Application of Chemometrics in Biosensing: A Brief Review
Abstract
1. Introduction
2. Typical Chemometric Tools Employed in Biosensor Research
3. Application of Chemometrics in Different Fields of Biosensing
3.1. Environmental Monitoring
3.2. Water Quality
3.3. Food and Beverages Analysis
3.4. Biological and Medical Chemistry
4. Experimental Design and Mathematical Modeling
5. Future Perspectives
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AChE | Acetylcholinesterase |
| ANN | Artificial Neural Network |
| FIA | Flow Injection Analysis |
| ET | Electronic Tongue |
| GFP | Green Fluorescent Protein |
| GOx | Glucose Oxidase |
| HSA | Human Serum Albumin |
| HCA | Hierarchical Cluster Analysis |
| LDA | Linear Discriminant Analysis |
| MAB | Monoclonal Antibody |
| MCR-ALS | Multivariate Curve Resolution Alternating Least Squares |
| NPs | Nanoparticles |
| PCA | Principle Component Analysis |
| PLS | Partial Least Squares |
| RMSEP | Root-Mean-Square Error of Prediction |
| SIA | Sequential Injection Analysis |
| SVM | Supported Vector Machines |
| QSAR | Quantitative Structure-Activity Relationship |
| QSPR | Quantitative Structure-Property Relationship |
References
- Thvenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification (Technical Report). Pure Appl. Chem. 1999, 71, 2333–2348. [Google Scholar] [CrossRef]
- Wang, J. Electrochemical glucose biosensors. Chem. Rev. 2008, 108, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, Z.; Feng, Y. Construction of bienzyme biosensors based on combination of the one-step electrodeposition and covalent-coupled sol-gel process. Sci. China Ser. B Chem. 2009, 52, 2269–2274. [Google Scholar] [CrossRef]
- del Valle, M. Electronic Tongues Employing Electrochemical Sensors. Electroanalysis 2010, 22, 1539–1555. [Google Scholar] [CrossRef]
- Neves, B.J.; Braga, R.C.; Melo-Filho, C.C.; Moreira-Filho, J.T.; Muratov, E.N.; Andrade, C.H. QSAR-Based Virtual Screening: Advances and Applications in Drug Discovery. Front. Pharmacol. 2018, 9, 1275. [Google Scholar] [CrossRef]
- Liu, H.; Sun, P.; Liu, H.; Yang, S.; Wang, L.; Wang, Z. Acute toxicity of benzophenone-type UV filters for Photobacterium phosphoreum and Daphnia magna: QSAR analysis, interspecies relationship and integrated assessment. Chemosphere 2015, 135, 182–188. [Google Scholar] [CrossRef]
- Su, L.; Zhang, X.; Yuan, X.; Zhao, Y.; Zhang, D.; Qin, W. Evaluation of joint toxicity of nitroaromatic compounds and copper to Photobacterium phosphoreum and QSAR analysis. J. Hazard. Mater. 2012, 241–242, 450–455. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal Component Analysis; Royal Society of Chemistry: London, UK, 2014; Volume 6. [Google Scholar]
- Tønning, E.; Sapelnikova, S.; Christensen, J.; Carlsson, C.; Winther-Nielsen, M.; Dock, E.; Solna, R.; Skladal, P.; Nørgaard, L.; Ruzgas, T.; et al. Chemometric exploration of an amperometric biosensor array for fast determination of wastewater quality. Biosens. Bioelectron. 2005, 21, 608–617. [Google Scholar] [CrossRef]
- Wold, S.; Sjöström, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Raud, M.; Kikas, T. Bioelectronic tongue and multivariate analysis: A next step in BOD measurements. Water Res. 2013, 47, 2555–2562. [Google Scholar]
- Marini, F.; Bucci, R.; Magrì, A.L.; Magrì, A.D. Artificial neural networks in chemometrics: History, examples and perspectives. Microchem. J. 2008, 88, 178–185. [Google Scholar] [CrossRef]
- Brown, S.; Tauler, R.; Walczak, B. (Eds.) Comprehensive Chemometrics: Chemical and Biochemical Data Analysis; Elsevier: Amsterdam, The Netherlands, 2020; ISBN 978-0-444-64166-3. [Google Scholar]
- Cortina, M.; Del Valle, M.; Marty, J.L. Electronic tongue using an enzyme inhibition biosensor array for the resolution of pesticide mixtures. Electroanalysis 2008, 20, 54–60. [Google Scholar] [CrossRef]
- Gutés, A.; Céspedes, F.; Alegret, S.; Del Valle, M. Determination of phenolic compounds by a polyphenol oxidase amperometric biosensor and artificial neural network analysis. Biosens. Bioelectron. 2005, 20, 1668–1673. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.K.; Dantas, M.V.C.; Nogueira, A.B.; Etchegaray, A.; Filgueiras, P.R.; Poppi, R.J. Simultaneous determination of different phenolic compounds using electrochemical biosensor and multivariate calibration. J. Braz. Chem. Soc. 2018, 29, 482–489. [Google Scholar] [CrossRef]
- Ejeian, F.; Etedali, P.; Mansouri-Tehrani, H.A.; Soozanipour, A.; Low, Z.X.; Asadnia, M.; Taheri-Kafrani, A.; Razmjou, A. Biosensors for wastewater monitoring: A review. Biosens. Bioelectron. 2018, 118, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, T.T.; Leca, B.; Vilatte, F.; Marty, J.L.; Fournier, D.; Schmid, R.D. Improved multianalyte detection of organophosphates and carbamates with disposable multielectrode biosensors using recombinant mutants of Drosophila acetylcholinesterase and artificial neural networks. Biosens. Bioelectron. 2000, 15, 193–201. [Google Scholar] [CrossRef]
- Valdés-Ramírez, G.; Gutiérrez, M.; del Valle, M.; Ramírez-Silva, M.T.; Fournier, D.; Marty, J.L. Automated resolution of dichlorvos and methylparaoxon pesticide mixtures employing a Flow Injection system with an inhibition electronic tongue. Biosens. Bioelectron. 2009, 24, 1103–1108. [Google Scholar] [CrossRef]
- Alonso, G.A.; Istamboulie, G.; Noguer, T.; Marty, J.L.; Muñoz, R. Rapid determination of pesticide mixtures using disposable biosensors based on genetically modified enzymes and artificial neural networks. Sens. Actuators B Chem. 2012, 164, 22–28. [Google Scholar] [CrossRef]
- Ni, Y.; Cao, D.; Kokot, S. Simultaneous enzymatic kinetic determination of pesticides, carbaryl and phoxim, with the aid of chemometrics. Anal. Chim. Acta 2007, 588, 131–139. [Google Scholar] [CrossRef]
- Crew, A.; Lonsdale, D.; Byrd, N.; Pittson, R.; Hart, J.P. A screen-printed, amperometric biosensor array incorporated into a novel automated system for the simultaneous determination of organophosphate pesticides. Biosens. Bioelectron. 2011, 26, 2847–2851. [Google Scholar] [CrossRef]
- Ferentinos, K.P.; Yialouris, C.P.; Blouchos, P.; Moschopoulou, G.; Kintzios, S. Pesticide residue screening using a novel artificial neural network combined with a bioelectric cellular biosensor. Biomed Res. Int. 2013, 2013, 813519. [Google Scholar] [CrossRef] [PubMed]
- Dock, E.; Christensen, J.; Olsson, M.; Tønning, E.; Ruzgas, T.; Emnéus, J. Multivariate data analysis of dynamic amperometric biosensor responses from binary analyte mixtures—Application of sensitivity correction algorithms. Talanta 2005, 65, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Freire, R.S.; Ferreira, M.M.C.; Durán, N.; Kubota, L.T. Dual amperometric biosensor device for analysis of binary mixtures of phenols by multivariate calibration using partial least squares. Anal. Chim. Acta 2003, 485, 263–269. [Google Scholar] [CrossRef]
- Cetó, X.; González-Calabuig, A.; del Valle, M. Use of a Bioelectronic Tongue for the Monitoring of the Photodegradation of Phenolic Compounds. Electroanalysis 2015, 27, 225–233. [Google Scholar] [CrossRef]
- Ebrahimi, D.; Chow, E.; Gooding, J.J.; Hibbert, D.B. Multi-analyte sensing: A chemometrics approach to understanding the merits of electrode arrays versus single electrodes. Analyst 2008, 133, 1090–1096. [Google Scholar] [CrossRef] [PubMed]
- Serrano, N.; Prieto-Simón, B.; Cetó, X.; Del Valle, M. Array of peptide-modified electrodes for the simultaneous determination of Pb(II), Cd(II) and Zn(II). Talanta 2014, 125, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Zhuang, Q.; Wang, Y.; Ni, Y. Label-free fluorescent catalytic biosensor for highly sensitive and selective detection of the ferrous ion in water samples using a layered molybdenum disulfide nanozyme coupled with an advanced chemometric model. Analyst 2016, 141, 1822–1829. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Xiang, L.; Zhang, Y.; Wang, Y.; Nie, Z. Simultaneous quantitative analysis of K+ and Tl+ in serum and drinking water based on UV–Vis spectra and chemometrics. Spectrochim. Acta—Part A Mol. Biomol. Spectrosc. 2020, 238, 118392. [Google Scholar] [CrossRef] [PubMed]
- Czolkos, I.; Dock, E.; Tønning, E.; Christensen, J.; Winther-Nielsen, M.; Carlsson, C.; Mojzíková, R.; Skládal, P.; Wollenberger, U.; Nørgaard, L.; et al. Prediction of wastewater quality using amperometric bioelectronic tongues. Biosens. Bioelectron. 2016, 75, 375–382. [Google Scholar] [CrossRef]
- Feng, Y.; Kayode, O.; Harper, W.F. Using microbial fuel cell output metrics and nonlinear modeling techniques for smart biosensing. Sci. Total Environ. 2013, 449, 223–228. [Google Scholar] [CrossRef]
- Bro, R. Multiway calibration. Multilinear PLS. J. Chemom. 1996, 10, 47–61. [Google Scholar] [CrossRef]
- Chandra, S.; Chapman, J.; Power, A.; Roberts, J.; Cozzolino, D. The application of state-of-the-art analytic tools (biosensors and spectroscopy) in beverage and food fermentation process monitoring. Fermentation 2017, 3, 50. [Google Scholar] [CrossRef]
- Scognamiglio, V.; Arduini, F.; Palleschi, G.; Rea, G. Biosensing technology for sustainable food safety. TrAC—Trends Anal. Chem. 2014, 62, 1–10. [Google Scholar] [CrossRef]
- Neethirajan, S.; Ragavan, V.; Weng, X.; Chand, R. Biosensors for Sustainable Food Engineering: Challenges and Perspectives. Biosensors 2018, 8, 23. [Google Scholar] [CrossRef] [PubMed]
- Narsaiah, K.; Jha, S.N.; Bhardwaj, R.; Sharma, R.; Kumar, R. Optical biosensors for food quality and safety assurance-A review. J. Food Sci. Technol. 2012, 49, 383–406. [Google Scholar] [CrossRef]
- Skov, T.; Honoré, A.H.; Jensen, H.M.; Næs, T.; Engelsen, S.B. Chemometrics in foodomics: Handling data structures from multiple analytical platforms. TrAC—Trends Anal. Chem. 2014, 60, 71–79. [Google Scholar] [CrossRef]
- Bianchi, F.; Giannetto, M.; Careri, M. Analytical systems and metrological traceability of measurement data in food control assessment. TrAC—Trends Anal. Chem. 2018, 107, 142–150. [Google Scholar] [CrossRef]
- Mishra, R.K.; Alonso, G.A.; Istamboulie, G.; Bhand, S.; Marty, J.L. Automated flow based biosensor for quantification of binary organophosphates mixture in milk using artificial neural network. Sens. Actuators B Chem. 2015, 208, 228–237. [Google Scholar] [CrossRef]
- Nesakumar, N.; Sethuraman, S.; Krishnan, U.M.; Rayappan, J.B.B. Cyclic voltammetric acetylcholinesterase biosensor for the detection of captan in apple samples with the aid of chemometrics. Anal. Bioanal. Chem. 2015, 407, 4863–4868. [Google Scholar] [CrossRef]
- Asadollahi-Baboli, M.; Mani-Varnosfaderani, A. Rapid and simultaneous determination of tetracycline and cefixime antibiotics by mean of gold nanoparticles-screen printed gold electrode and chemometrics tools. Meas. J. Int. Meas. Confed. 2014, 47, 145–149. [Google Scholar] [CrossRef]
- Cetó, X.; Céspedes, F.; Pividori, M.I.; Gutiérrez, J.M.; Del Valle, M. Resolution of phenolic antioxidant mixtures employing a voltammetric bio-electronic tongue. Analyst 2012, 137, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Cetó, X.; Céspedes, F.; del Valle, M. Assessment of Individual Polyphenol Content in Beer by Means of a Voltammetric BioElectronic Tongue. Electroanalysis 2013, 25, 68–76. [Google Scholar] [CrossRef]
- Cetó, X.; Apetrei, C.; Del Valle, M.; Rodríguez-Méndez, M.L. Evaluation of red wines antioxidant capacity by means of a voltammetric e-tongue with an optimized sensor array. Electrochim. Acta 2014, 120, 180–186. [Google Scholar] [CrossRef]
- Medina-Plaza, C.; García-Hernández, C.; de Saja, J.A.; Fernández-Escudero, J.A.; Barajas, E.; Medrano, G.; García-Cabezón, C.; Martin-Pedrosa, F.; Rodriguez-Mendez, M.L. The advantages of disposable screen-printed biosensors in a bioelectronic tongue for the analysis of grapes. LWT—Food Sci. Technol. 2015, 62, 940–947. [Google Scholar] [CrossRef]
- Kim, G.; Moon, J.H.; Morgan, M. Multivariate data analysis of impedimetric biosensor responses from Salmonella typhimurium. Anal. Methods 2013, 5, 4074–4080. [Google Scholar] [CrossRef]
- Espinoza, M.A.; Istamboulie, G.; Chira, A.; Noguer, T.; Stoytcheva, M.; Marty, J.L. Detection of glycoalkaloids using disposable biosensors based on genetically modified enzymes. Anal. Biochem. 2014, 457, 85–90. [Google Scholar] [CrossRef]
- Gutés, A.; Ibáñez, A.B.; Del Valle, M.; Céspedes, F. Automated SIA e-tongue employing a voltammetric biosensor array for the simultaneous determination of glucose and ascorbic acid. Electroanalysis 2006, 18, 82–88. [Google Scholar] [CrossRef]
- Ezhilan, M.; Gumpu, M.B.; Ramachandra, B.L.; Nesakumar, N.; Babu, K.J.; Krishnan, U.M.; Rayappan, J.B.B. Design and development of electrochemical biosensor for the simultaneous detection of melamine and urea in adulterated milk samples. Sens. Actuators B Chem. 2017, 238, 1283–1292. [Google Scholar] [CrossRef]
- Wong, S.F.; Low, K.H.; Khor, S.M. Differential-based biosensor array for fluorescence-chemometric discrimination and the quantification of subtle chloropropanols by cross-reactive serum albumin scaffolding. Talanta 2020, 218, 121169. [Google Scholar] [CrossRef]
- Salamanca-Neto, C.A.R.; Marcheafave, G.G.; Scremin, J.; Barbosa, E.C.M.; Camargo, P.H.C.; Dekker, R.F.H.; Scarminio, I.S.; Barbosa-Dekker, A.M.; Sartori, E.R. Chemometric-assisted construction of a biosensing device to measure chlorogenic acid content in brewed coffee beverages to discriminate quality. Food Chem. 2020, 315, 126306. [Google Scholar] [CrossRef]
- Higson, S. Biosensors for Medical Applications; Elsevier: Amsterdam, the Netherlands, 2012. [Google Scholar]
- Umali, A.P.; Anslyn, E.V. A general approach to differential sensing using synthetic molecular receptors. Curr. Opin. Chem. Biol. 2010, 14, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Olivares, D.; Kaoud, T.S.; Zeng, L.; Pridgen, J.R.; Zhuang, D.L.; Ekpo, Y.E.; Nye, J.R.; Telles, M.; Anslyn, E.V.; Dalby, K.N. Quantification of ERK Kinase Activity in Biological Samples Using Differential Sensing. ACS Chem. Biol. 2020, 15, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Zamora-Olivares, D.; Kaoud, T.S.; Jose, J.; Ellington, A.; Dalby, K.N.; Anslyn, E.V. Differential sensing of MAP kinases using SOX-peptides. Angew. Chem. Int. Ed. 2014, 53, 14064–14068. [Google Scholar] [CrossRef] [PubMed]
- Miranda, O.R.; Chen, H.T.; You, C.C.; Mortenson, D.E.; Yang, X.C.; Bunz, U.H.F.; Rotello, V.M. Enzyme-amplified array sensing of proteins in solution and in biofluids. J. Am. Chem. Soc. 2010, 132, 5285–5289. [Google Scholar] [CrossRef] [PubMed]
- De, M.; Rana, S.; Akpinar, H.; Miranda, O.R.; Arvizo, R.; Bunz, U.W.E.H.F.; Rotello, V.M. Sensing of Proteins in Human Serum using Nanoparticle-Green Fluorescent Protein Conjugates. Nat. Chem. 2010, 1, 461–465. [Google Scholar] [CrossRef]
- Motiei, L.; Pode, Z.; Koganitsky, A.; Margulies, D. Targeted protein surface sensors as a tool for analyzing small populations of proteins in biological mixtures. Angew. Chem. Int. Ed. 2014, 53, 9289–9293. [Google Scholar] [CrossRef]
- Barroso, T.G.; Martins, R.C.; Fernandes, E.; Cardoso, S.; Rivas, J.; Freitas, P.P. Detection of BCG bacteria using a magnetoresistive biosensor: A step towards a fully electronic platform for tuberculosis point-of-care detection. Biosens. Bioelectron. 2018, 100, 259–265. [Google Scholar] [CrossRef]
- Bajaj, A.; Rana, S.; Miranda, O.R.; Yawe, J.C.; Jerry, D.J.; Bunz, U.H.F.; Rotello, V.M. Cell surface-based differentiation of cell types and cancer states using a gold nanoparticle-GFP based sensing array. Chem. Sci. 2010, 1, 134–138. [Google Scholar] [CrossRef]
- Tomita, S.; Sakao, M.; Kurita, R.; Niwa, O.; Yoshimoto, K. A polyion complex sensor array for markerless and noninvasive identification of differentiated mesenchymal stem cells from human adipose tissue. Chem. Sci. 2015, 6, 5831–5836. [Google Scholar] [CrossRef]
- Muthu, P.; Lutz, S. Quantitative Detection of Nucleoside Analogues by Multi-enzyme Biosensors using Time-Resolved Kinetic Measurements. ChemMedChem 2016, 11, 660–666. [Google Scholar] [CrossRef]
- Diehl, K.L.; Ivy, M.A.; Rabidoux, S.; Petry, S.M.; Müller, G.; Anslyn, E.V. Differential sensing for the regio- and stereoselective identification and quantitation of glycerides. Proc. Natl. Acad. Sci. USA 2015, 112, E3977–E3986. [Google Scholar] [CrossRef] [PubMed]
- Mazafi, A.; Shukla, R.P.; Shukla, S.K.; Lavon, A.; Ben-Yoav, H.; Silue, A.; McDowell, B.; Peixoto, N. Intelligent Multi-Electrode Arrays as the Next Generation of Electrochemical Biosensors for Real-Time Analysis of Neurotransmitters. In Proceedings of the MeMeA 2018—2018 IEEE International Symposium on Medical Measurements and Applications, Rome, Italy, 11–13 June 2018; pp. 1–5. [Google Scholar]
- Liu, R.; Chen, W.; Xu, K. The influence of experiment design on the model precision in the noninvasive glucose sensing by near-infrared spectroscopy. Opt. Health Care Biomed. Opt. III 2007, 6826, 682626. [Google Scholar]
- Ashley, J.; Shahbazi, M.A.; Kant, K.; Chidambara, V.A.; Wolff, A.; Bang, D.D.; Sun, Y. Molecularly imprinted polymers for sample preparation and biosensing in food analysis: Progress and perspectives. Biosens. Bioelectron. 2017, 91, 606–615. [Google Scholar] [CrossRef] [PubMed]
- Curk, T.; Dobnikar, J.; Frenkel, D. Rational design of molecularly imprinted polymers. Soft Matter 2015, 12, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Muzyka, K.; Karim, K.; Guerreiro, A.; Poma, A.; Piletsky, S. Optimisation of the synthesis of vancomycinselective molecularly imprinted polymer nanoparticles using automatic photoreactor. Nanoscale Res. Lett. 2014, 9, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Mijangos, I.; Navarro-Villoslada, F.; Guerreiro, A.; Piletska, E.; Chianella, I.; Karim, K.; Turner, A.; Piletsky, S. Influence of initiator and different polymerisation conditions on performance of molecularly imprinted polymers. Biosens. Bioelectron. 2006, 22, 381–387. [Google Scholar] [CrossRef]
- Kempe, H.; Kempe, M. Novel Method for the Synthesis of Molecularly Imprinted Polymer Bead Libraries. Macromol. Rapid Commun. 2004, 25, 315–320. [Google Scholar] [CrossRef]
- Kryscio, D.R.; Shi, Y.; Ren, P.; Peppas, N.A. Molecular docking simulations for macromolecularly imprinted polymers. Ind. Eng. Chem. Res. 2011, 50, 13877–13884. [Google Scholar] [CrossRef]
- Khan, M.S.; Wate, P.S.; Krupadam, R.J. Combinatorial screening of polymer precursors for preparation of benzo[α] pyrene imprinted polymer: An ab initio computational approach. J. Mol. Model. 2012, 18, 1969–1981. [Google Scholar] [CrossRef]
- Nicholls, I.A.; Chavan, S.; Golker, K.; Karlsson, B.C.G.; Olsson, G.D.; Rosengren, A.M.; Suriyanarayanan, S.; Wiklander, J.G. Theoretical and computational strategies for the study of the molecular imprinting process and polymer performance. Adv. Biochem. Eng. Biotechnol. 2015, 150, 25–50. [Google Scholar]
- Choulier, L.; Andersson, K.; Hämäläinen, M.D.; Van Regenmortel, M.H.V.; Malmqvist, M.; Altschuh, D. QSAR studies applied to the prediction of antigen-antibody interaction kinetics as measured by BIACORE. Protein Eng. 2002, 15, 373–382. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, M.; Ni, N.; Wang, B.; Zhang, Y. Modeling the excitation wavelengths (Λex) of boronic acids. J. Mol. Model. 2008, 14, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Martynko, E.; Solov’ev, V.; Varnek, A.; Legin, A.; Kirsanov, D. QSPR Modeling of Potentiometric Mg2+/Ca2+ Selectivity for PVC-plasticized Sensor Membranes. Electroanalysis 2020, 32, 792–798. [Google Scholar] [CrossRef]
- Soloviev, V.; Varnek, A.; Babain, V.; Polukeev, V.; Ashina, J.; Legin, E.; Legin, A.; Kirsanov, D. QSPR modeling of potentiometric sensitivity towards heavy metal ions for polymeric membrane sensors. Sens. Actuators, B Chem. 2019, 301, 126941. [Google Scholar] [CrossRef]
- Panchuk, V.; Lvova, L.; Kirsanov, D.; Gonçalves, C.G.; Di Natale, C.; Paolesse, R.; Legin, A. Extending electronic tongue calibration lifetime through mathematical drift correction: Case study of microcystin toxicity analysis in waters. Sens. Actuators B Chem. 2016, 237, 962–968. [Google Scholar] [CrossRef]
- Rudnitskaya, A. Calibration update and drift correction for electronic noses and tongues. Front. Chem. 2018, 6, 433. [Google Scholar] [CrossRef]
- Olivieri, A.C. Analytical figures of merit: From univariate to multiway calibration. Chem. Rev. 2014, 114, 5358–5378. [Google Scholar] [CrossRef]
- Parastar, H.; Kirsanov, D. Analytical Figures of Merit for Multisensor Arrays. ACS Sens. 2020, 5, 580–587. [Google Scholar] [CrossRef]

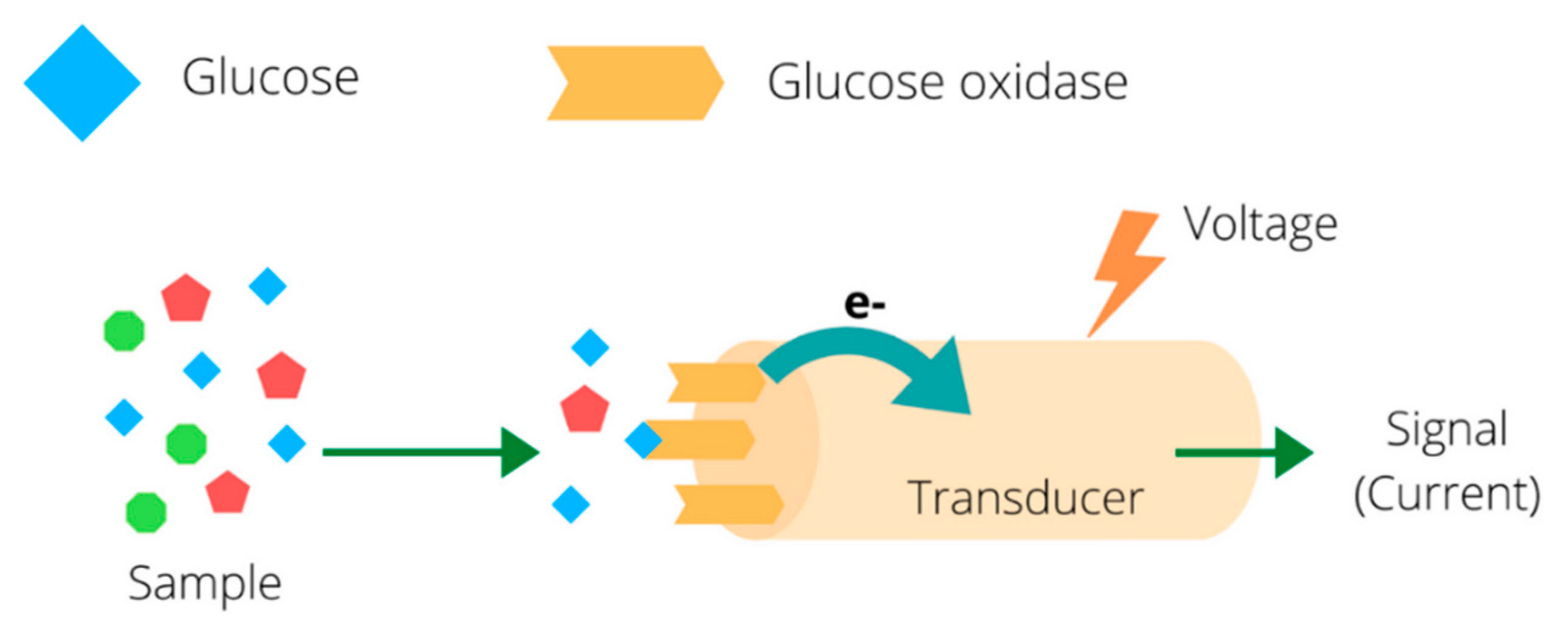

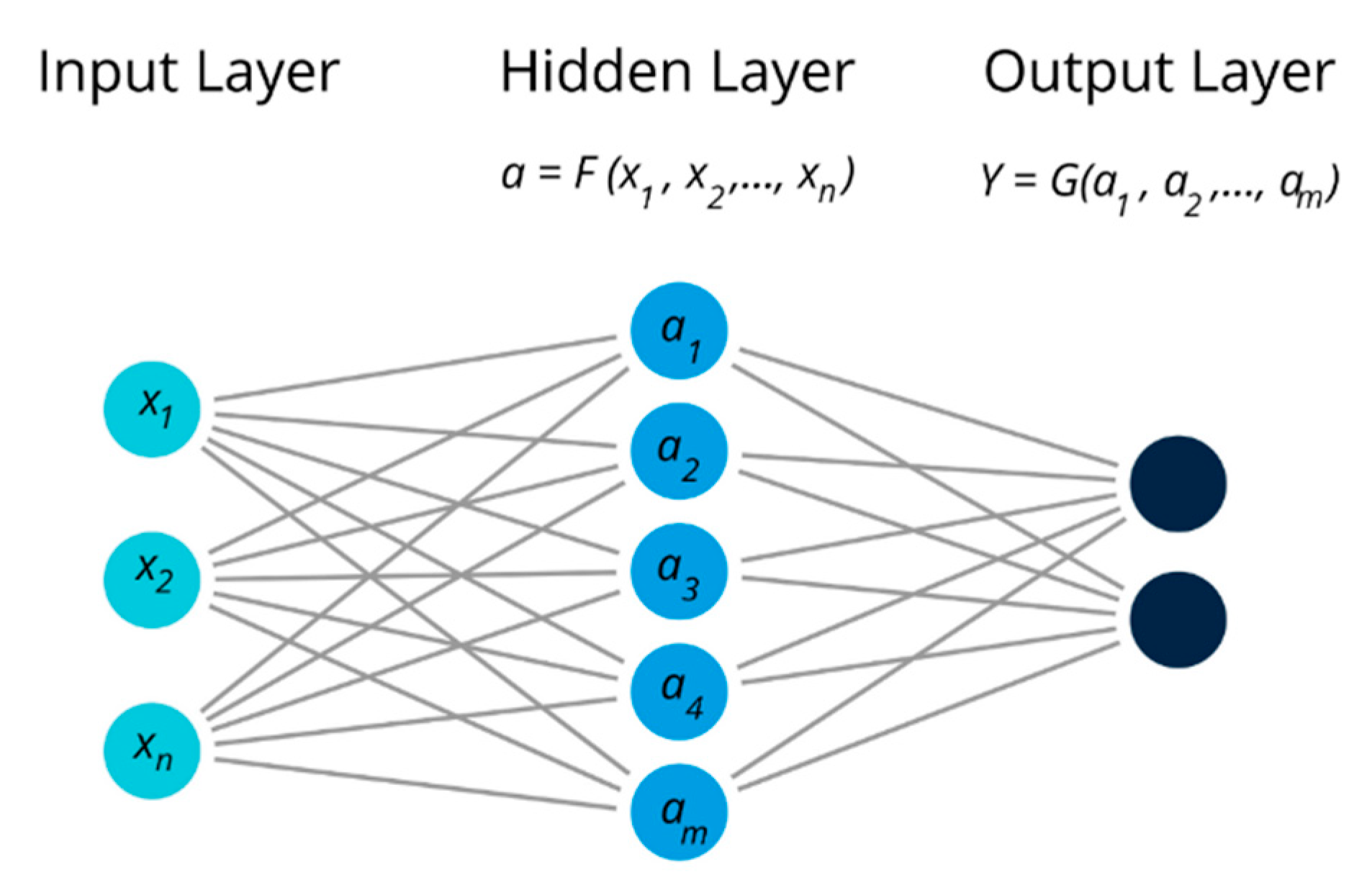
| Analytes | Transduction Principle | Data Analysis | Bioreceptor Type | Description | # of Sensors (Channels) | Reference |
|---|---|---|---|---|---|---|
| Pesticides detection | ||||||
| Paraoxon and carbofuran | Amperometry | ANNs | Enzyme | AChE inhibition | 4 | [18] |
| Dichlorvos and carbofuran | 3 | [14] | ||||
| Dichlorvos and methylparaoxon | AChE inhibition, FIA system | 3 | [19] | |||
| Chlorpyriphos oxon, chlorfenvinphos and azinphos-methyl oxon | AChE inhibition | 2 | [20] | |||
| Carbaryl, phoxim | Spectrophotometry | AChE inhibition, subsequent reaction of thiocholine with 5,5-dithiobis(2-nitrobenzoic) acid | 1 | [21] | ||
| Dichlorvos, malaoxon, chlorpyrifos-oxon, chlorpyrifos-methyl-oxon, chlorfenvinphos, pirimiphos-methyl-oxon | Chrono-amperometry | AChE inhibition in an automated system | 6 | [22] | ||
| Classification of pesticides residues into three groups (carbamates, pyrethroids, organophosphates) | Potentiometry | Cells | Cellular sensors based on bioelectric recognition assay | 1 | [23] | |
| Phenolic compounds | ||||||
| Catechol and 4-chlorophenol | Amperometry | PLS | Enzyme | A tyrosinase-based sensor in a FIA system | 1 | [24] |
| Phenol, catechol, m-cresol | Linear sweep voltammetry | ANNs | Polyphenol oxidase-based sensor in a SIA system | 1 | [15] | |
| Binary mixtures: phenol/chlorophenol, cathecol/phenol, cresol/chlorocresol, phenol/cresol | PLS | Tyrosinase- and laccase-based sensors | 2 | [25] | ||
| Catechol, m-cresol, guaiacol (in artificial wastewater) | Cyclic voltammetry | FFT *, ANNs | Sensors based on Tyr, Lac, and Cu NPs | 4 | [26] | |
| Metal ions | ||||||
| Cu2+, Cd2+, Pb2+ | Square wave voltammetry | nPLS ** | Peptide | Single Au electrode modified with three different peptides | 1 | [27] |
| Cd2+, Pb2+, Zn2+ | Differential pulse adsorptive stripping voltammetry | FFT, ANNs | Peptide | Array of peptide-modified electrodes | 3 | [28] |
| Fe2+ | Spectrophotometry | PARAFAC x | Nanosheet MoS2 | Fe2+/MoS2 oxidation catalysis to form highly fluorescent compound (DAPN) | 1 | [29] |
| K+, Tl+ | Spectrophotometry | PLS | DNA and NPs | ssDNA-AuNPs catalysis of the oxidation of TMB with H2O2 to generate fluorescent compounds | 8 | [30] |
| Water quality metrics | ||||||
| General wastewater quality | Amperometry | PCA | Enzyme | An array of enzymes immobilized onto C and Pt working electrodes | 8 | [9] |
| Organic pollutant indexes in wastewater | PCA, PLS | Enzyme | Immobilized enzymes onto working electrodes | 16 | [31] | |
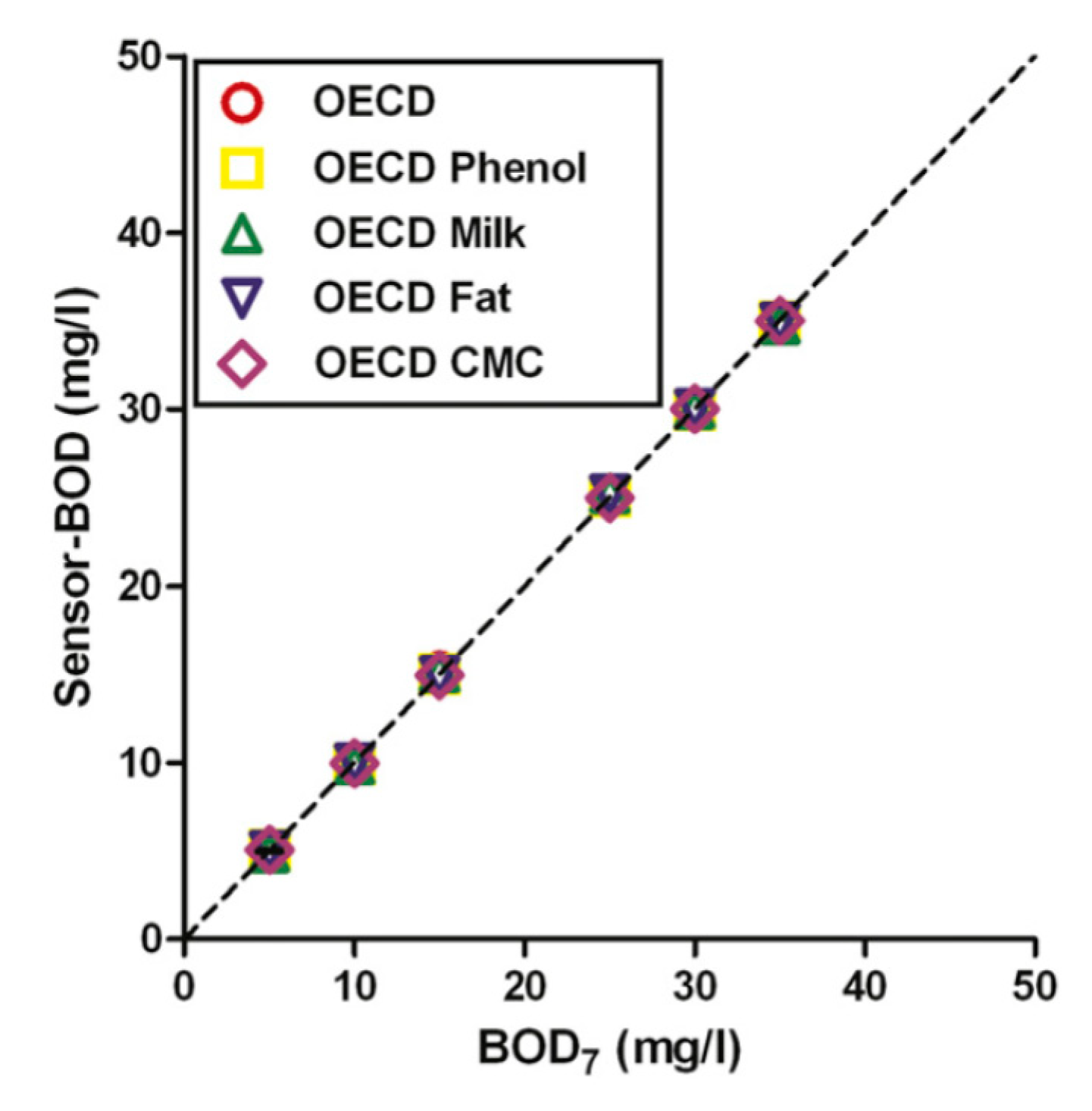
| Biochemical oxygen demand (BOD) in wastewater | PLS | Microorganism | Microorganisms immobilized on the surface of a Clark-type electrode | 7 | [11] | |
| Chemical oxygen demand (COD) | Generated current | ANNs | Microorganism | Microbial fuel cells-based sensors | 1 | [32] |
| Analytes | Sample | Transduction Principle | Data Analysis | Bioreceptor Type | Description | # of Sensors (Channels) | Reference |
|---|---|---|---|---|---|---|---|
| Pesticides: chlorpyriphos-oxon and malaoxon | Milk | Amperometry | ANNs | Enzyme | AChE inhibition in an FIA system | 2 | [40] |
| Insecticides: captan | Apples | Cyclic voltammetry | PCA, regression analysis | AChE inhibition | 1 | [41] | |
| Antibiotics: tetracycline and cefixime | Milk | Square wave voltammetry | PCA, ANNs | Amino acid monolayer | Screen-printed Au electrode modified with Au NPs and a self-assembled monolayer of cysteine | 1 | [42] |
| Phenolic compounds | |||||||
| Catechol, caffeic acid, catechin | Wine | Cyclic voltammetry | ANNs | Enzyme | Tyrosinase and laccase biosensors combined with Cu NPs | 4 | [43] |
| Ferulic, gallic, sinapic acids | Beer | ANNs, PCA | [44] | ||||
| Total phenolic content | Wine | ANNs, DWT * + PLS | Tyrosinase and laccase sensors with electron mediators | 12 | [45] | ||
| Total phenolic content and discrimination of grape varieties | PCA | Tyrosinase and GOx biosensors with electron mediators | 6 | [46] | |||
| Bacteria: Salmonella typhimurium | Pork | Electrochemical impedance spectroscopy | PLS | Antibody | Salmonella antibodies immobilized on the surface of a gold microelectrode | 1 | [47] |
| Glycoalkaloids: α-solanine and α-chaconine | Potato | Amperometry | ANNs | Enzyme | AChE inhibition | 2 | [48] |
| Glucose and ascorbic acid | Fruit juice | Linear sweep voltammetry | ANNs | Enzyme | GOx biosensors with metal catalysts in an SIA system | 3 | [49] |
| Melamine and urea | Milk | Amperometry | LR ** | AChE inhibition on the Pt/ZnO/Chitosan bioelectrode | 1 | [50] | |
| Chloropropanols | Soy sauce | Spectrophotometry | LDA, PLS | Protein | Differential optical sensing with serum albumins coupled with a fluorophore | 3 | [51] |
| Chlorogenic acid | Coffee | Square wave voltammetry | PCA | Fungus | The measurement of laccase production by funghi in different conditions | 1 | [52] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martynko, E.; Kirsanov, D. Application of Chemometrics in Biosensing: A Brief Review. Biosensors 2020, 10, 100. https://doi.org/10.3390/bios10080100
Martynko E, Kirsanov D. Application of Chemometrics in Biosensing: A Brief Review. Biosensors. 2020; 10(8):100. https://doi.org/10.3390/bios10080100
Chicago/Turabian StyleMartynko, Ekaterina, and Dmitry Kirsanov. 2020. "Application of Chemometrics in Biosensing: A Brief Review" Biosensors 10, no. 8: 100. https://doi.org/10.3390/bios10080100
APA StyleMartynko, E., & Kirsanov, D. (2020). Application of Chemometrics in Biosensing: A Brief Review. Biosensors, 10(8), 100. https://doi.org/10.3390/bios10080100





