Electrochemical DNA Sensor for Sensitive BRCA1 Detection Based on DNA Tetrahedral-Structured Probe and Poly-Adenine Mediated Gold Nanoparticles
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Instruments
2.2. Synthesis of Tetrahedral-Structured Probes (TSPs)
2.3. Synthesis the Nanocomposites of Gold Nanoparticles (AuNPs)-DNA-r
2.4. Development of Electrochemical DNA Sensor
3. Results and Discussion
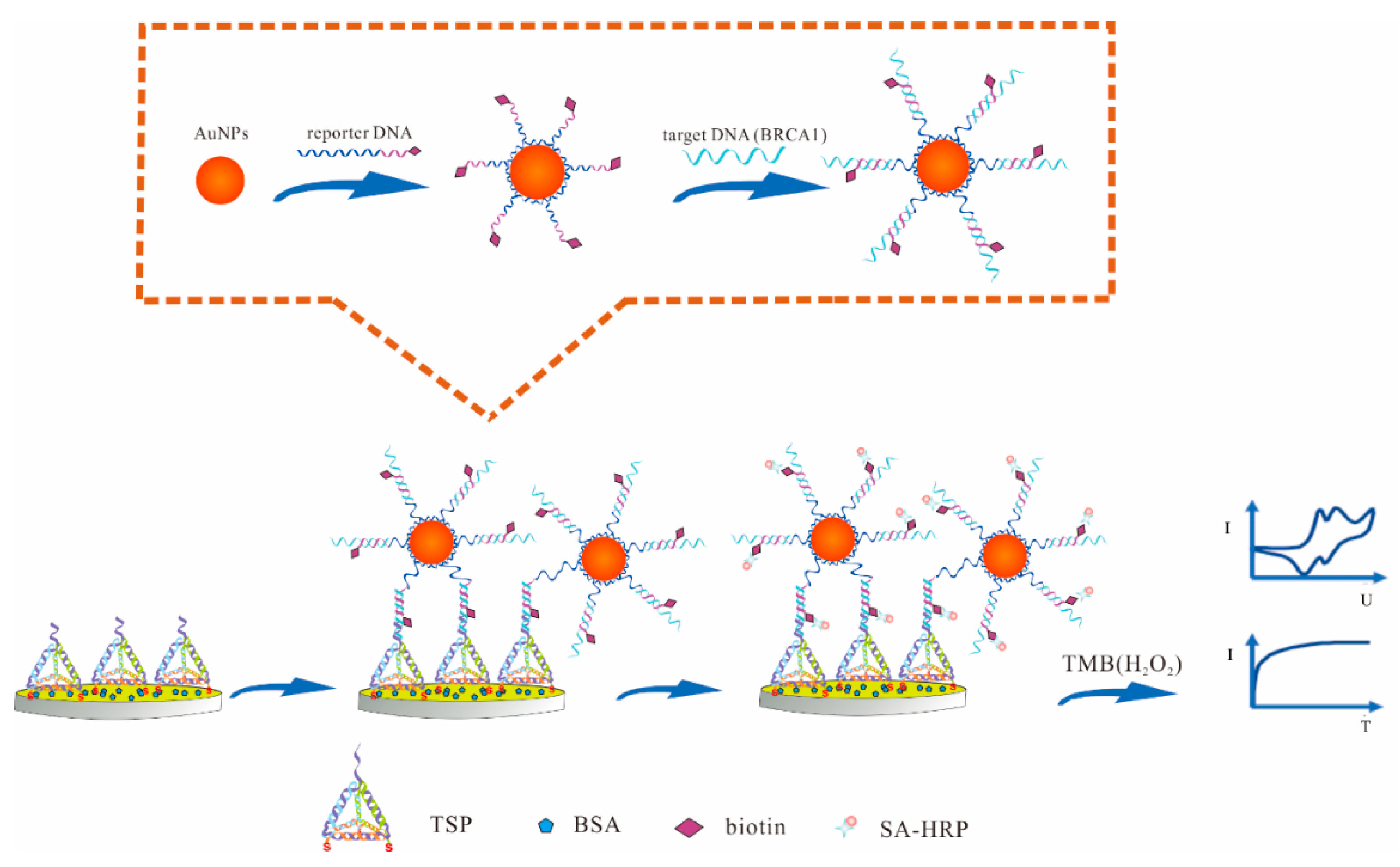
3.1. Principle of the Electrochemical DNA Sensor
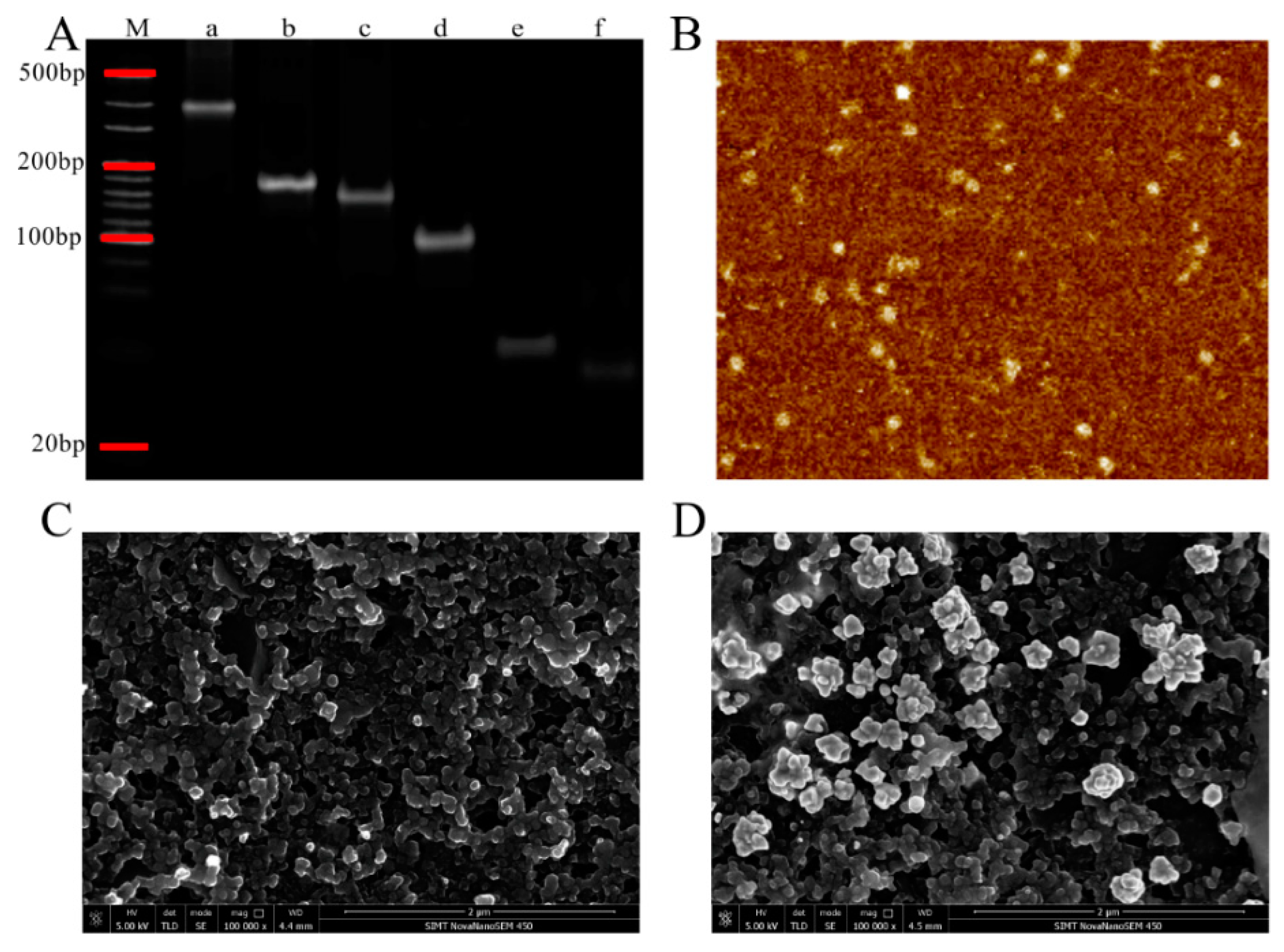
3.2. Characterization of TSPs and AuNPs Electrode
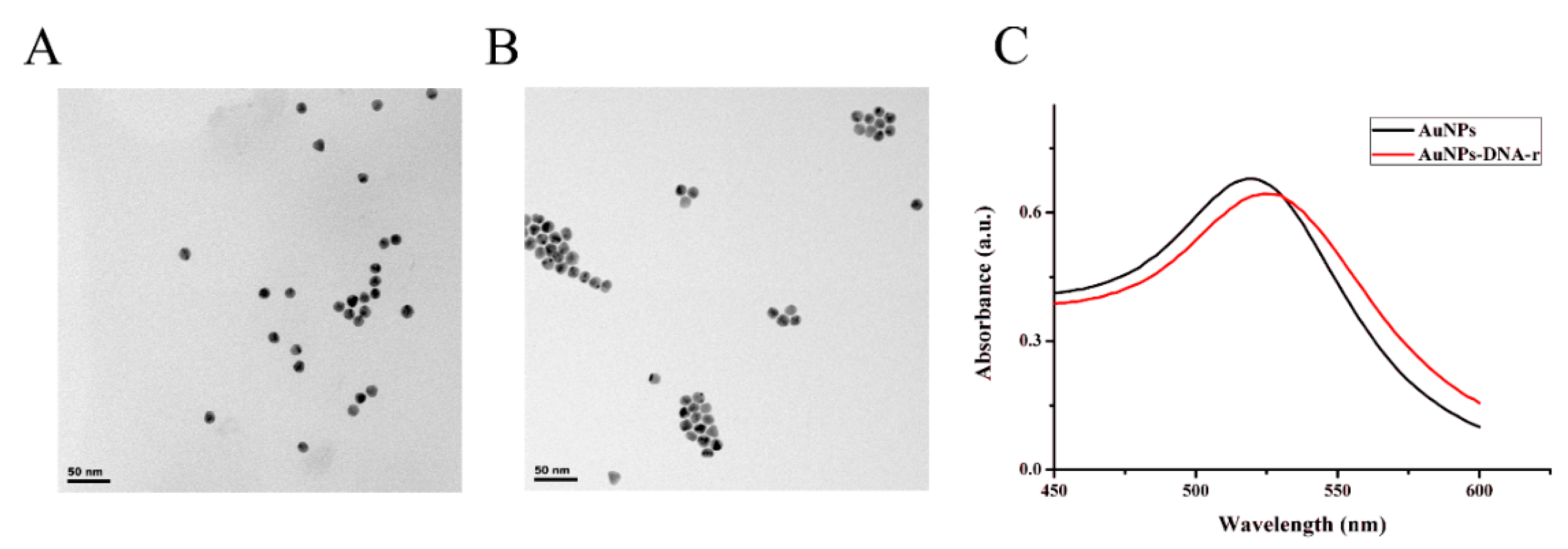
3.3. Characterization of the AuNPs-DNA-r
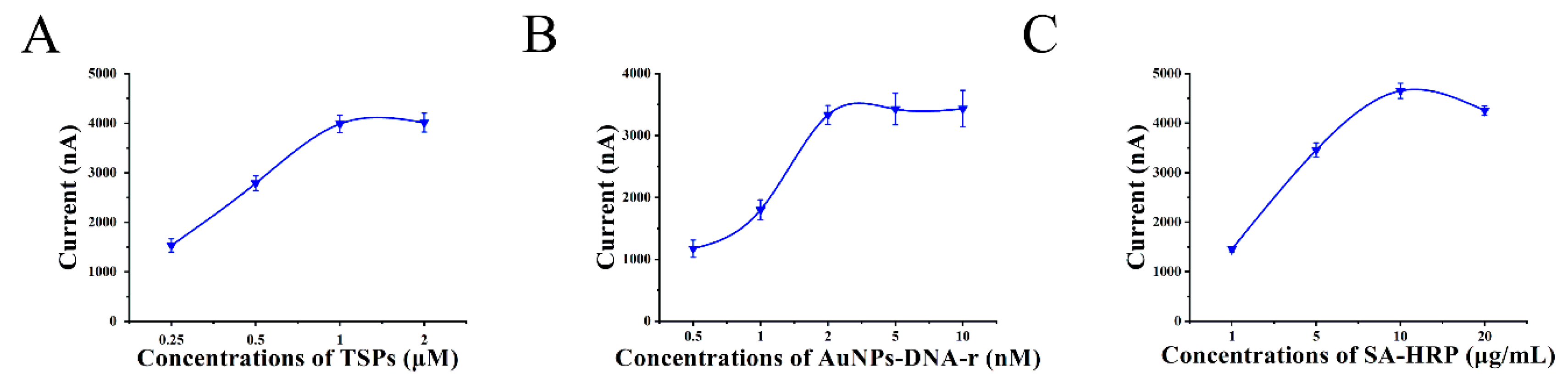
3.4. Optimization of the Experimental Conditions
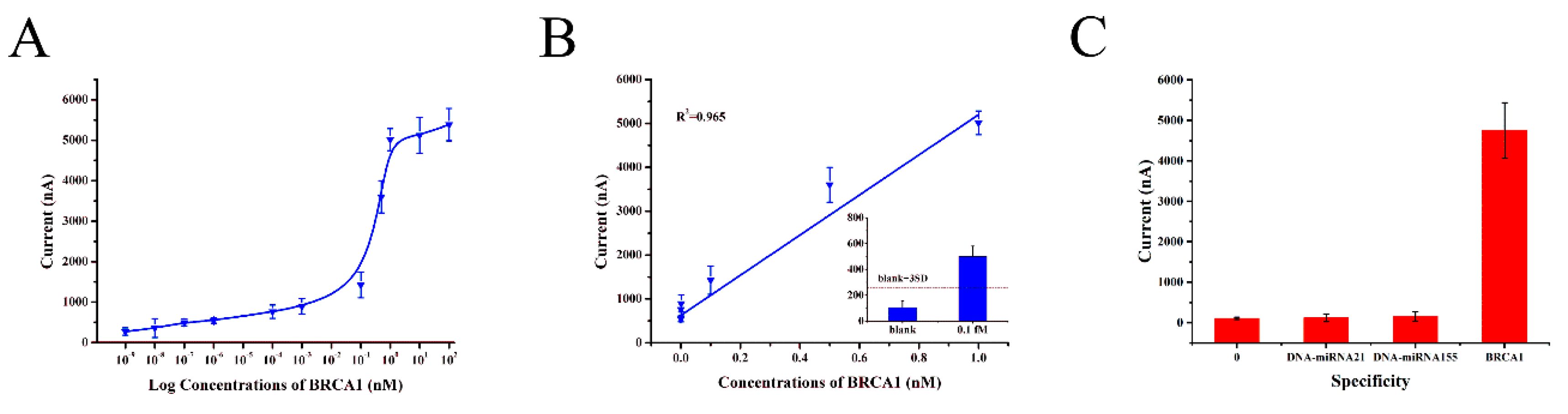
3.5. Performance of the Electrochemical DNA Sensor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Tutt, A.; Ashworth, A. The relationship between the roles of BRCA genes in DNA repair and cancer predisposition. Trends Mol. Med. 2002, 8, 571–576. [Google Scholar] [CrossRef]
- Miki, Y.; Swensen, J.; Shattuck-Eidens, D.; Futreal, P.A.; Harshman, K.; Tavtigian, S.; Liu, Q.; Cochran, C.; Bennett, L.M.; Ding, W.; et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 1994, 266, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Ford, D.; Easton, D.F.; Bishop, D.T.; Narod, S.A.; Goldgar, D.E. Risks of cancer in BRCA1-mutation carriers. Lancet 1994, 343, 692–695. [Google Scholar] [CrossRef]
- Han, S.H.; Lee, K.R.; Lee, D.G.; Kim, B.Y.; Lee, K.E.; Chung, W.S. Mutation analysis of BRCA1 and BRCA2 from 793 Korean patients with sporadic breast cancer. Clin. Genet. 2006, 70, 496–501. [Google Scholar] [CrossRef]
- Li, N.; Zhang, X.; Cai, Y.; Xu, X.; Zhang, L.; Pan, K.F.; Wu, L.Y.; Wang, M.R. BRCA1 germline mutations in Chinese patients with hereditary breast and ovarian cancer. Int. J. Gynecol. Cancer 2006, 16, 172–178. [Google Scholar] [CrossRef]
- Friedman, L.S.; Ostermeyer, E.A.; Szabo, C.I.; Dowd, P.; Lynch, E.D.; Rowell, S.E.; King, M.C. Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat. Genet. 1994, 8, 399–404. [Google Scholar] [CrossRef]
- Rashid, M.U.; Zaidi, A.; Torres, D.; Sultan, F.; Benner, A.; Naqvi, B.; Shakoori, A.R.; Seidel-Renkert, A.; Farooq, H.; Narod, S.; et al. Prevalence of BRCA1 and BRCA2 mutations in Pakistani breast and ovarian cancer patients. Int. J. Gynecol. Cancer 2006, 119, 2832–2839. [Google Scholar] [CrossRef]
- Chen, L.; Zeng, G.; Zhang, Y.; Tang, L.; Huang, D.; Liu, C.; Pang, Y.; Luo, J. Trace detection of picloram using an electrochemical immunosensor based on three-dimensional gold nanoclusters. Anal. Biochem. 2010, 407, 172–179. [Google Scholar] [CrossRef]
- Huang, Y.; Tao, M.; Luo, S.; Zhang, Y.; Situ, B.; Ye, X.; Chen, P.; Jiang, X.; Wang, Q.; Zheng, L. A novel nest hybridization chain reaction based electrochemical assay for sensitive detection of circulating tumor DNA. Anal. Chim. Acta 2020, 1107, 40–47. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, W.; Lin, Z.; Liu, B.; Zhou, X. Dual-output toehold-mediated strand displacement amplification for sensitive homogeneous electrochemical detection of specie-specific DNA sequences for species identification. Biosens. Bioelectron. 2020, 161, 112256. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-C.; Wang, E.; Tai, C.-S.; Chiu, Y.-C.; Li, C.-W.; Lin, Y.-R.; Lee, T.-H.; Huang, C.-W.; Chen, J.-C.; Chen, W.L. Improving the reproducibility, accuracy, and stability of an electrochemical biosensor platform for point-of-care use. Biosens. Bioelectron. 2020, 155, 112111. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Plaxco, K.W.; Heeger, A.J. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc. Natl. Acad. Sci. USA 2003, 100, 9134–9137. [Google Scholar] [CrossRef]
- Diculescu, V.C.; Chiorcea-Paquim, A.-M.; Oliveira-Brett, A.M. Applications of a DNA-electrochemical biosensor. TrAC Trends Anal. Chem. 2016, 79, 23–36. [Google Scholar] [CrossRef]
- Goodman, R.P.; Berry, R.M.; Turberfield, A.J. The single-step synthesis of a DNA tetrahedron. Chem. Commun. 2004, 12, 1372–1373. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.P.; Schaap, I.A.T.; Tardin, C.F.; Erben, C.M.; Berry, R.M.; Schmidt, C.F.; Turberfield, A.J. Rapid Chiral Assembly of Rigid DNA Building Blocks for Molecular Nanofabrication. Science 2005, 310, 1661–1665. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.; Lu, N.; Wen, Y.; Song, S.; Liu, Y.; Yan, H.; Fan, C. A DNA Nanostructure-based Biomolecular Probe Carrier Platform for Electrochemical Biosensing. Adv. Mater. 2010, 22, 4754–4758. [Google Scholar] [CrossRef]
- Pei, H.; Wan, Y.; Li, J.; Hu, H.; Su, Y.; Huang, Q.; Fan, C. Regenerable electrochemical immunological sensing at DNA nanostructure-decorated gold surfaces. Chem. Commun. 2011, 47, 6254–6256. [Google Scholar] [CrossRef]
- Wen, Y.; Pei, H.; Shen, Y.; Xi, J.; Lin, M.; Lu, N.; Shen, X.; Li, J.; Fan, C. DNA Nanostructure-based Interfacial engineering for PCR-free ultrasensitive electrochemical analysis of microRNA. Sci. Rep. 2012, 2, 867. [Google Scholar] [CrossRef]
- Zeng, D.; Zhang, H.; Zhu, D.; Li, J.; San, L.; Wang, Z.; Wang, C.; Wang, Y.; Wang, L.; Zuo, X.; et al. A novel ultrasensitive electrochemical DNA sensor based on double tetrahedral nanostructures. Biosens. Bioelectron. 2015, 71, 434–438. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C. DNA in a material world. Nature 2003, 421, 427–431. [Google Scholar] [CrossRef]
- Jayakumar, K.; Rajesh, R.; Dharuman, V.; Venkatasan, R.; Hahn, J.H.; Karutha Pandian, S. Gold nano particle decorated graphene core first generation PAMAM dendrimer for label free electrochemical DNA hybridization sensing. Biosens. Bioelectron. 2012, 31, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Senel, M.; Dervisevic, M.; Kokkokoğlu, F. Electrochemical DNA biosensors for label-free breast cancer gene marker detection. Anal. Bioanal. Chem. 2019, 411, 2925–2935. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zhang, S.; Fang, X.; Kong, J. Double signal amplification strategy for ultrasensitive electrochemical biosensor based on nuclease and quantum dot-DNA nanocomposites in the detection of breast cancer 1 gene mutation. Biosens. Bioelectron. 2019, 142, 111544. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, P.A.; Sandhyarani, N. Graphene-DNA electrochemical sensor for the sensitive detection of BRCA1 gene. Sens. Actuators B Chem. 2014, 204, 777–782. [Google Scholar] [CrossRef]
- Abdul Rasheed, P.; Sandhyarani, N. Attomolar detection of BRCA1 gene based on gold nanoparticle assisted signal amplification. Biosens. Bioelectron. 2015, 65, 333–340. [Google Scholar] [CrossRef]
- Pei, H.; Zuo, X.; Zhu, D.; Huang, Q.; Fan, C. Functional DNA nanostructures for theranostic applications. Acc. Chem. Res. 2014, 47, 550–559. [Google Scholar] [CrossRef]
- Schreiner, S.M.; Shudy, D.F.; Hatch, A.L.; Opdahl, A.; Whitman, L.J.; Petrovykh, D.Y. Controlled and efficient hybridization achieved with DNA probes immobilized solely through preferential DNA-substrate interactions. Anal. Chem. 2010, 82, 2803–2810. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, H.; Zeng, D.; Sun, W.; Zhang, H.; Aldalbahi, A.; Wang, Y.; San, L.; Fan, C.; Zuo, X.; et al. Elaborately designed diblock nanoprobes for simultaneous multicolor detection of microRNAs. Nanoscale 2015, 7, 15822–15829. [Google Scholar] [CrossRef]
- Liu, Z.; Lei, S.; Zou, L.; Li, G.; Xu, L.; Ye, B. Highly ordered 3D electrochemical DNA biosensor based on dual orientation controlled rolling motor and graftable tetrahedron DNA. Biosens. Bioelectron. 2020, 147, 111759. [Google Scholar] [CrossRef]
- Wen, Y.; Pei, H.; Wan, Y.; Su, Y.; Huang, Q.; Song, S.; Fan, C. DNA Nanostructure-Decorated Surfaces for Enhanced Aptamer-Target Binding and Electrochemical Cocaine Sensors. Anal. Chem. 2011, 83, 7418–7423. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhao, B.; Wang, D.; Wen, Y.; Liu, G.; Dong, H.; Song, S.; Fan, C. DNA Nanostructure-Based Universal Microarray Platform for High-Efficiency Multiplex Bioanalysis in Biofluids. ACS Appl. Mater. Interfaces 2014, 6, 17944–17953. [Google Scholar] [CrossRef] [PubMed]
- Hui, N.; Sun, X.; Niu, S.; Luo, X. PEGylated Polyaniline Nanofibers: Antifouling and Conducting Biomaterial for Electrochemical DNA Sensing. ACS Appl. Mater. Interfaces 2017, 9, 2914–2923. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, L.; Ye, H.; Yu, L.; Zhu, X.; Lin, Z.; Wu, G.; Li, X.; Liu, X.; Chen, G. An ultrasensitive electrochemical impedance sensor for a special BRCA1 breast cancer gene sequence based on lambda exonuclease assisted target recycling amplification. Chem. Commun. 2012, 48, 6390–6392. [Google Scholar] [CrossRef]
- Rasheed, P.A.; Sandhyarani, N. Femtomolar level detection of BRCA1 gene using a gold nanoparticle labeled sandwich type DNA sensor. Colloids Surf. B Biointerfaces 2014, 117, 7–13. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, D.; Su, J.; He, G.; Xu, Y.; Wang, C.; Zheng, M.; Qian, Q.; Mi, X. Electrochemical DNA Sensor for Sensitive BRCA1 Detection Based on DNA Tetrahedral-Structured Probe and Poly-Adenine Mediated Gold Nanoparticles. Biosensors 2020, 10, 78. https://doi.org/10.3390/bios10070078
Feng D, Su J, He G, Xu Y, Wang C, Zheng M, Qian Q, Mi X. Electrochemical DNA Sensor for Sensitive BRCA1 Detection Based on DNA Tetrahedral-Structured Probe and Poly-Adenine Mediated Gold Nanoparticles. Biosensors. 2020; 10(7):78. https://doi.org/10.3390/bios10070078
Chicago/Turabian StyleFeng, Dezhi, Jing Su, Guifang He, Yi Xu, Chenguang Wang, Mengmeng Zheng, Qiuling Qian, and Xianqiang Mi. 2020. "Electrochemical DNA Sensor for Sensitive BRCA1 Detection Based on DNA Tetrahedral-Structured Probe and Poly-Adenine Mediated Gold Nanoparticles" Biosensors 10, no. 7: 78. https://doi.org/10.3390/bios10070078
APA StyleFeng, D., Su, J., He, G., Xu, Y., Wang, C., Zheng, M., Qian, Q., & Mi, X. (2020). Electrochemical DNA Sensor for Sensitive BRCA1 Detection Based on DNA Tetrahedral-Structured Probe and Poly-Adenine Mediated Gold Nanoparticles. Biosensors, 10(7), 78. https://doi.org/10.3390/bios10070078





