Newly Developed System for Acetamiprid Residue Screening in the Lettuce Samples Based on a Bioelectric Cell Biosensor
Abstract
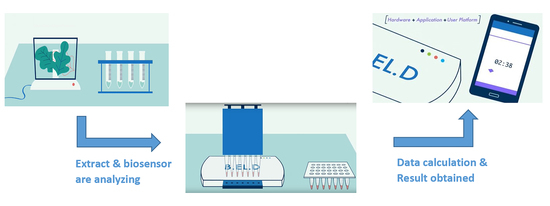
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture and Sensor Fabrication from Vero Cells
2.3. Sample Preparation
2.4. Spiking Lettuce Extract Samples
2.5. Assay Procedure
2.5.1. Biosensor Device
2.5.2. The Algorithm for Data Acquisition and Signal Processing
2.5.3. The Software Interface
2.6. Conventional Sample Analysis
3. Results and Discussion
3.1. Cells with or without Antibody Response to the Presence of Acetamiprid
3.2. Biosensor Response to Spiking Lettuce Extract Samples
3.3. Database creation
3.4. Biosensor Response and Conventional Analysis of Lettuce Samples Provided from Market
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Stevens, T.; Kilmer, R. Pesticide Residues on Fresh Tomatoes and Strawberries. IFAS Ext. 1999, 331, 1–22. [Google Scholar]
- Mestres, C.; Colonna, P.; Buleon, A. Characteristics of starch networks within rice flour noodles and mungbean starch vermicelli. J. Food Sci. 1988, 53, 1809–1812. [Google Scholar] [CrossRef]
- US Environmental Protection Agency. Code of Federal Regulations (CFR); US Environmental Protection Agency: Washington, DC, USA, 2017; Part 180; p. 40.
- Marın, A.; Vidal, J.M.N.; Gonzalez, F.E.; Frenich, A.G.; Glass, C.; Sykes, M. Assessment of potential (inhalation and dermal) and actual exposure to acetamiprid by greenhouse applicators using liquid chromatography–tandem mass spectrometry. J. Chromatogr. B 2004, 804, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Xu, Q.; Yu, L.; Mao, A.; Hu, X. A novel sensor for the detection of acetamiprid in vegetables based on its photocatalytic degradation compound. Food Chem. 2016, 194, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.-H.; Min, H.; Lü, Z.-H.; Yuan, H.-P. Influence of acetamiprid on soil enzymatic activities and respiration. Eur. J. Soil Biol. 2006, 42, 120–126. [Google Scholar] [CrossRef]
- Kocaman, A.; Topaktas, M. Genotoxic effects of a particular mixture of acetamiprid and alpha-cypermethrin on chromosome aberration, sister chromatid exchange, and micronucleus formation in human peripheral blood lymphocytes. Environ. Toxicol. 2010, 25, 157–168. [Google Scholar] [PubMed]
- Kaur, R.P.; Gupta, V.; Christopher, A.F.; Bansal, P. Potential pathways of pesticide action on erectile function—A contributory factor in male infertility. Asia Pac. J. Reprod. 2015, 4, 322–330. [Google Scholar] [CrossRef]
- Zhang, X.; Mobley, N.; Zhang, J.; Zheng, X.; Lu, L.; Ragin, O.; Smith, C.J. Analysis of agricultural residues on tea using d-SPE sample preparation with GC-NCI-MS and UHPLC-MS/MS. J. Agric. Food Chem. 2010, 58, 11553–11560. [Google Scholar] [CrossRef]
- Zhang, B.; Pan, X.; Venne, L.; Dunnum, S.; Mcmurry, S.T.; Cobb, G.P.; Anderson, T.A. Development of a method for the determination of 9 currently used cotton pesticides by gas chromatography with electron capture detection. Talanta 2008, 75, 1055–1060. [Google Scholar] [CrossRef]
- Watanabe, E.; Miyake, S.; Baba, K.; Eun, H.; Endo, S. Immunoassay for acetamiprid detection: Application to residue analysis and comparison with liquid chromatography. Anal. Bioanal. Chem. 2006, 386, 1441–1448. [Google Scholar] [CrossRef]
- Mateu-Sanchez, M.; Moreno, M.; Arrebola, F.J.; Vidal, J.L.M. Analysis of acetamiprid in vegetables using gas chromatography-tandem mass spectrometry. Anal. Sci. 2003, 19, 701–704. [Google Scholar] [CrossRef] [PubMed]
- Wanatabe, S.; Ito, S.; Kamata, Y.; Omoda, N.; Yamazaki, T.; Munakata, H.; Kaneko, T.; Yuasa, Y. Development of competitive enzyme-linked immunosorbent assays (ELISAs) based on monoclonal antibodies for chloronicotinoid insecticides imidacloprid and acetamiprid. Anal. Chim. Acta 2001, 427, 211–219. [Google Scholar] [CrossRef]
- Hassani, S.; Momtaz, S.; Vakhshiteh, F.; Maghsoudi, A.S.; Ganjali, M.R.; Norouzi, P.; Abdollahi, M. Biosensors and their applications in detection of organophosphorus pesticides in the environment. Arch. Toxicol. 2017, 91, 109–130. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mozaz, S.; Lopez de Alda, M.J.; Barceló, D. Biosensors as useful tools for environmental analysis and monitoring. Anal. Bioanal. Chem. 2006, 386, 1025–1041. [Google Scholar] [CrossRef] [PubMed]
- Scognamiglio, V.; Pezzotti, G.; Pezzotti, I.; Cano, J.; Buonasera, K.; Giannini, D.; Giardi, M.T. Biosensors for effective environmental and agrifood protection and commercialization: From research to market. Microchim. Acta 2010, 170, 215–225. [Google Scholar] [CrossRef]
- Luque de Castro, M.D.; Herrera, M.C. Enzyme Inhibition Based Biosensors and Biosensing Systems: Questionable Analytical Devices. Biosens. Bioelectron. 2003, 18, 279–294. [Google Scholar] [CrossRef]
- Jaffrezic-Renault, N. New Trends in Biosensors for Organophosphorus Pesticides. Sensors 2001, 1, 60–74. [Google Scholar] [CrossRef]
- Grieshaber, D.; MacKenzie, R.; Vöros, J.; Reimhult, E. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- Jiang, X.; Li, D.; Xu, X.; Ying, Y.; Li, Y.; Ye, Z.; Wang, J. Immunosensors for Detection of Pesticides Residues. Biosens. Bioelectron. 2008, 23, 1577–1587. [Google Scholar] [CrossRef]
- Suri, C.R.; Boro, R.; Nangia, Y.; Gandhi, S.; Sharma, P.; Wangoo, N.; Rajesh, K.; Shekhawat, G.S. Immunoanalytical Techniques for Analyzing Pesticides in the Environment. Trends Anal. Chem. 2009, 28, 29–39. [Google Scholar] [CrossRef]
- Kintzios, S. Cell-based sensors in clinical chemistry. Mini-Rev. Clin. Chem. 2007, 7, 1019–1026. [Google Scholar]
- Mavrikou, S.; Flampouri, K.; Moschopoulou, G.; Mangana, O.; Michaelides, A.; Kintzios, S. Assessment of organophosphate and carbamate pesticide residues in cigarette tobacco with a novel cell biosensor. Sensors 2008, 8, 2818–2832. [Google Scholar] [CrossRef]
- Flampouri, K.; Mavrikou, S.; Kintzios, S.; Miliadis, G.; Aplada-Sarlis, P. Development and validation of a cellular biosensor detecting pesticide residues in tomatoes. Talanta 2010, 80, 1799–1804. [Google Scholar] [CrossRef] [PubMed]
- Moschopoulou, G.; Vitsa, K.; Bem, F.; Vassilakos, N.; Perdikaris, A.; Blouhos, P.; Yialouris, C.; Frossiniotis, D.; Anthopoulos, I.; Maggana, O.; et al. Engineering of the membrane of fibroblast cells with virus-specific antibodies: A novel biosensor tool for virus detection. Biosens. Bioelectron. 2008, 24, 1027–1030. [Google Scholar] [CrossRef] [PubMed]
- Moschopoulou, G.; Kintzios, S. Application of membrane-engineering to bioelectric recognition cell sensors for the detection of picomole concentrations of superoxide radical: A novel biosensor principle. Anal. Chim. Acta 2006, 573, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Zeira, M.; Tosi, P.F.; Mouneimne, Y.; Lazarte, J.; Sneed, L.; Volsky, D.L.; Nikolau, C. Full-length CD4 electroinserted in the erythrocyte membrane as a long-lived inhibitor of infection by human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 1991, 88, 4409. [Google Scholar] [CrossRef]
- Anastassiades, M.; Lehotay, S.J. Fast and easy multiresidue methods employing acetonitrile extraction/partitioning and dispersive solid-phase extraction for the determination of pesticide residues in produce. J. AOAC Int. 2003, 86, 412–413. [Google Scholar] [CrossRef]
- Apostolou, T.; Pascual, N.; Marco, M.P.; Moschos, A.; Petropoulos, A.; Kaltsas, G.; Kintzios, S. Extraction-less, rapid assay for the direct detection of 2,4,6-trichloroanisole (TCA) in cork samples. Talanta 2014, 125, 336–340. [Google Scholar] [CrossRef]
- Dujaković, N.; Grujić, S.; Radišić, M.; Vasiljević, T.; Laušević, M. Determination of pesticides in surface and ground waters by liquid chromatography–electrospray–tandem mass spectrometry. Anal. Chim. Acta 2010, 678, 63–72. [Google Scholar]
- Tanner, G.; Czerwenka, C. LC-MS/MS analysis of neonicotinoid insecticides in honey: Methodology and residue findings in Austrian honeys. J. Agric. Food Chem. 2011, 59, 12271–12277. [Google Scholar] [CrossRef] [PubMed]





| Sample | Concentration (μg mL−1) b |
|---|---|
| LETTUCE 1 | 0.0002 |
| LETTUCE 2 | 0.0024 |
| LETTUCE 3 | 0.0065 |
| LETTUCE 4 | 0.0129 |
| LETTUCE 5 | 0.0022 |
| LETTUCE 6 | 0.0029 |
| LETTUCE 7 | 0.0027 |
| LETTUCE 8 | 0.0007 |
| LOLLO ROSSO1 | 0.001 |
| ICEBERG 1 | 0.0001 |
| ORGANIC LETTUCE 1 | 0 |
| ORGANIC LETTUCE 2 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Apostolou, T.; Loizou, K.; Hadjilouka, A.; Inglezakis, A.; Kintzios, S. Newly Developed System for Acetamiprid Residue Screening in the Lettuce Samples Based on a Bioelectric Cell Biosensor. Biosensors 2020, 10, 8. https://doi.org/10.3390/bios10020008
Apostolou T, Loizou K, Hadjilouka A, Inglezakis A, Kintzios S. Newly Developed System for Acetamiprid Residue Screening in the Lettuce Samples Based on a Bioelectric Cell Biosensor. Biosensors. 2020; 10(2):8. https://doi.org/10.3390/bios10020008
Chicago/Turabian StyleApostolou, Theofylaktos, Konstantinos Loizou, Agni Hadjilouka, Antonios Inglezakis, and Spyridon Kintzios. 2020. "Newly Developed System for Acetamiprid Residue Screening in the Lettuce Samples Based on a Bioelectric Cell Biosensor" Biosensors 10, no. 2: 8. https://doi.org/10.3390/bios10020008
APA StyleApostolou, T., Loizou, K., Hadjilouka, A., Inglezakis, A., & Kintzios, S. (2020). Newly Developed System for Acetamiprid Residue Screening in the Lettuce Samples Based on a Bioelectric Cell Biosensor. Biosensors, 10(2), 8. https://doi.org/10.3390/bios10020008