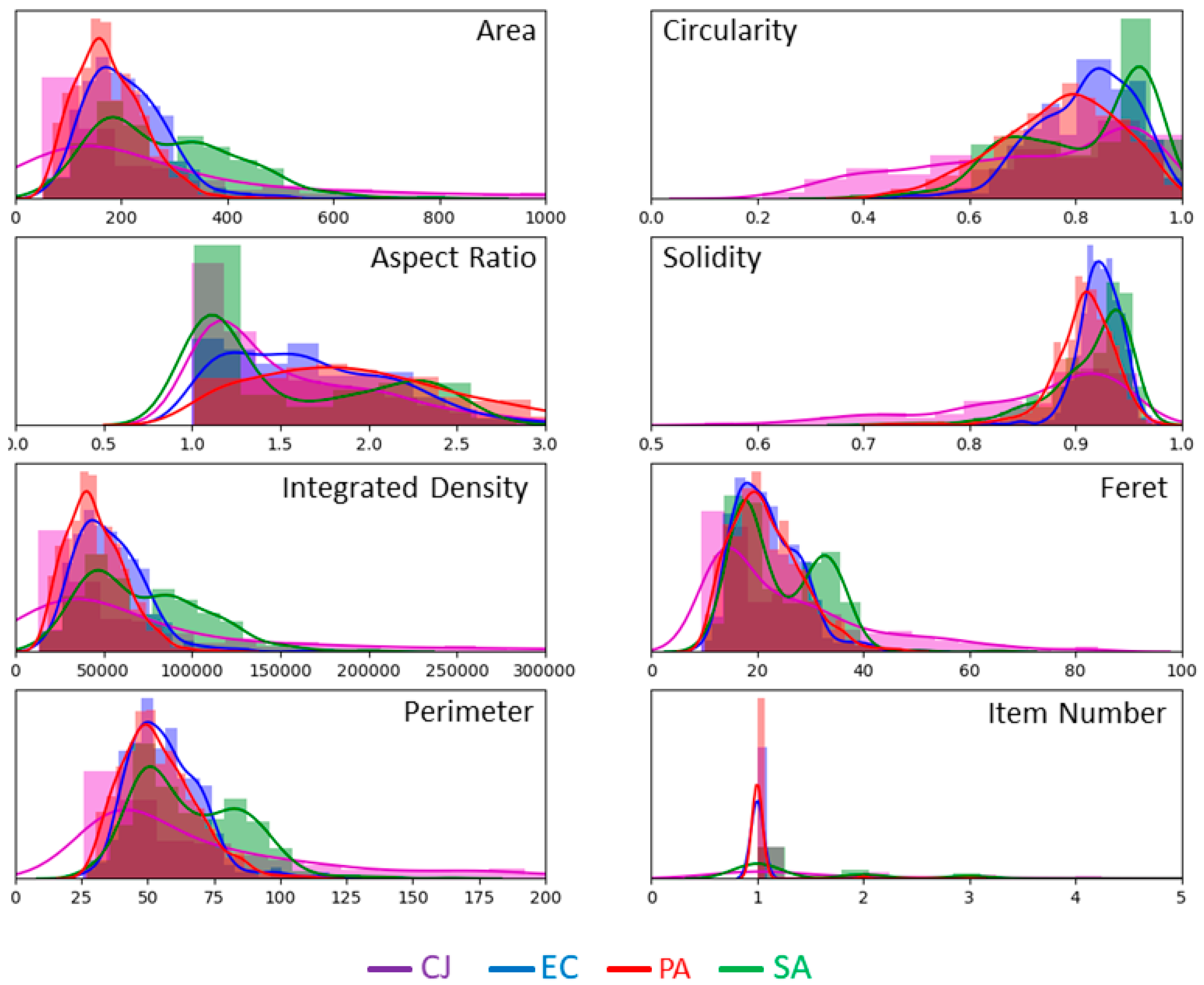
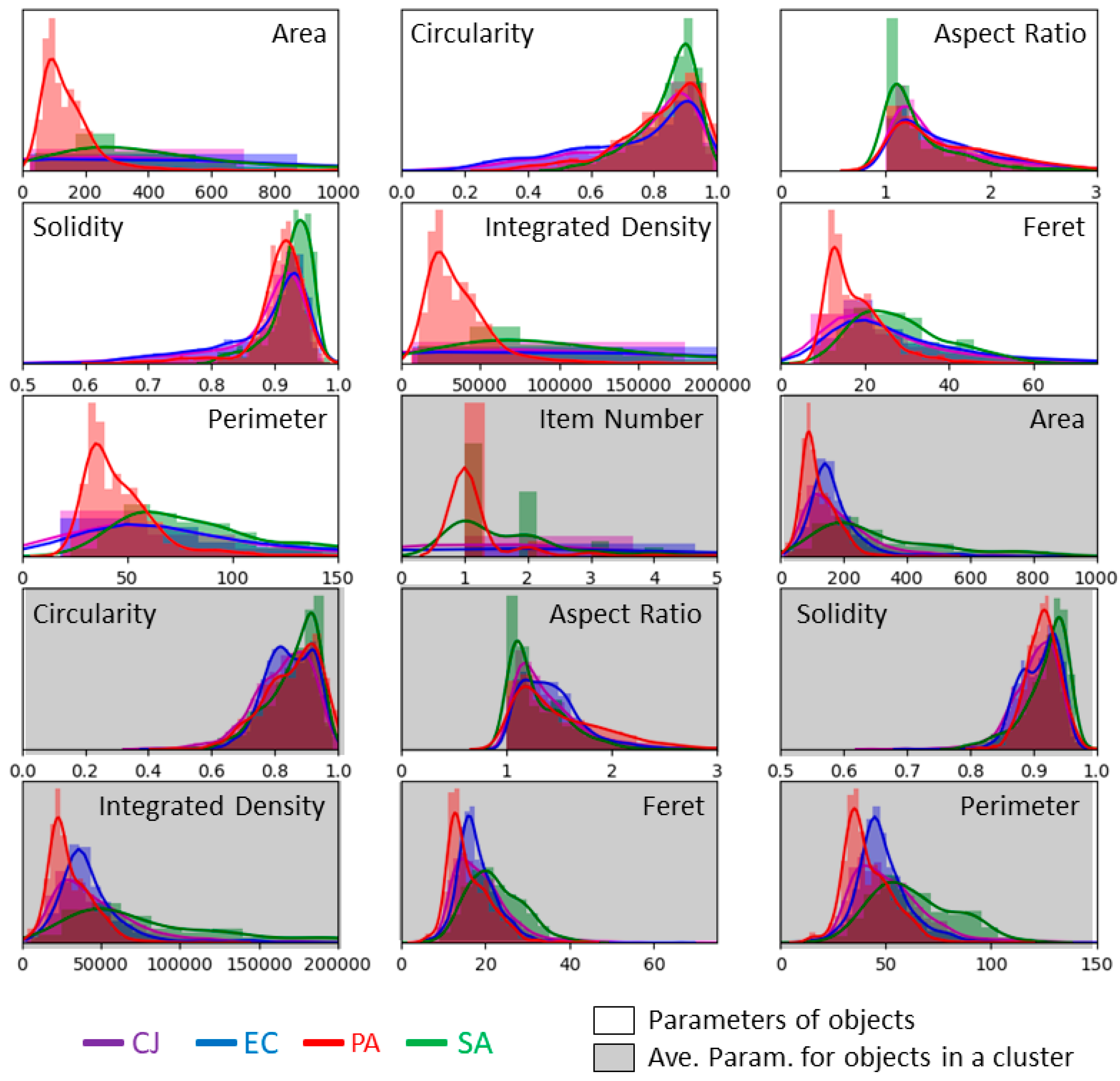
| Morphological Parameter | Symbol | Definition |
|---|---|---|
| Area | A | The number of pixels inside the detected object. |
| Perimeter | P | The number of pixels that compose the boundary of the object. |
| Circularity | C | Defined by the equation . |
| Aspect Ratio | AR | The ratio of the major axis and the minor axis of the ellipse that fits the object outline. AR = 1 means that the object is perfectly round. AR tends to 0 for skinny objects. |
| Solidity | S | Defined as the ratio between the area of the object and the area of the smallest convex shape that encompasses the object. S = 1 defines a perfectly convex object. S tends to 0 for rough objects or objects with erratic outlines. |
| Integrated Density | D | The sum of the intensity of the pixels inside the detected object. |
| Feret diameter | F | The maximal distance between two points of the outline of the detected object. |
| Actual | Bacteria in | Predicted | 1-over-4 | Binary | Tournament | |||
|---|---|---|---|---|---|---|---|---|
| Label | the Dataset | Label | Number | % | Number | % | Number | % |
| CJ | 489 | CJ | 378 | 77.3 | 365.5 | 74.7 | 413.3 | 84.6 |
| EC | 29 | 5.9 | 14 | 2.9 | 21.3 | 4.4 | ||
| PA | 34 | 7.0 | 17 | 3.5 | 27.3 | 5.6 | ||
| SA | 48 | 9.8 | 15.5 | 3.2 | 27 | 5.4 | ||
| Others | 0 | 0 | 77 | 15.7 | 0 | 0 | ||
| EC | 560 | CJ | 34 | 6.0 | 9 | 1.6 | 21.8 | 3.9 |
| EC | 427 | 76.3 | 417 | 74.5 | 462.3 | 82.6 | ||
| PA | 50 | 8.9 | 27.5 | 4.9 | 40 | 7.1 | ||
| SA | 49 | 8.8 | 27.5 | 4.9 | 35.8 | 6.4 | ||
| Others | 0 | 0 | 79 | 14.1 | 0 | 0 | ||
| PA | 1000 | CJ | 75 | 7.5 | 29 | 2.9 | 79.6 | 8.0 |
| EC | 109 | 10.9 | 75 | 7.5 | 67.6 | 6.8 | ||
| PA | 804 | 80.4 | 785 | 78.5 | 842 | 84.3 | ||
| SA | 12 | 1.2 | 5 | 0.5 | 10 | 1.0 | ||
| Others | 0 | 0 | 106 | 10.6 | 0 | 0 | ||
| SA | 444 | CJ | 32 | 7.2 | 16.5 | 3.7 | 12.6 | 2.8 |
| EC | 33 | 7.4 | 17.5 | 4.0 | 16.6 | 3.8 | ||
| PA | 18 | 4.0 | 9 | 2.0 | 5 | 1.1 | ||
| SA | 361 | 81.3 | 369 | 83.1 | 409 | 92.2 | ||
| Others | 0 | 0 | 32 | 7.2 | 0 | 0 | ||
| Actual | Bacteria in | Predicted | 1-over-4 | Binary | Tournament | |||
|---|---|---|---|---|---|---|---|---|
| Label | the dataset | Label | Number | % | Number | % | Number | % |
| CJ | 1537 | CJ | 798 | 51.9 | 798 | 51.9 | 796 | 51.8 |
| EC | 217 | 14.1 | 222 | 14.5 | 231 | 15.0 | ||
| PA | 197 | 12.8 | 272 | 17.6 | 231 | 15.0 | ||
| SA | 325 | 21.2 | 237 | 15.5 | 279 | 18.2 | ||
| Others | 0 | 0 | 8 | 0.5 | 0 | 0 | ||
| EC | 4608 | CJ | 609 | 13.2 | 830 | 18.0 | 599 | 13.0 |
| EC | 2890 | 62.7 | 2352 | 51.1 | 2751 | 59.7 | ||
| PA | 257 | 5.6 | 613 | 13.3 | 474 | 10.3 | ||
| SA | 852 | 18.5 | 782 | 17.0 | 782 | 17.0 | ||
| Others | 0 | 0 | 29 | 0.6 | 0 | 0 | ||
| PA | 2109 | CJ | 439 | 20.8 | 540 | 25.6 | 455 | 21.6 |
| EC | 356 | 17.0 | 316 | 15.0 | 345 | 16.4 | ||
| PA | 867 | 41.1 | 916 | 43.4 | 890 | 42.2 | ||
| SA | 444 | 21.1 | 326 | 15.5 | 417 | 19.8 | ||
| Others | 0 | 0 | 11 | 0.5 | 0 | 0 | ||
| SA | 1111 | CJ | 78 | 7.0 | 164 | 14.8 | 105 | 9.4 |
| EC | 133 | 12.0 | 135 | 12.1 | 135 | 12.2 | ||
| PA | 81 | 7.3 | 103 | 9.2 | 73 | 6.7 | ||
| SA | 819 | 73.7 | 704 | 63.4 | 796 | 71.7 | ||
| Others | 0 | 0 | 5 | 0.5 | 0 | 0 | ||
| No Hit | Single Hit | Double Hit | Triple Hit | Quadruple Hit | |
|---|---|---|---|---|---|
| PBS | 12.1% | 86.4% | 1.3% | 0.0% | 0.0% |
| Isopropanol | 0.7% | 39.1% | 50.4% | 9.7% | 0.0% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Claudinon, J.; Steltenkamp, S.; Fink, M.; Sych, T.; Verreman, B.; Römer, W.; Madec, M. A Label-Free Optical Detection of Pathogens in Isopropanol as a First Step towards Real-Time Infection Prevention. Biosensors 2021, 11, 2. https://doi.org/10.3390/bios11010002
Claudinon J, Steltenkamp S, Fink M, Sych T, Verreman B, Römer W, Madec M. A Label-Free Optical Detection of Pathogens in Isopropanol as a First Step towards Real-Time Infection Prevention. Biosensors. 2021; 11(1):2. https://doi.org/10.3390/bios11010002
Chicago/Turabian StyleClaudinon, Julie, Siegfried Steltenkamp, Manuel Fink, Taras Sych, Benoît Verreman, Winfried Römer, and Morgan Madec. 2021. "A Label-Free Optical Detection of Pathogens in Isopropanol as a First Step towards Real-Time Infection Prevention" Biosensors 11, no. 1: 2. https://doi.org/10.3390/bios11010002
APA StyleClaudinon, J., Steltenkamp, S., Fink, M., Sych, T., Verreman, B., Römer, W., & Madec, M. (2021). A Label-Free Optical Detection of Pathogens in Isopropanol as a First Step towards Real-Time Infection Prevention. Biosensors, 11(1), 2. https://doi.org/10.3390/bios11010002





