Newly Developed System for the Robust Detection of Listeria monocytogenes Based on a Bioelectric Cell Biosensor
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Cell Culture and Antibody Electroinsertion
2.3. Bacteria Culturing and Sample Preparation
2.4. Experimental Design and Assay Procedure
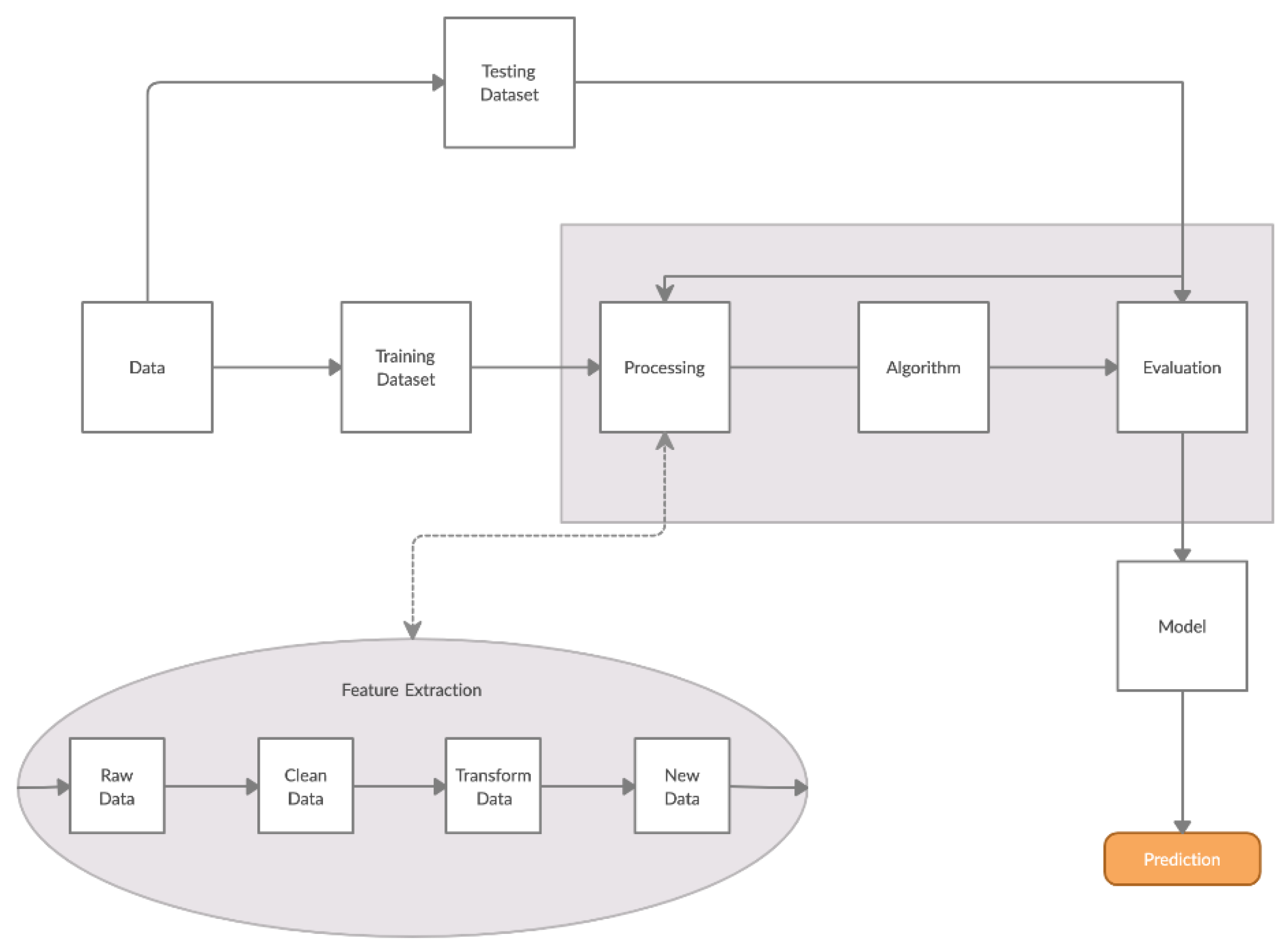
2.5. Algorithm for Response Processing
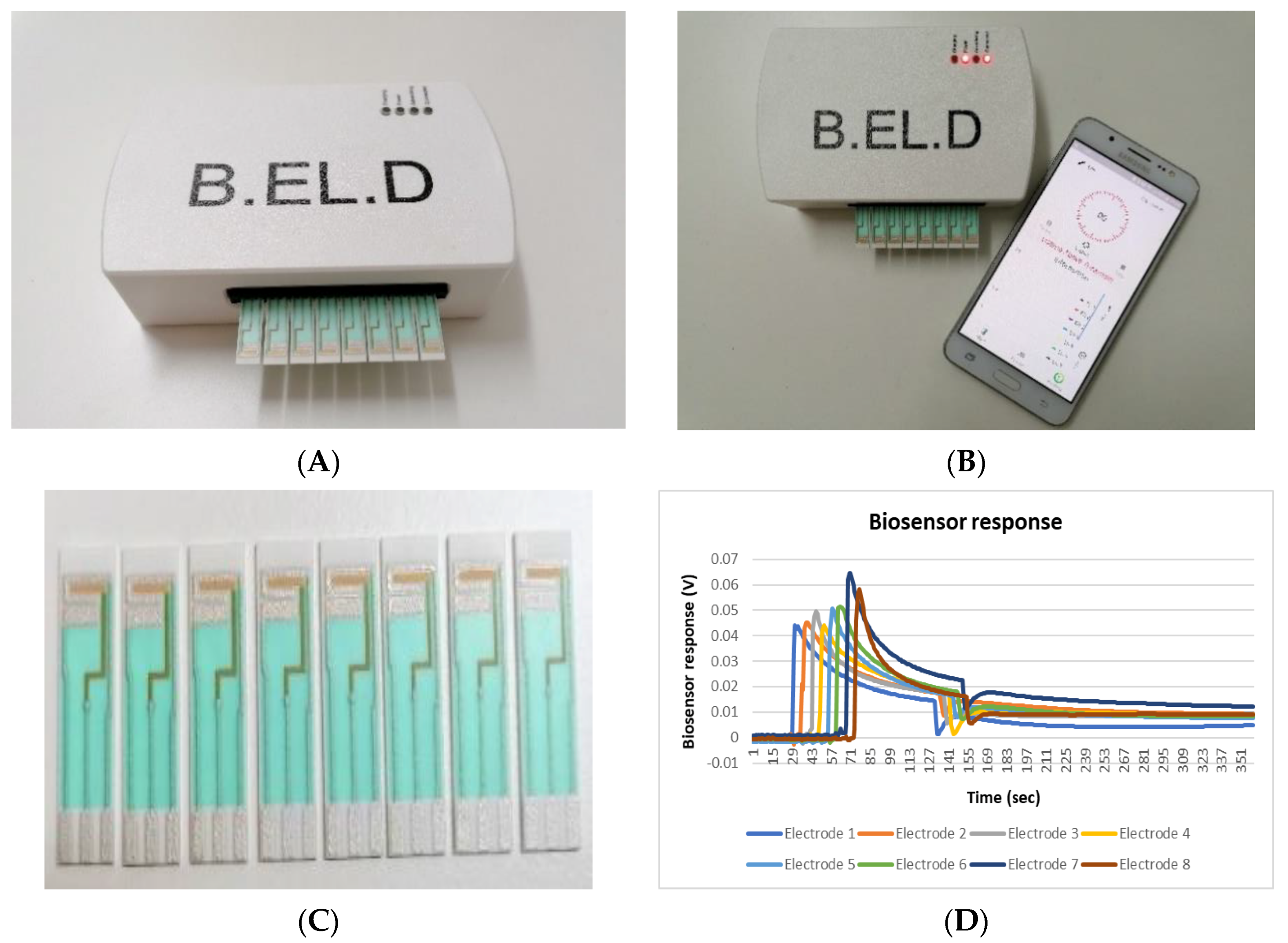
- Dataset: The dataset contained 200 measurements from 100 negative and 100 positive samples. Each measurement consisted of the data obtained by the 8 screen-printed electrodes that recorded the cell response as a time series of potentiometric measurements (in Volts) and comprised 360 values per electrode (sampling rate of 2 Hz). The detected measurements were visualized through a voltage/time graph (Figure 1D).
- Training/Testing Dataset: The obtained dataset was split into training and testing dataset as follows: 30% was used for training (60 measurements) and 70% was used for testing (140 measurements). The training dataset was utilized to determine the algorithm thresholds and the testing dataset was utilized for the algorithm evaluation.
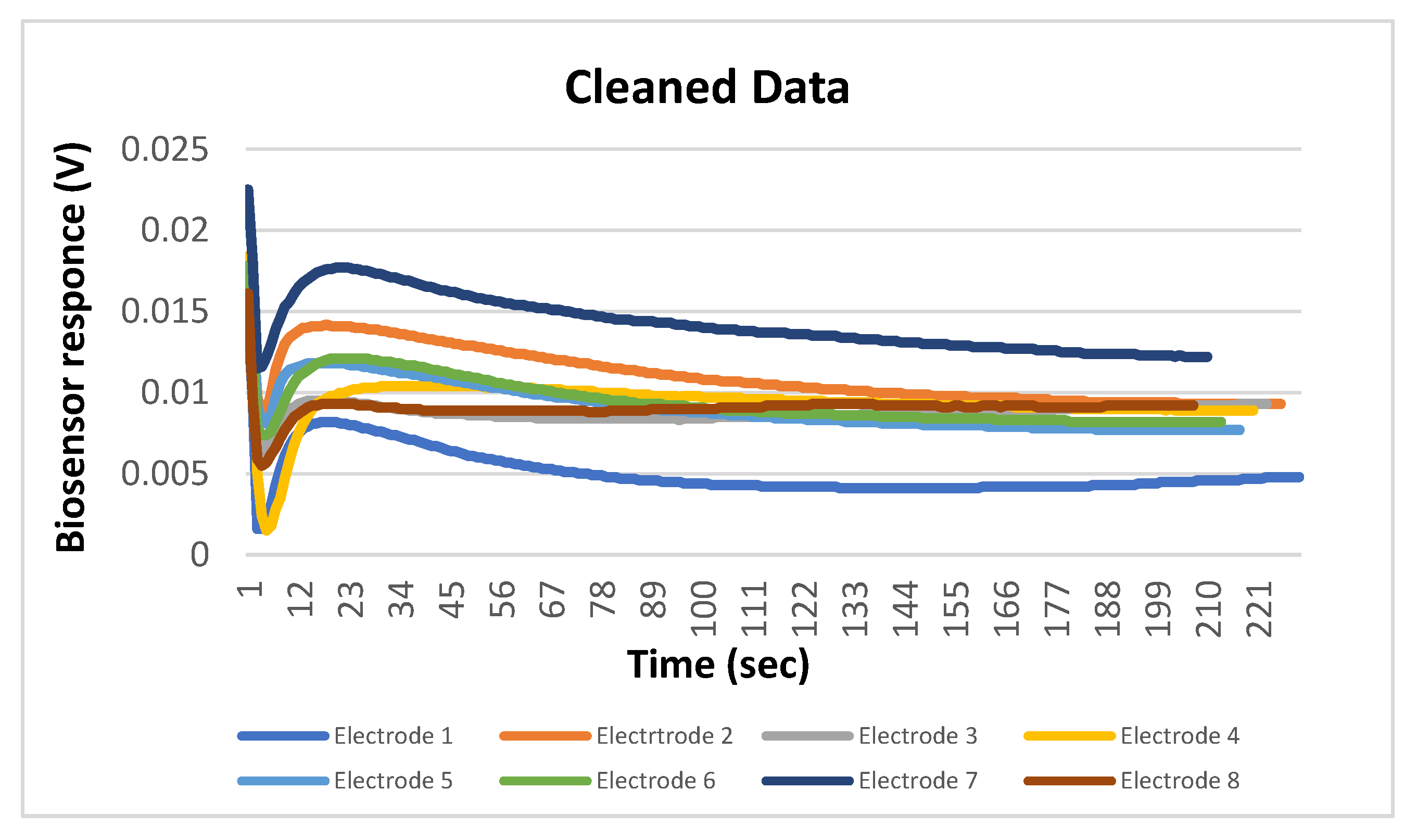
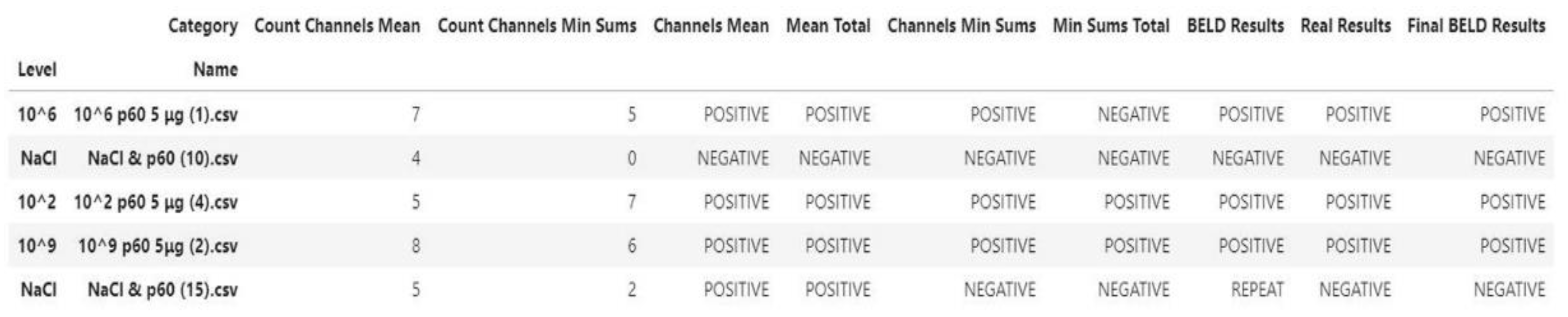
- Processing/Feature Extraction: The processing of the dataset was performed in a two-step procedure. In the first step, peaks were determined and starting noise was cleaned to smooth and calibrate the signal before the first peak. Then, all values were shifted to start at y = 0 and from each experimental dataset values from 120 to 200 were shifted at x = 0 and kept for further analysis (Figure 3). In the second step, feature vectors were extracted from the cleaned data and used as input to develop an algorithm able to detect L. monocytogenes in sterile saline samples. Each feature vector was calculated based on the following: (a) the average values (mean) for each cleaned dataset and (b) the rolling average with rolling window size 50 (minimum sums) [30]. This procedure was applied in each electrode channel (channel 1, channel 2, etc.) and the overall test dataset (mean average and minimum sums average for all 8 electrodes) (Figure 4). Hence, from the initial experimental raw dataset with 360 × 8 values, only 1 × 18 (1 values for each channel (8 values in total) + 1 overall value/(a) and (b)) feature values were used for the sample discrimination and the algorithm development.
2.6. Statiatical Analysis
3. Results and Discussion
3.1. Antibody Selection and Biosensor Response in the Presence of L. monocytogenes
Calculation of Method’s Performance Characteristics
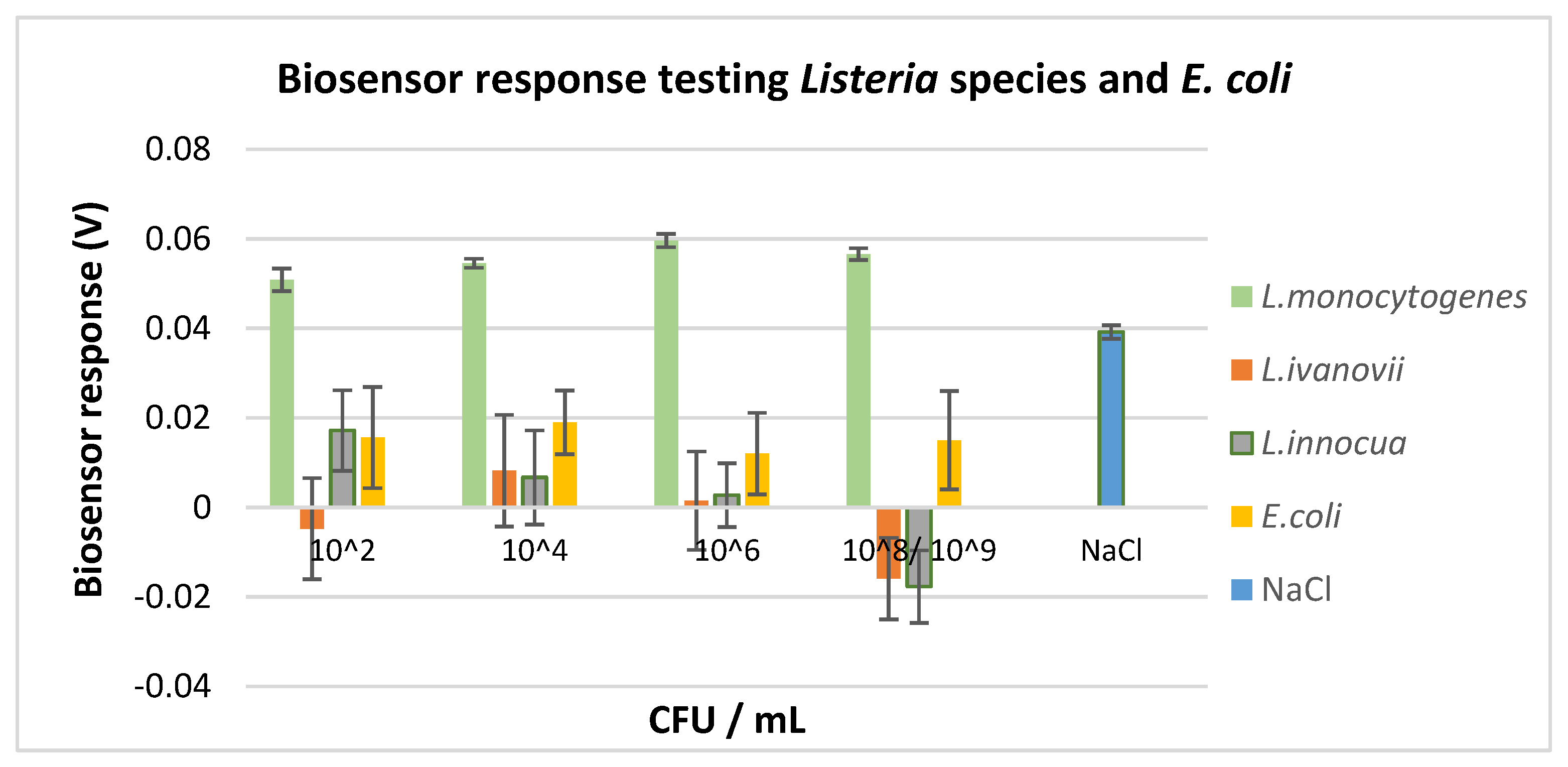
3.2. Selectivity Assay
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization (WHO). Available online: https://www.who.int/NEWS-ROOM/FACT-SHEETS/DETAIL/FOOD-SAFETY/ (accessed on 29 June 2020).
- Centers for Disease Control and Prevention (CDC). Listeria (Listeriosis). 2020. Available online: https://www.cdc.gov/listeria/index.html/ (accessed on 29 June 2020).
- International Organization for Standardization (ISO). IS0 11290-1:2017: Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes and of Listeria spp.—Part 1: Detection Method; ISO: Geneva, Switzerland, 2017; Available online: https://www.iso.org/standard/60313.html (accessed on 1 November 2020).
- Ali, S.; Hassan, A.; Hassan, G.; Eun, C.; Bae, J.; Lee, C.H.; Kim, I. Disposable all-printed electronic biosensor for instantaneous detection and classification of pathogens. Sci. Rep. 2018, 8, 5920. [Google Scholar] [CrossRef] [PubMed]
- Kanayeva, D.A.; Wang, R.; Rhoads, D.; Erf, G.F.; Slavik, M.F.; Tung, S.; Li, Y. Efficient separation and sensitive detection of Listeria monocytogenes using an impedance immunosensor based on magnetic nanoparticles, a microfluidic chip, and an interdigitated microelectrode. J. Food Prot. 2012, 75, 1951–1959. [Google Scholar] [CrossRef] [PubMed]
- Scognamiglio, V.; Pezzotti, G.; Pezzotti, I.; Cano, J.; Buonasera, K.; Giannini, D.; Giardi, M.T. Biosensors for effective environmental and agrifood protection and commercialization: From research to market. Microchim. Acta 2010, 170, 215–225. [Google Scholar] [CrossRef]
- Thevenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical Biosensors: Recommended Definitions and Classification. Pure Appl. Chem. 1999, 7, 2333–2348. [Google Scholar] [CrossRef]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar]
- Bhalla, N.; Jolly, P.; Formisano, N.; Estrela, P. Introduction to biosensors. Essays Biochem. 2016, 60, 1–8. [Google Scholar]
- Zhang, Z.; Cong, Y.; Huang, Y.; Du, X. Nanomaterials-based Electrochemical Immunosensors. Micromachines 2019, 10, 397. [Google Scholar] [CrossRef]
- Asal, M.; Özen, Ö.; Şahinler, M.; Polatoğlu, İ. Recent developments in enzyme, DNA and immuno-based biosensors. Sensors 2018, 18, 1924. [Google Scholar] [CrossRef]
- Gupta, N.; Renugopalakrishnan, V.; Liepmann, D.; Paulmurugan, R.; Malhotra, B.D. Cell-based biosensors: Recent trends, challenges and future perspectives. Biosens. Bioelectron. 2019, 141, 111435. [Google Scholar] [CrossRef]
- Inda, M.E.; Mimee, M.; Lu, T.K. Cell-based biosensors for immunology, inflammation, and allergy. J. Allergy Clin. Immunol. 2019, 144, 645–647. [Google Scholar] [CrossRef]
- Curtis, T.; Naal, R.M.Z.G.; Batt, C.; Tabb, J.; Holowka, D. Development of a mast cell-based biosensor. Biosens. Bioelectron. 2008, 23, 1024–1031. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Lenz, D.; Robinson, J.P.; Rickus, J.L.; Bhunia, A.K. A novel and simple cell-based detection system with a collagen-encapsulated B-lymphocyte cell line as a biosensor for rapid detection of pathogens and toxins. Lab. Investig. 2008, 88, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Bhunia, A.K. Cell-based biosensor for rapid screening of pathogens and toxins. Biosens. Bioelectron. 2010, 26, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Kintzios, S. Cell-based sensors in clinical chemistry. Mini Rev. Med. Chem. 2007, 7, 1019–1026. [Google Scholar] [CrossRef]
- Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a portable, ultra-rapid and ultra-sensitive cell-based biosensor for the direct detection of the SARS-CoV-2 S1 spike protein antigen. Biosensors 2020, 20, 3121. [Google Scholar]
- Moschopoulou, G.; Valero, T.; Kintzios, S. Molecular Identification through Membrane Engineering as a revolutionary concept for the construction of cell sensors with customized target recognition properties: The example of superoxide detection. Procedia Eng. 2011, 25, 1541–1544. [Google Scholar] [CrossRef][Green Version]
- Moschopoulou, G.; Vitsa, K.; Bem, F.; Vassilakos, N.; Perdikaris, A.; Blouhos, P.; Yialouris, C.; Frossiniotis, D.; Anthopoulos, I.; Maggana, O.; et al. Engineering of the membrane of fibroblast cells with virus-specific antibodies: A novel biosensor tool for virus detection. Biosens. Bioelectron. 2008, 24, 1033–1036. [Google Scholar] [CrossRef]
- Perdikaris, A.; Alexandropoulos, N.; Kintzios, S. Development of a novel, ultra-rapid biosensor for the qualitative detection of hepatitis b virus-associated antigens and anti-HBV, based on “membrane-engineered” fibroblast cells with virus-specific antibodies and antigens. Sensors 2009, 9, 2176–2186. [Google Scholar] [CrossRef]
- Perdikaris, A.; Vassilakos, N.; Yiakoumettis, I.; Kektsidou, O.; Kintzios, S. Development of a portable, high throughput biosensor system for rapid plant virus detection. J. Virol. Methods 2011, 177, 94–99. [Google Scholar] [CrossRef]
- Moschopoulou, G.; Kintzios, S. Application of “membrane-engineering” to bioelectric recognition cell sensors for the detection of picomole concentrations of superoxide radical: A novel biosensor principle. Anal. Chim. Acta 2006, 573–574, 90–96. [Google Scholar] [CrossRef]
- Moschopoulou, G.; Valero, T.; Kintzios, S. Superoxide determination using membrane engineered cells: An example of a novel concept for the construction of cell sensors with customized target recognition properties. Sens. Actuators B Chem. 2012, 175, 88–94. [Google Scholar] [CrossRef]
- Kintzios, S.; Pistola, E.; Konstas, J.; Bem, F.; Matakiadis, T.; Alexandropoulos, N.; Bem, F.; Ekonomou, G.; Biselis, J.; Levin, R. Bioelectric recognition assay (BERA). Biosens. Bioelectron. 2001, 16, 467–480. [Google Scholar] [CrossRef]
- Apostolou, T.; Loizou, K.; Hadjilouka, A.; Inglezakis, A.; Kintzios, S. Newly developed system for acetamiprid residue screening in the lettuce samples based on a bioelectric biosensor. Biosensors 2020, 10, 8. [Google Scholar] [CrossRef]
- Zeira, M.; Tosi, P.F.; Mouneimne, Y.; Lazarte, J.; Sneed, L.; Volsky, D.L.; Nikolau, C. Full-length CD4 electroinserted in the erythrocyte membrane as a long-lived inhibitor of infection by human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 1991, 88, 4409–4413. [Google Scholar] [CrossRef]
- Kokla, A.; Blouchos, P.; Livaniou, E.; Zikos, C.; Kakabakos, S.E.; Petrou, P.S.; Kintzios, S. Visualization of the membrane-engineering concept: Evidence for the specific orientation of electroinserted antibodies and selective binding of target analytes. J. Mol. Recognit. 2013, 26, 627–632. [Google Scholar] [CrossRef]
- Glas, A.S.; Lijmer, J.G.; Prins, M.H.; Bonsel, G.J.; Bossuyt, P.M.M. The diagnostic odds ratio: A single indicator of test performance. J. Clin. Epidemiol. 2003, 56, 1129–1135. [Google Scholar] [CrossRef]
- Van der Walt, S.; Colbert, S.C.; Varoquaux, G. The NumPy Array: A Structure for Efficient Numerical Computation, Computing in Science & Engineering. Comput. Sci. Eng. 2011, 13, 22–30. [Google Scholar]
- Paramithiotis, S.; Hadjilouka, A.; Drosinos, E.H. Listeria pathogenicity island 1. Structure and function. In Listeria Monocytogenes: Food Sources, Prevalence and Management Strategies; Hambrick, E.C., Ed.; Nova Publishers: New York, NY, USA, 2014; pp. 265–282. [Google Scholar]
- Koubova, V.; Brynda, E.; Karasova, L.; Skvor, J.; Homola, J.; Dostalek, J.; Tobiska, P.; Rosicky, J. Detection of foodborne pathogens using surface plasmon resonance biosensors. Sens. Actuators B Chem. 2001, 74, 100–105. [Google Scholar] [CrossRef]
- Leonard, P.; Hearty, S.; Quinn, J.; O’Kennedy, R. A generic approach for the detection of whole Listeria monocytogenes cells in contaminated samples using surface plasmon resonance. Biosens. Bioelectron. 2004, 19, 1331–1335. [Google Scholar] [CrossRef]
- Sharma, H.; Mutharasan, R. Rapid and sensitive immunodetection of Listeria monocytogenes in milk using a novel piezoelectric cantilever sensor. Biosens. Bioelectron. 2013, 45, 158–162. [Google Scholar] [CrossRef]
- Cheng, C.; Peng, Y.; Bai, J.; Zhang, X.; Liu, Y.; Fan, X.; Ning, B.; Gao, Z. Rapid detection of Listeria monocytogenes in milk by self-assembled electrochemical immunosensor. Sens. Actuators B Chem. 2014, 190, 900–906. [Google Scholar] [CrossRef]
- Wang, D.; Chen, Q.; Huo, H. Efficient separation and quantitative detection of Listeria monocytogenes based on screen-printed interdigitated electrode, urease and magnetic nanoparticles. Food Control 2017, 73, 555–561. [Google Scholar] [CrossRef]
- De Michele, C.; De Los Rios, P.; Foffi, G.; Piazza, F. Simulation and Theory of Antibody Binding to Crowded Antigen-Covered Surfaces. PLoS Comput. Biol. 2016, 12, e1004752. [Google Scholar] [CrossRef]
- Cho, I.H.; Paek, E.H.; Lee, H.; Kang, J.Y.; Kim, T.S.; Paek, S.H. Site-directed biotinylation of antibodies for controlled immobilization on solid surfaces. Anal. Biochem. 2007, 365, 14–23. [Google Scholar] [CrossRef]
- Kwon, Y.; Han, Z.; Karatan, E.; Mrksich, M.; Kay, B.K. Antibody Arrays Prepared by Cutinase-Mediated Immobilization on Self-Assembled Monolayers. Anal. Chem. 2004, 76, 5713–5720. [Google Scholar] [CrossRef]
- Benarroch, J.M.; Asally, M. The Microbiologist’s Guide to Membrane Potential Dynamics. Trends Microbiol. 2020, 28, 304–314. [Google Scholar] [CrossRef]


| Results 1 | Performance Indices 2 | ||
|---|---|---|---|
| TP | 75 | Se | 97.4% |
| FP | 2 | Sp | 84.13% |
| TN | 53 | PPV | 88.24% |
| FN | 10 | NPV | 96.36% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hadjilouka, A.; Loizou, K.; Apostolou, T.; Dougiakis, L.; Inglezakis, A.; Tsaltas, D. Newly Developed System for the Robust Detection of Listeria monocytogenes Based on a Bioelectric Cell Biosensor. Biosensors 2020, 10, 178. https://doi.org/10.3390/bios10110178
Hadjilouka A, Loizou K, Apostolou T, Dougiakis L, Inglezakis A, Tsaltas D. Newly Developed System for the Robust Detection of Listeria monocytogenes Based on a Bioelectric Cell Biosensor. Biosensors. 2020; 10(11):178. https://doi.org/10.3390/bios10110178
Chicago/Turabian StyleHadjilouka, Agni, Konstantinos Loizou, Theofylaktos Apostolou, Lazaros Dougiakis, Antonios Inglezakis, and Dimitrios Tsaltas. 2020. "Newly Developed System for the Robust Detection of Listeria monocytogenes Based on a Bioelectric Cell Biosensor" Biosensors 10, no. 11: 178. https://doi.org/10.3390/bios10110178
APA StyleHadjilouka, A., Loizou, K., Apostolou, T., Dougiakis, L., Inglezakis, A., & Tsaltas, D. (2020). Newly Developed System for the Robust Detection of Listeria monocytogenes Based on a Bioelectric Cell Biosensor. Biosensors, 10(11), 178. https://doi.org/10.3390/bios10110178







