Label-Free MicroRNA Optical Biosensors
Abstract
1. Introduction
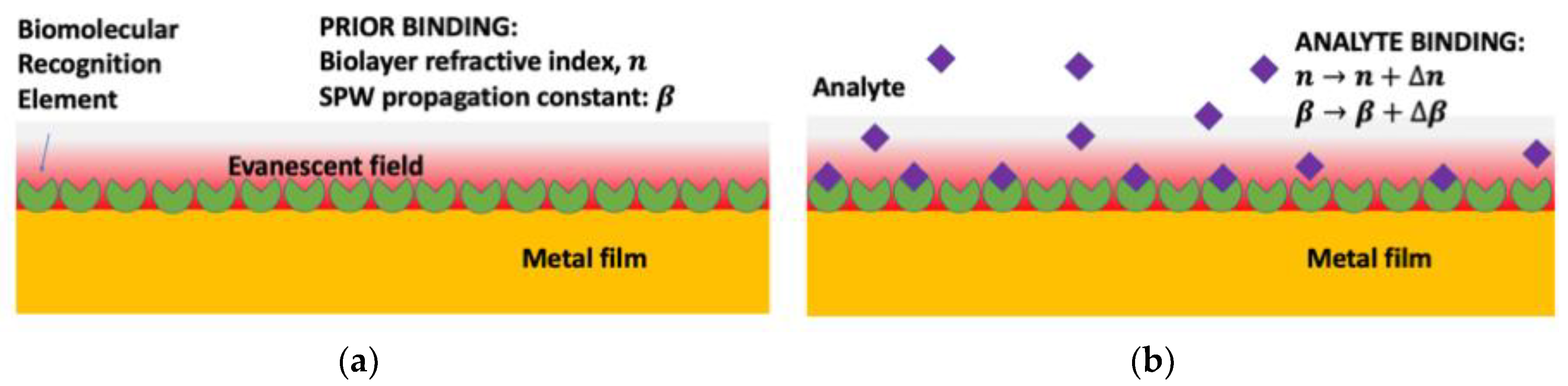
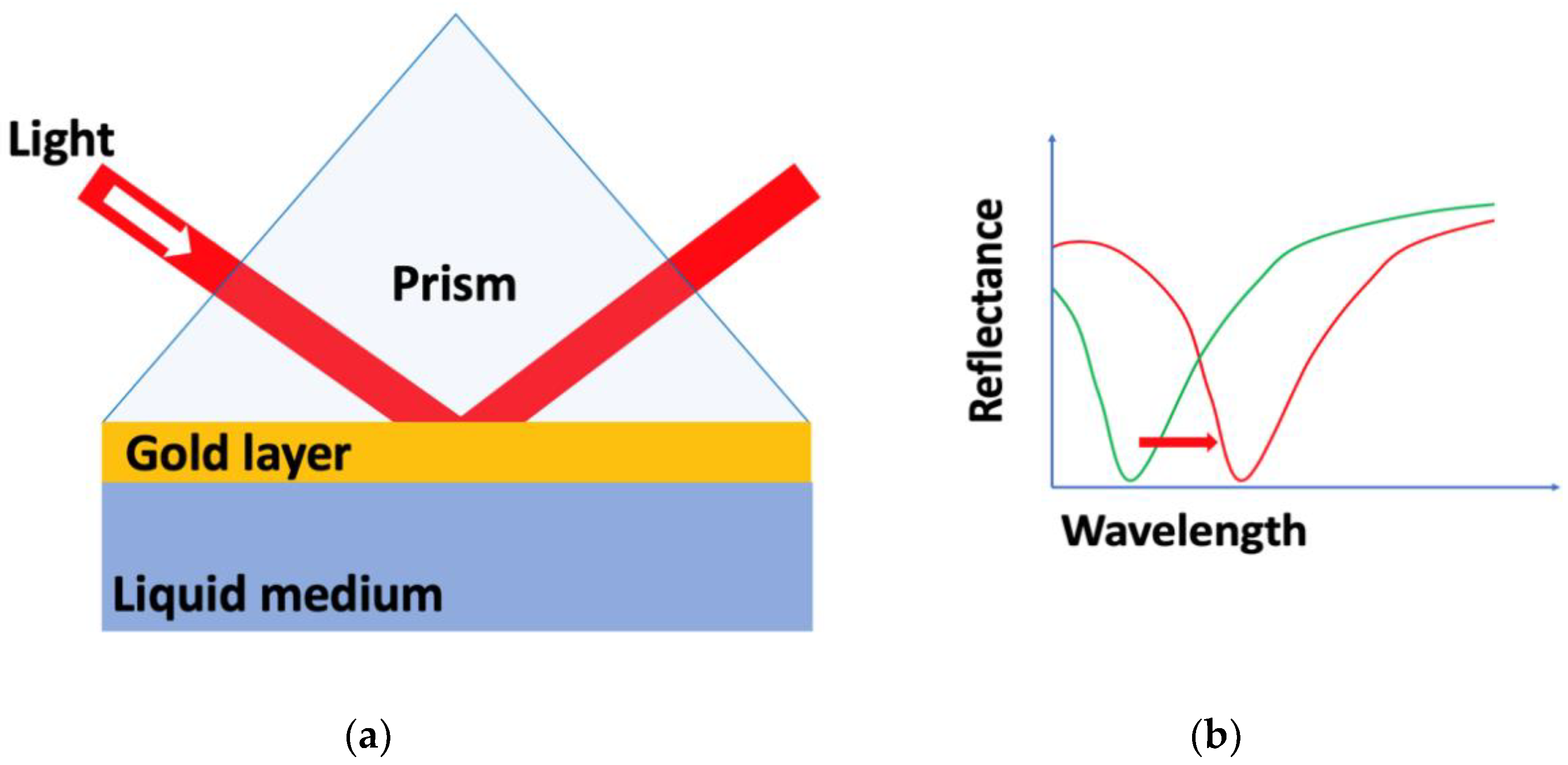
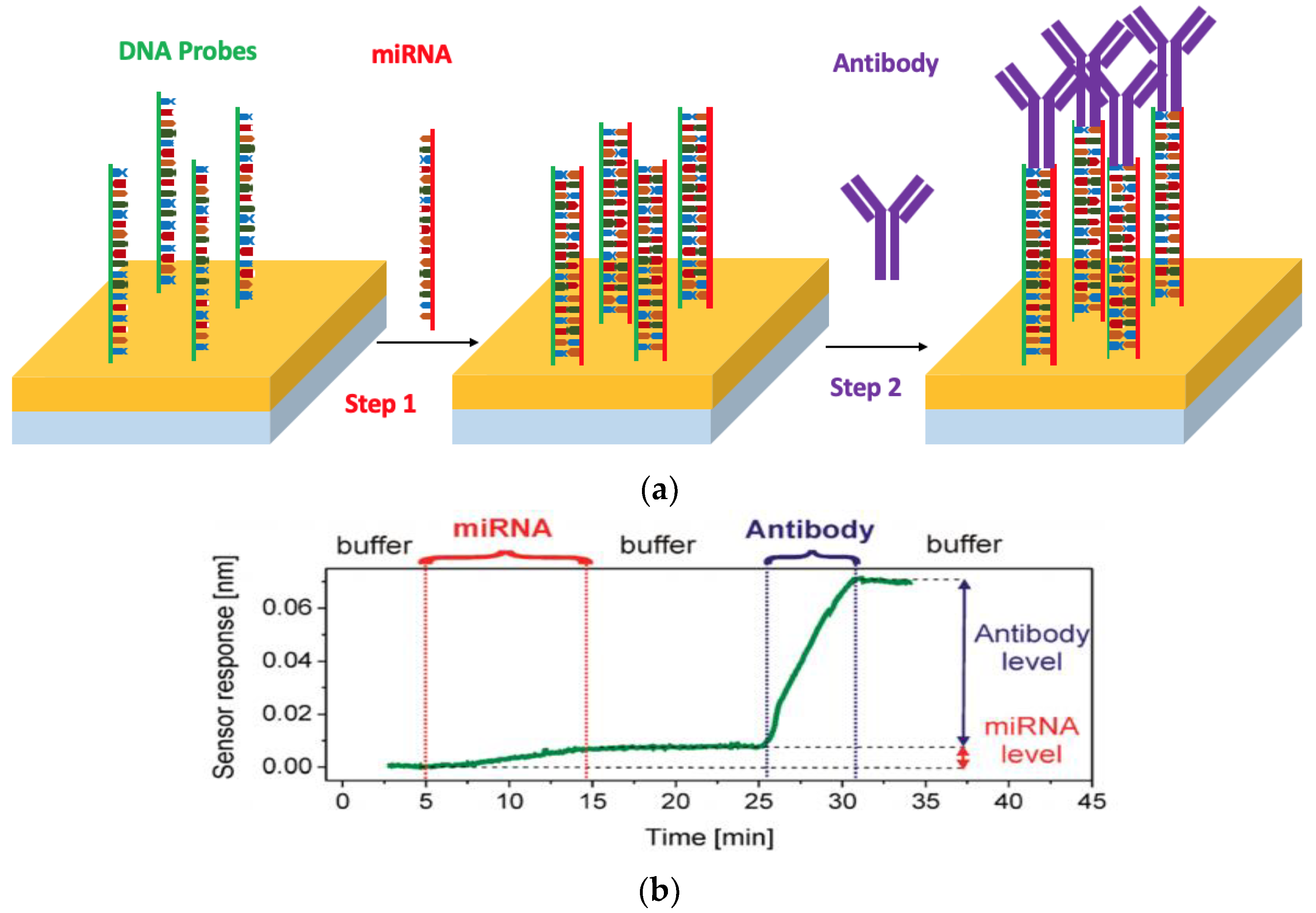
2. Surface Plasmon Resonance (SPR) Biosensors
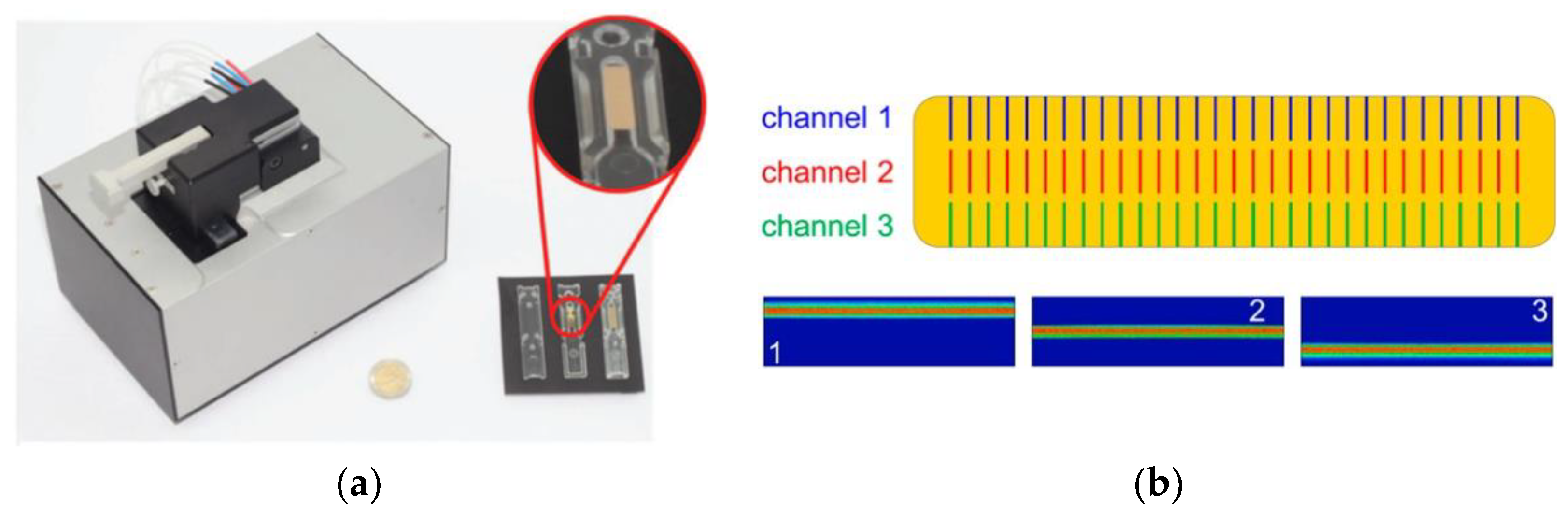
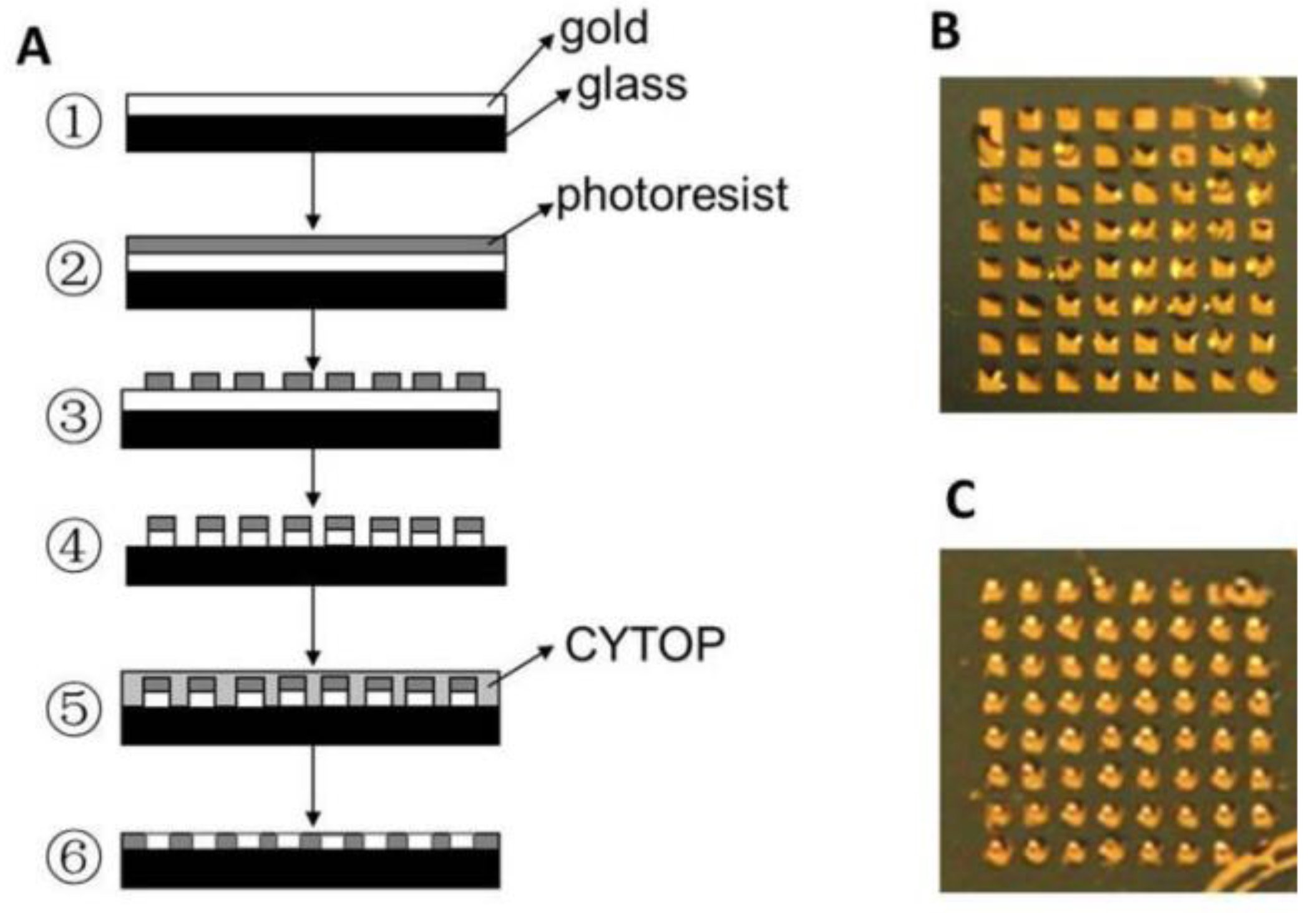
2.1. SPR Imaging (SPRi)
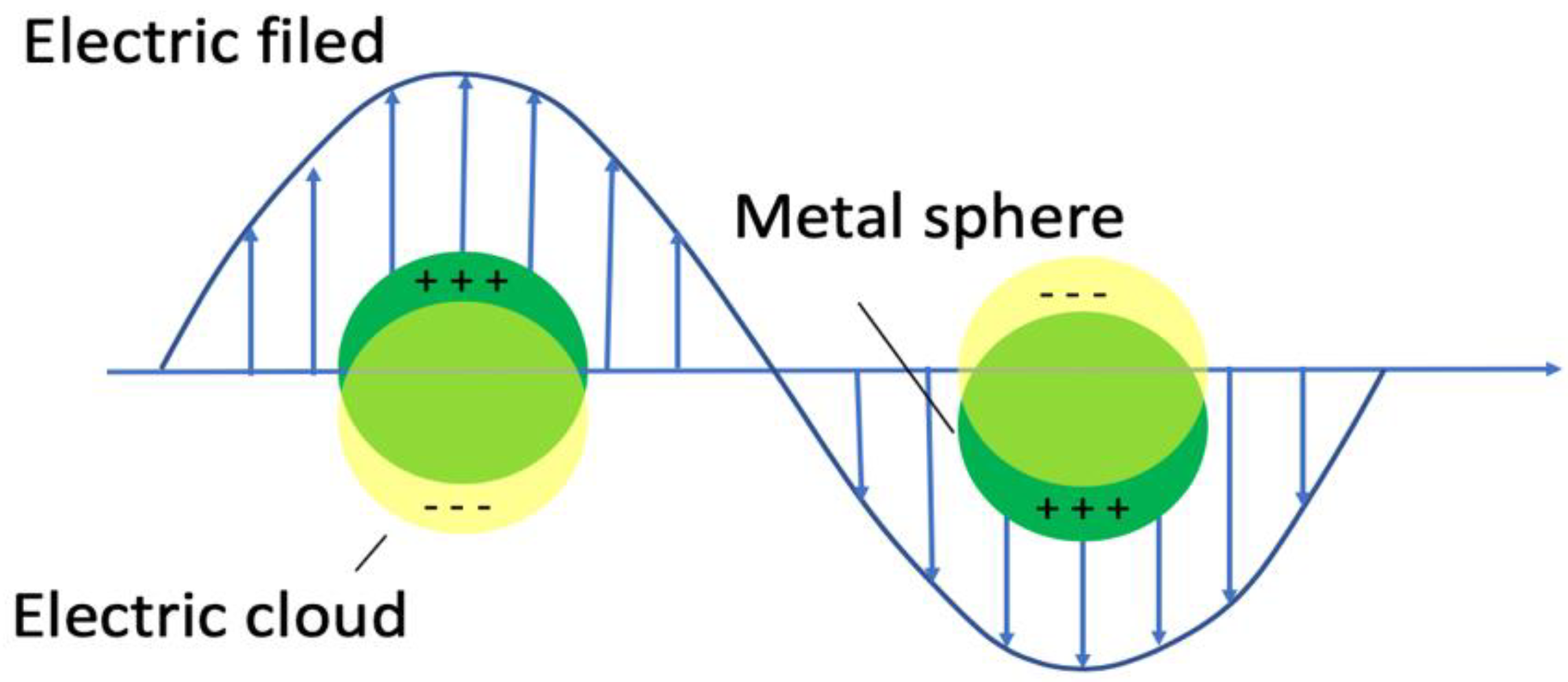
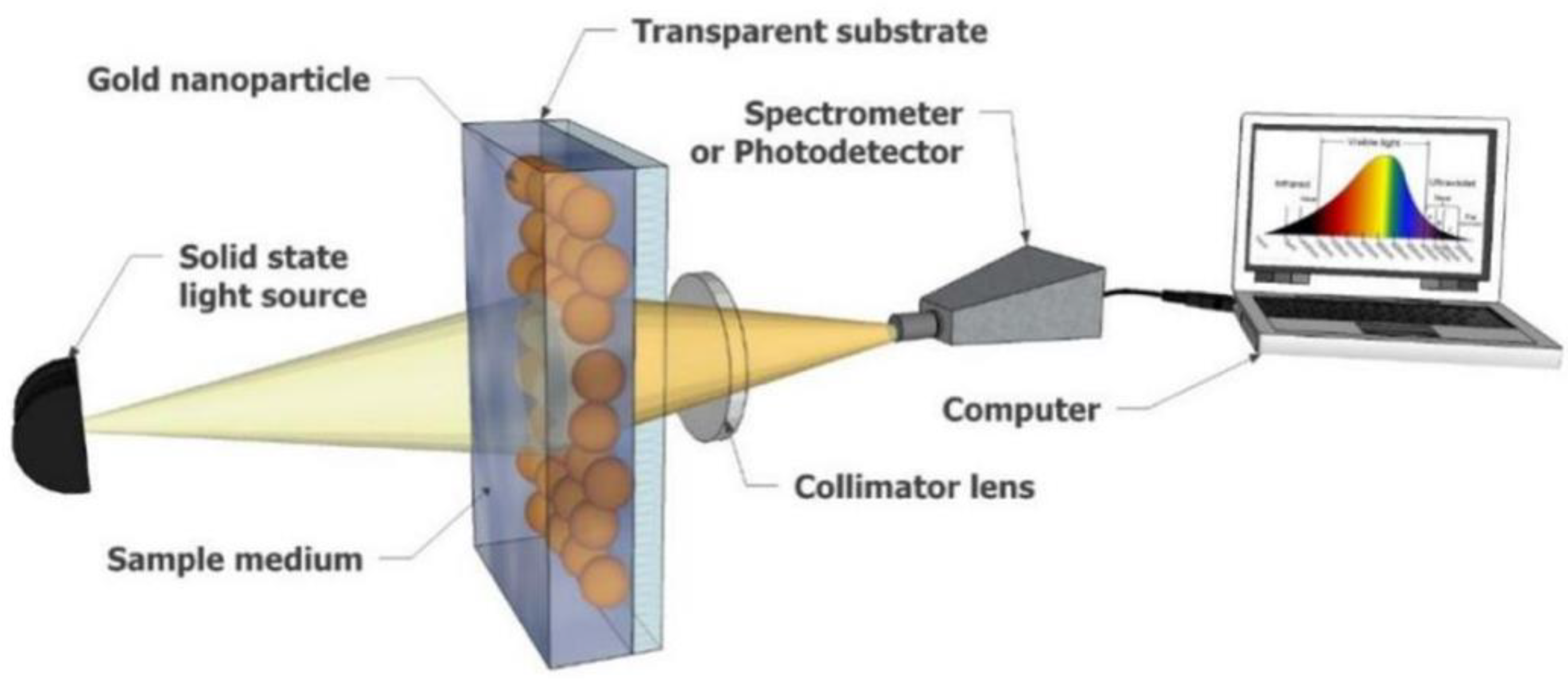
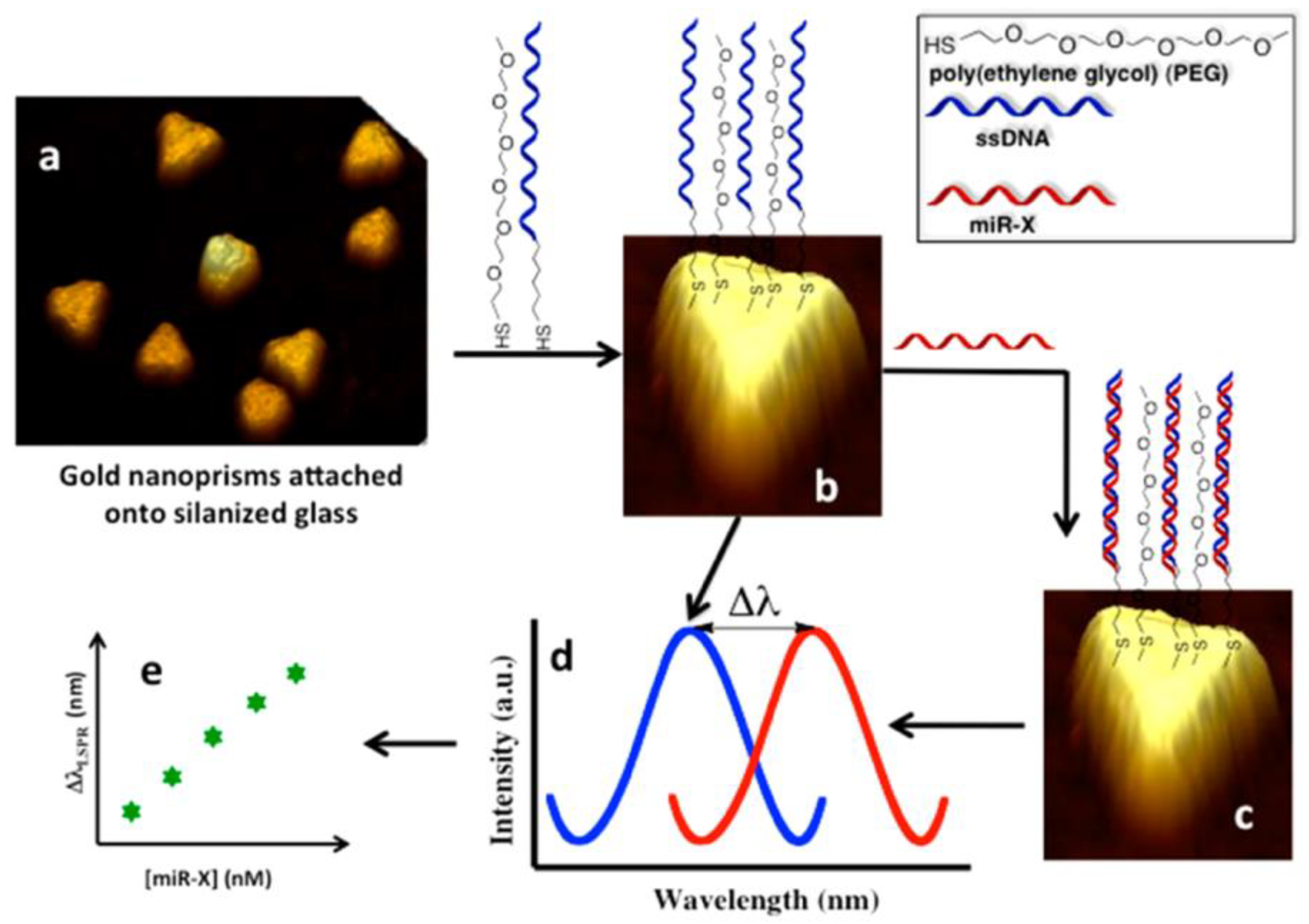
2.2. Localized Surface Plasmon Resonance (LSPR)
3. Integrated Planar Optical Waveguide Interferometer Biosensors
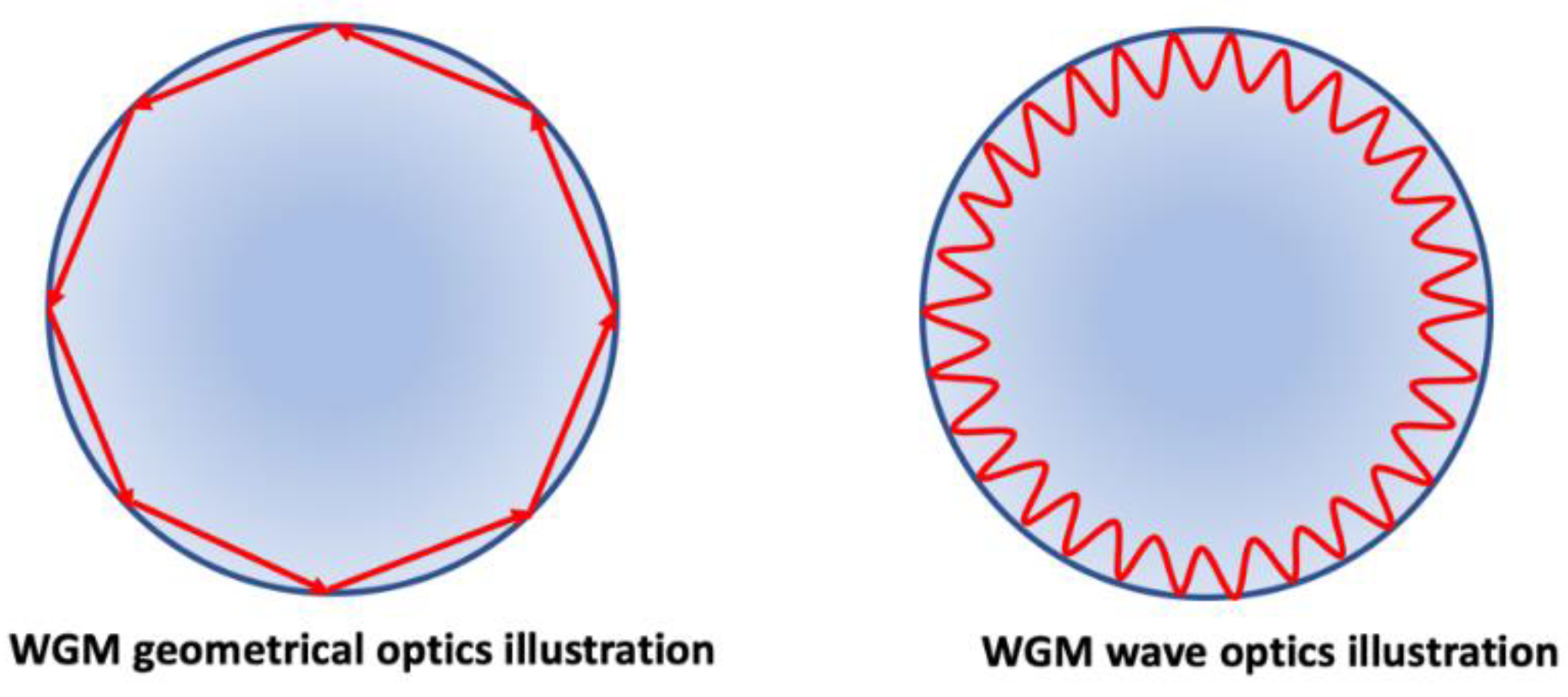
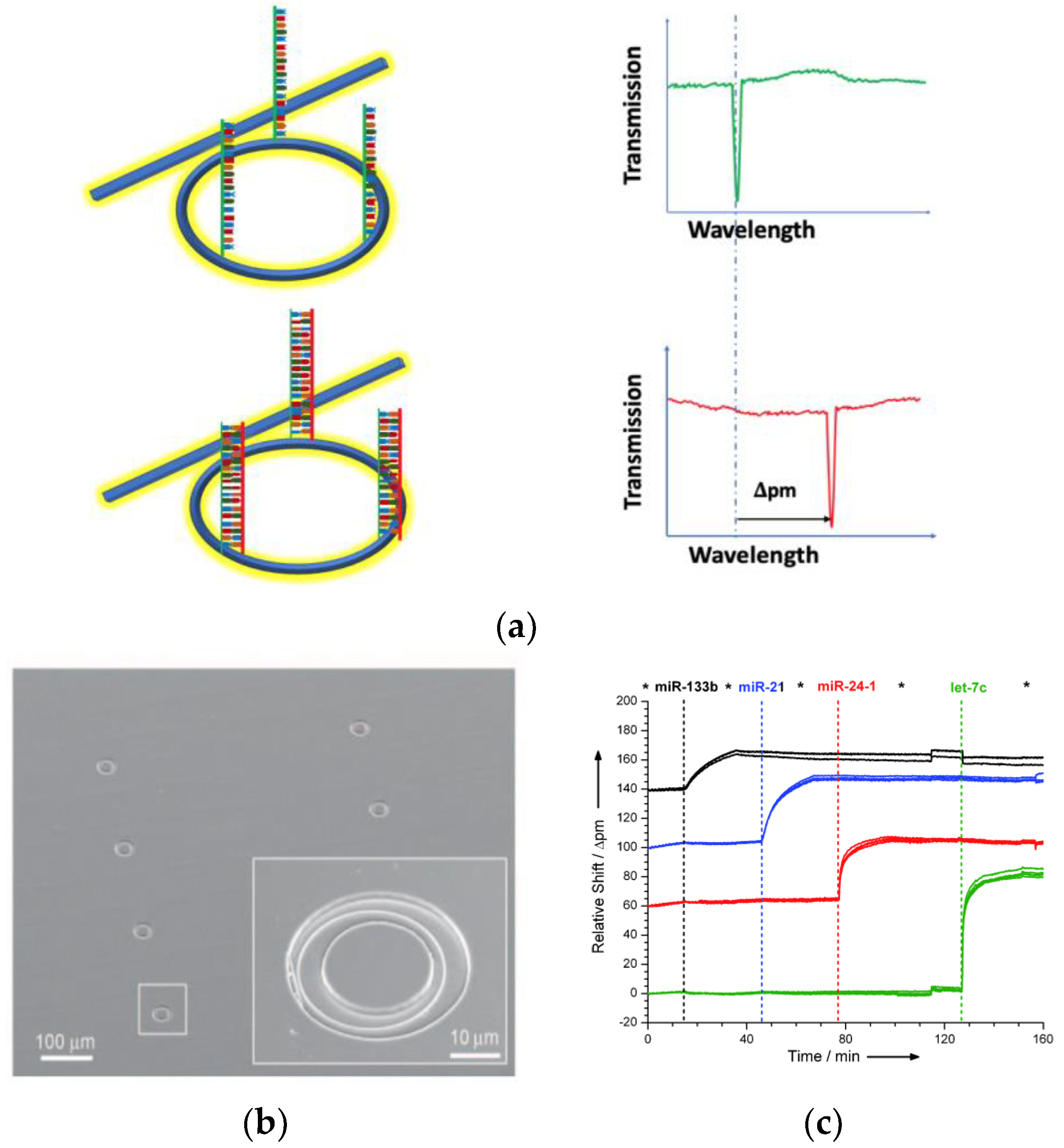
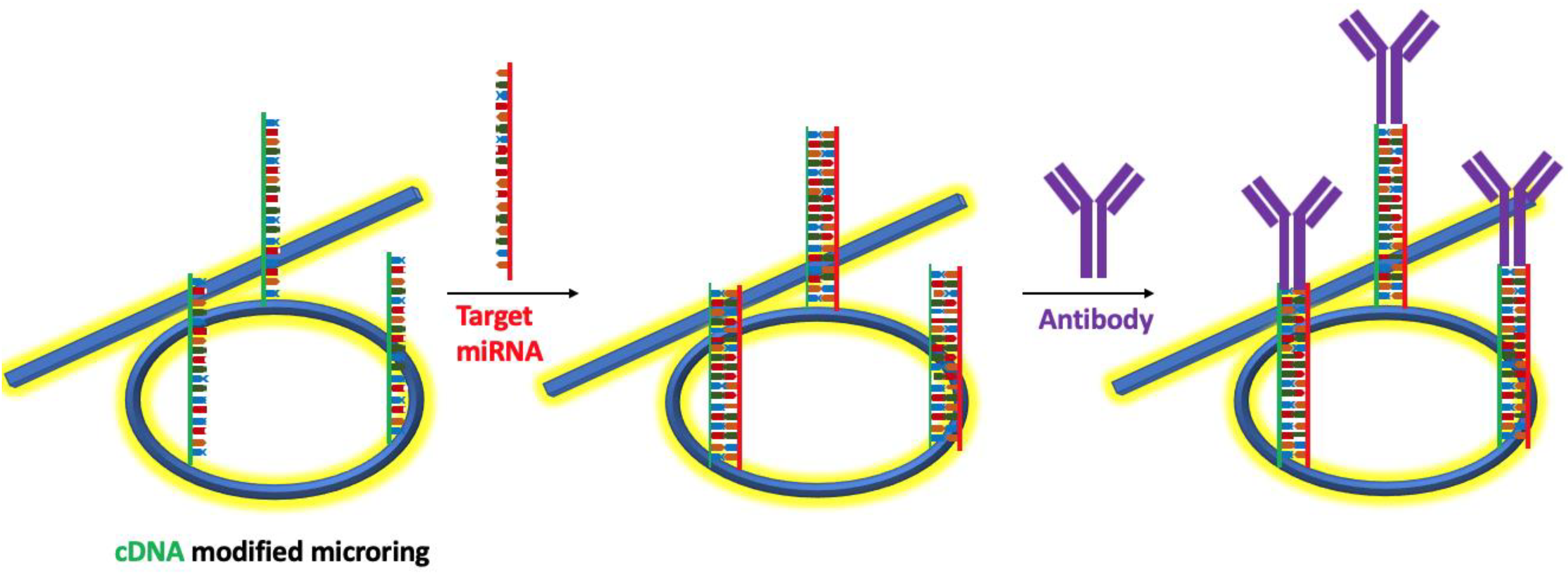
4. Whispering Gallery Modes (WGMs) Resonators Biosensors
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ambros, V. microRNAs: Tiny regulators with great potential. Cell 2001, 107, 823–826. [Google Scholar] [CrossRef]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Saliminejad, K.; Khorram Khorshid, H.R.; Soleymani Fard, S.; Ghaffari, S.H. An overview of microRNAs: Biology, functions, therapeutics, and analysis methods. J. Cell. Physiol. 2019, 234, 5451–5465. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef]
- Reinhart, B.J.; Weinstein, E.G.; Rhoades, M.W.; Bartel, B.; Bartel, D.P. MicroRNAs in plants. Trends Plant Sci. 2002, 7, 1616–1626. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and Host Interactions. Annu. Rev. Microbiol. 2010, 64, 123–141. [Google Scholar] [CrossRef]
- Wahid, F.; Shehzad, A.; Khan, T.; Kim, Y.Y. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta Mol. Cell Res. 2010, 1803, 1231–1243. [Google Scholar] [CrossRef]
- Sekar, D.; Venugopal, B.; Sekar, P.; Ramalingam, K. Role of microRNA 21 in diabetes and associated/related diseases. Gene 2016, 582, 14–18. [Google Scholar] [CrossRef]
- Feng, J.; Xing, W.; Xie, L. Regulatory Roles of MicroRNAs in Diabetes. Int. J. Mol. Sci. 2016, 17, 1729. [Google Scholar] [CrossRef]
- Tang, X.; Tang, G.; Özcan, S. Role of microRNAs in diabetes. Biochim. Biophys. Acta Gene Regul. Mech. 2008, 1779, 697–701. [Google Scholar] [CrossRef] [PubMed]
- Bronze-Da-Rocha, E. MicroRNAs expression profiles in cardiovascular diseases. Biomed. Res. Int. 2014, 2014, 985408. [Google Scholar] [CrossRef]
- Zhong, X.; Coukos, G.; Zhang, L. miRNAs in human cancer. Methods Mol. Biol. 2012, 822, 295–306. [Google Scholar] [PubMed]
- Chin, L.J.; Slack, F.J. A truth serum for cancer—MicroRNAs have major potential as cancer biomarkers. Cell Res. 2008, 18, 983–984. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Ferracin, M.; Liu, C.-G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed]
- Vettori, S.; Gay, S.; Distler, O. Role of MicroRNAs in Fibrosis. Open Rheumatol. J. 2012, 6, 130–139. [Google Scholar] [CrossRef] [PubMed]
- O’Reilly, S. MicroRNAs in fibrosis: Opportunities and challenges. Arthritis Res. Ther. 2016, 18, 11. [Google Scholar] [CrossRef]
- Jiang, X.; Tsitsiou, E.; Herrick, S.E.; Lindsay, M.A. MicroRNAs and the regulation of fibrosis. FEBS J. 2010, 277, 2015–2021. [Google Scholar] [CrossRef]
- Rajasekaran, S.; Rajaguru, P.; Gandhi, P.S.S. MicroRNAs as potential targets for progressive pulmonary fibrosis. Front. Pharmacol. 2015, 6, 473. [Google Scholar] [CrossRef]
- Pauley, K.M.; Cha, S.; Chan, E.K. MicroRNA in autoimmunity and autoimmune diseases. J. Autoimmun. 2009, 32, 189–194. [Google Scholar] [CrossRef]
- Hébert, S.S.; De Strooper, B. Alterations of the microRNA network cause neurodegenerative disease. Trends Neurosci. 2009, 32, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Satoh, J. Molecular network of microRNA targets in Alzheimer’s disease brains. Exp. Neurol. 2012, 235, 436–446. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Bonini, N.M. MicroRNAs and neurodegeneration: Role and impact. Trends Cell Biol. 2013, 23, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Chen, J. MicroRNAs, signaling pathways and diseases. Ann. Transl. Med. 2015, 3, 329. [Google Scholar]
- Schmieder, S.; Weißpflog, J.; Danz, N.; Klotzbach, U.; Sonntag, F. Detection of miRNA using a surface plasmon resonance biosensor and antibody amplification. Curr. Dir. Biomed. Eng. 2016, 2, 135–138. [Google Scholar] [CrossRef]
- Brase, J.C.; Wuttig, D.; Kuner, R.; Sültmann, H. Serum microRNAs as non-invasive biomarkers for cancer. Mol. Cancer 2010, 9, 306. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Zendjabil, M. Circulating microRNAs as novel biomarkers of Alzheimer’s disease. Clin. Chim. Acta 2018, 484, 99–104. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of Novel Genes Coding for Small Expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Liu, Q.; Shin, Y.; Kee, J.S.; Kim, K.W.; Rafei, S.R.M.; Perera, A.P.; Tu, X.; Lo, G.-Q.; Ricci, E.; Colombel, M.; et al. Mach–Zehnder interferometer (MZI) point-of-care system for rapid multiplexed detection of microRNAs in human urine specimens. Biosens. Bioelectron. 2015, 71, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Cheng, H.H.; Tewari, M. MicroRNA profiling: Approaches and considerations. Nat. Rev. Genet. 2012, 13, 358. [Google Scholar] [CrossRef] [PubMed]
- Cohen, L.; Hartman, M.R.; Amardey-Wellington, A.; Walt, D.R. Digital direct detection of microRNAs using single molecule arrays. Nucleic Acids Res. 2017, 45, e137. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.N.; Mutharasan, R. Biosensor-based microRNA detection: Techniques, design, performance, and challenges. Analyst 2014, 139, 1576. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Lei, J.; Ding, L.; Wen, Y.; Ju, H.; Zhang, X. MicroRNA: Function, Detection, and Bioanalysis. Chem. Rev. 2013, 113, 6207–6233. [Google Scholar] [CrossRef]
- Catuogno, S.; Esposito, C.L.; Quintavalle, C.; Cerchia, L.; Condorelli, G.; De Franciscis, V. Recent Advance in Biosensors for microRNAs Detection in Cancer. Cancers 2011, 3, 1877–1898. [Google Scholar] [CrossRef]
- Chandrasekaran, A.R.; MacIsaac, M.; Dey, P.; Levchenko, O.; Zhou, L.; Andres, M.; Dey, B.K.; Halvorsen, K. Cellular microRNA detection with miRacles: MicroRNA-activated conditional looping of engineered switches. Sci. Adv. 2019, 5, 9443. [Google Scholar] [CrossRef]
- Git, A.; Dvinge, H.; Salmon-Divon, M.; Osborne, M.; Kutter, C.; Hadfield, J. Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA 2010, 16, 991–1006. [Google Scholar] [CrossRef]
- Várallyay, É.; Burgyán, J.; Havelda, Z. MicroRNA detection by northern blotting using locked nucleic acid probes. Nat. Protoc. 2008, 3, 190. [Google Scholar] [CrossRef]
- Koscianska, E.; Starega-Roslan, J.; Sznajder, L.J.; Olejniczak, M.; Galka-Marciniak, P.; Krzyzosiak, W.J. Northern blotting analysis of microRNAs, their precursors and RNA interference triggers. BMC Mol. Biol. 2011, 12, 14. [Google Scholar] [CrossRef]
- Liu, C.-G.; Calin, G.A.; Meloon, B.; Gamliel, N.; Sevignani, C.; Ferracin, M.; Dumitru, C.D.; Shimizu, M.; Zupo, S.; Dono, M.; et al. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc. Natl. Acad. Sci. USA 2004, 101, 9740–9744. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Xi, Y. Challenges for MicroRNA Microarray Data Analysis. Microarrays 2013, 2, 34–50. [Google Scholar] [CrossRef] [PubMed]
- Fiammengo, R. Can nanotechnology improve cancer diagnosis through miRNA detection? Biomark. Med. 2017, 11, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Liang, W.; Li, Y.; Sun, Y.; Xiang, Y.; Zhang, Y.; Dai, Z.; Duo, Y.; Wu, L.; Qi, K.; et al. Ultrasensitive detection of miRNA with an antimonene-based surface plasmon resonance sensor. Nat. Commun. 2019, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- La, M.; Liu, L.; Zhou, B.-B. Nanomaterials-Based Fluorimetric Methods for MicroRNAs Detection. Materials 2015, 8, 2809–2829. [Google Scholar] [CrossRef]
- Carrascosa, L.G.; Huertas, C.S.; Lechuga, L.M. Prospects of optical biosensors for emerging label-free RNA analysis. TrAC Trends Anal. Chem. 2016, 80, 177–189. [Google Scholar] [CrossRef]
- Borisov, S.M.; Wolfbeis, O.S. Optical Biosensors. Chem. Rev. 2008, 108, 423–461. [Google Scholar] [CrossRef]
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta 2008, 620, 8–26. [Google Scholar] [CrossRef]
- Cooper, M.A. Optical biosensors in drug discovery. Nat. Rev. Drug Discov. 2002, 1, 515–528. [Google Scholar] [CrossRef]
- Wolfbeis, O.S. Materials for fluorescence-based optical chemical sensors. J. Mater. Chem. 2005, 15, 2657. [Google Scholar] [CrossRef]
- Giuliano, K.A.; Taylor, D.L. Fluorescent-protein biosensors: New tools for drug discovery. Trends Biotechnol. 1998, 16, 135–140. [Google Scholar] [CrossRef]
- Girigoswami, K.; Akhtar, N. Nanobiosensors and fluorescence based biosensors: An overview. Int. J. Nano Dimens. 2019, 10, 1–17. [Google Scholar]
- Khansili, N.; Rattu, G.; Krishna, P.M. Label-free optical biosensors for food and biological sensor applications. Sens. Actuators B Chem. 2018, 265, 35–49. [Google Scholar] [CrossRef]
- Sang, S.; Wang, Y.; Feng, Q.; Wei, Y.; Ji, J.; Zhang, W. Progress of New Label-Free Techniques for Biosensors: A Review. Crit. Rev. Biotechnol. 2016, 36, 465–481. [Google Scholar] [CrossRef] [PubMed]
- Liedberg, B.; Lundström, I.; Stenberg, E. Principles of biosensing with an extended coupling matrix and surface plasmon resonance. Sens. Actuators B Chem. 1993, 11, 63–72. [Google Scholar] [CrossRef]
- Bo, L.; Claes, N.; Ingemar, L. Surface plasmon resonance for gas detection and biosensing. Sens. Actuators 1983, 4, 299–304. [Google Scholar]
- Homola, J. Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef]
- Nguyen, H.H.; Park, J.; Kang, S.; Kim, M. Surface Plasmon Resonance: A Versatile Technique for Biosensor Applications. Sensors 2015, 15, 10481–10510. [Google Scholar] [CrossRef]
- Meza-Sánchez, D.E.; Maravillas-Montero, J.L. Clinical and Biomedical Applications of Surface Plasmon Resonance Systems. Rev. Investig. Clin. 2019, 71, 85–90. [Google Scholar] [CrossRef]
- Szunerits, S.; Spadavecchia, J.; Boukherroub, R. Surface plasmon resonance: Signal amplification using colloidal gold nanoparticles for enhanced sensitivity. Rev. Anal. Chem. 2014, 33, 153–164. [Google Scholar] [CrossRef]
- Homola, J. Present and future of surface plasmon resonance biosensors. Anal. Bioanal. Chem. 2003, 377, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Joshi, G.K.; Deitz-McElyea, S.; Johnson, M.; Mali, S.; Korc, M.; Sardar, R. Highly Specific Plasmonic Biosensors for Ultrasensitive MicroRNA Detection in Plasma from Pancreatic Cancer Patients. Nano Lett. 2014, 14, 6955–6963. [Google Scholar] [CrossRef] [PubMed]
- Joshi, G.K.; Deitz-McElyea, S.; Liyanage, T.; Lawrence, K.; Mali, S.; Sardar, R.; Korc, M. Label-Free Nanoplasmonic-Based Short Noncoding RNA Sensing at Attomolar Concentrations Allows for Quantitative and Highly Specific Assay of MicroRNA-10b in Biological Fluids and Circulating Exosomes. ACS Nano 2015, 9, 11075–11089. [Google Scholar] [CrossRef] [PubMed]
- Helmerhorst, E.; Chandler, D.J.; Nussio, M.; Mamotte, C.D. Real-time and Label-free Bio-sensing of Molecular Interactions by Surface Plasmon Resonance: A Laboratory Medicine Perspective. Clin. Biochem. Rev. 2012, 33, 161–173. [Google Scholar] [PubMed]
- Willets, K.A.; Van Duyne, R.P. Localized Surface Plasmon Resonance Spectroscopy and Sensing. Annu. Rev. Phys. Chem. 2007, 58, 267–297. [Google Scholar] [CrossRef]
- Šípová, H.; Homola, J. Surface plasmon resonance sensing of nucleic acids: A review. Anal. Chim. Acta 2013, 773, 9–23. [Google Scholar] [CrossRef]
- Matsubara, K.; Kawata, S.; Minami, S. Optical chemical sensor based on surface plasmon measurement. Appl. Opt. 1988, 27, 1160–1163. [Google Scholar] [CrossRef]
- Mirabella, F.M. Internal Reflection Spectroscopy. Appl. Spectrosc. Rev. 1985, 21, 45–178. [Google Scholar] [CrossRef]
- Mirabella, F.M. Principles, Theory and Practice of Internal Reflection Spectroscopy. In Handbook of Vibrational Spectroscopy; American Cancer Society: New York, NY, USA, 2006. [Google Scholar]
- Woods, D.A.; Bain, C.D. Total internal reflection spectroscopy for studying soft matter. Soft Matter 2014, 10, 1071. [Google Scholar] [CrossRef]
- Ćípová, H.; Zhang, S.; Dudley, A.M.; Galas, D.; Wang, K.; Homola, J. Surface Plasmon Resonance Biosensor for Rapid Label-Free Detection of Microribonucleic Acid at Subfemtomole Level. Anal. Chem. 2010, 82, 10110–10115. [Google Scholar] [CrossRef]
- Danz, N.; Kick, A.; Sonntag, F.; Schmieder, S.; Hofer, B.; Klotzbach, U.; Mertig, M. Surface plasmon resonance platform technology for multi-parameter analyses on polymer chips. Eng. Life Sci. 2011, 11, 566–572. [Google Scholar] [CrossRef]
- Qavi, A.J.; Bailey, R.C. Multiplexed detection and label-free quantitation of microRNAs using arrays of silicon photonic microring resonators. Angew. Chem. Int. Ed. 2010, 49, 4608–4611. [Google Scholar] [CrossRef] [PubMed]
- Estevez, M.-C.; Otte, M.A.; Sepúlveda, B.; Lechuga, L.M. Trends and challenges of refractometric nanoplasmonic biosensors: A review. Anal. Chim. Acta 2014, 806, 55–73. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Xu, J.; Chen, Y. Surface Plasmon Resonance Imaging Detection of Sub-femtomolar MicroRNA. Anal. Chem. 2017, 89, 10071–10077. [Google Scholar] [CrossRef] [PubMed]
- Tokel, O.; Inci, F.; Demirci, U. Advances in Plasmonic Technologies for Point of Care Applications. Chem. Rev. 2014, 114, 5728–5752. [Google Scholar] [CrossRef]
- Fang, S.; Lee, H.J.; Wark, A.W.; Corn, R.M. Attomole Microarray Detection of MicroRNAs by Nanoparticle-Amplified SPR Imaging Measurements of Surface Polyadenylation Reactions. J. Am. Chem. Soc. 2006, 128, 14044–14046. [Google Scholar] [CrossRef]
- Qavi, A.J.; Kindt, J.T.; Gleeson, M.A.; Bailey, R.C. Anti-DNA:RNA Antibodies and Silicon Photonic Microring Resonators: Increased Sensitivity for Multiplexed microRNA Detection. Anal. Chem. 2011, 83, 5949–5956. [Google Scholar] [CrossRef]
- Liang, L.; Jin, L.; Ran, Y.; Sun, L.-P.; Guan, B.-O. Interferometric detection of microRNAs using a capillary optofluidic sensor. Sens. Actuators B Chem. 2017, 242, 999–1006. [Google Scholar] [CrossRef]
- Nicolosi, V.; Chhowalla, M.; Kanatzidis, M.G.; Strano, M.S.; Coleman, J.N. Liquid Exfoliation of Layered Materials. Science 2013, 340, 1226419. [Google Scholar] [CrossRef]
- Bender, M. The use of light scattering for determining particle size and molecular weight and shape. J. Chem. Educ. 1952, 29, 15. [Google Scholar] [CrossRef]
- Manera, M.G.; Montagna, G.; Ferreiro-Vila, E.; Gonzalez-Garcia, L.; Sanchez-Valencia, J.R.; Gonzalez-Elipe, A.; Cebollada, A.; Garcia-Martin, J.M.; Armelles, G.; Rella, R. Enhanced gas sensing performance of TiO2 functionalized magneto-optical SPR sensors. J. Mater. Chem. 2011, 21, 16049. [Google Scholar] [CrossRef]
- Ruan, B.; You, Q.; Zhu, J.; Wu, L.; Guo, J.; Dai, X.; Xiang, Y. Improving the Performance of an SPR Biosensor Using Long-Range Surface Plasmon of Ga-Doped Zinc Oxide. Sensors 2018, 18, 2098. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Lee, Y.H.; Koh, C.S.L.; Phan-Quang, G.C.; Han, X.; Lay, C.L.; Sim, H.Y.F.; Kao, Y.-C.; An, Q.; Ling, X.Y. Designing surface-enhanced Raman scattering (SERS) platforms beyond hotspot engineering: Emerging opportunities in analyte manipulations and hybrid materials. Chem. Soc. Rev. 2019, 48, 731–756. [Google Scholar] [CrossRef] [PubMed]
- Cardinal, M.F.; Ende, E.V.; Hackler, R.A.; McAnally, M.O.; Stair, P.C.; Schatz, G.C.; Van Duyne, R.P. Expanding applications of SERS through versatile nanomaterials engineering. Chem. Soc. Rev. 2017, 46, 3886–3903. [Google Scholar] [CrossRef] [PubMed]
- Yeatman, E.; Ash, E.A. Surface plasmon microscopy. Electron. Lett. 1987, 23, 1091–1092. [Google Scholar] [CrossRef]
- Rothenhäusler, B.; Knoll, W. Surface—Plasmon microscopy. Nature 1988, 332, 615–617. [Google Scholar] [CrossRef]
- Spoto, G.; Minunni, M. Surface Plasmon Resonance Imaging: What Next? J. Phys. Chem. Lett. 2012, 3, 2682–2691. [Google Scholar] [CrossRef]
- Puiu, M.; Bala, C. SPR and SPR Imaging: Recent Trends in Developing Nanodevices for Detection and Real-Time Monitoring of Biomolecular Events. Sensors 2016, 16, 870. [Google Scholar] [CrossRef]
- Haes, A.J.; Hall, W.P.; Chang, L.; Klein, W.L.; Van Duyne, R.P. A Localized Surface Plasmon Resonance Biosensor: First Steps toward an Assay for Alzheimer’s Disease. Nano Lett. 2004, 4, 1029–1034. [Google Scholar] [CrossRef]
- Petryayeva, E.; Krull, U.J. Localized surface plasmon resonance: Nanostructures, bioassays and biosensing—A review. Anal. Chim. Acta 2011, 706, 8–24. [Google Scholar] [CrossRef]
- Bozzola, A.; Perotto, S.; De Angelis, F. Hybrid plasmonic–photonic whispering gallery mode resonators for sensing: A critical review. Analyst 2017, 142, 883–898. [Google Scholar] [CrossRef] [PubMed]
- Kazuma, E.; Tatsuma, T. Localized surface plasmon resonance sensors based on wavelength-tunable spectral dips. Nanoscale 2014, 6, 2397–2405. [Google Scholar] [CrossRef] [PubMed]
- Prabowo, B.A.; Purwidyantri, A.; Liu, K.-C. Surface Plasmon Resonance Optical Sensor: A Review on Light Source Technology. Biosensors 2018, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zheng, D.; Tan, Q.; Wang, M.X.; Gu, L.-Q. Nanopore-Based Detection of Circulating MicroRNAs in Lung Cancer Patients. Nat. Nanotechnol. 2011, 6, 668–674. [Google Scholar] [CrossRef] [PubMed]
- Kozma, P.; Kehl, F.; Ehrentreich-Förster, E.; Stamm, C.; Bier, F.F. Integrated Planar Optical Waveguide Interferometer Biosensors: A Comparative Review. Biosens. Bioelectron. 2014, 58, 287–307. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Tu, X.; Kim, K.W.; Kee, J.S.; Shin, Y.; Han, K.; Yoon, Y.-J.; Lo, G.-Q.; Park, M.K. Highly Sensitive Mach–Zehnder Interferometer Biosensor Based on Silicon Nitride Slot Waveguide. Sensors Actuators B Chem. 2013, 188, 681–688. [Google Scholar] [CrossRef]
- Tu, X.; Song, J.; Liow, T.-Y.; Park, M.K.; Yiying, J.Q.; Kee, J.S.; Yu, M.; Lo, G.-Q. Thermal Independent Silicon-Nitride Slot Waveguide Biosensor with High Sensitivity. Opt. Express 2012, 20, 2640–2648. [Google Scholar] [CrossRef]
- Vahala, K.J. Optical Microcavities. Nature 2003, 424, 839–846. [Google Scholar] [CrossRef]
- Vollmer, F.; Arnold, S.; Keng, D. Single Virus Detection from the Reactive Shift of a Whispering-Gallery Mode. Proc. Natl. Acad. Sci. USA 2008, 105, 20701–20704. [Google Scholar] [CrossRef]
- Jiang, X.; Qavi, A.J.; Huang, S.H.; Yang, L. Whispering Gallery Microsensors: A Review. arXiv e-prints 2018, arXiv:1805.00062. [Google Scholar]
- Zhu, J.; Ozdemir, S.K.; Xiao, Y.-F.; Li, L.; He, L.; Chen, D.-R.; Yang, L. On-Chip Single Nanoparticle Detection and Sizing by Mode Splitting in an Ultrahigh-Q Microresonator. Nat. Photonics 2009, 4, 46. [Google Scholar] [CrossRef]
- Shao, L.; Jiang, X.-F.; Yu, X.-C.; Li, B.-B.; Clements, W.R.; Vollmer, F.; Wang, W.; Xiao, Y.-F.; Gong, Q. Detection of Single Nanoparticles and Lentiviruses Using Microcavity Resonance Broadening. Adv. Mater. 2013, 25, 5616–5620. [Google Scholar] [CrossRef] [PubMed]
- Guan, G.; Arnold, S.; Otugen, V. Temperature Measurements Using a Microoptical Sensor Based on Whispering Gallery Modes. AIAA J. 2006, 44, 2385–2389. [Google Scholar] [CrossRef]
- Frenkel, M.; Avellan, M.; Guo, Z. Whispering-Gallery Mode Composite Sensors for on-Chip Dynamic Temperature Monitoring. Meas. Sci. Technol. 2013, 24, 75103. [Google Scholar] [CrossRef]
- Ioppolo, T.; Ötügen, M.V. Pressure Tuning of Whispering Gallery Mode Resonators. J. Opt. Soc. Am. B 2007, 24, 2721–2726. [Google Scholar] [CrossRef]
- Mahmood, A.; Kavungal, V.; Ahmed, S.S.; Kopcansky, P.; Zavisova, V.; Farrell, G.; Semenova, Y. Magnetic Field Sensing Using Whispering-Gallery Modes in a Cylindrical Microresonator Infiltrated with Ferronematic Liquid Crystal. Opt. Express 2017, 25, 12195–12202. [Google Scholar] [CrossRef] [PubMed]
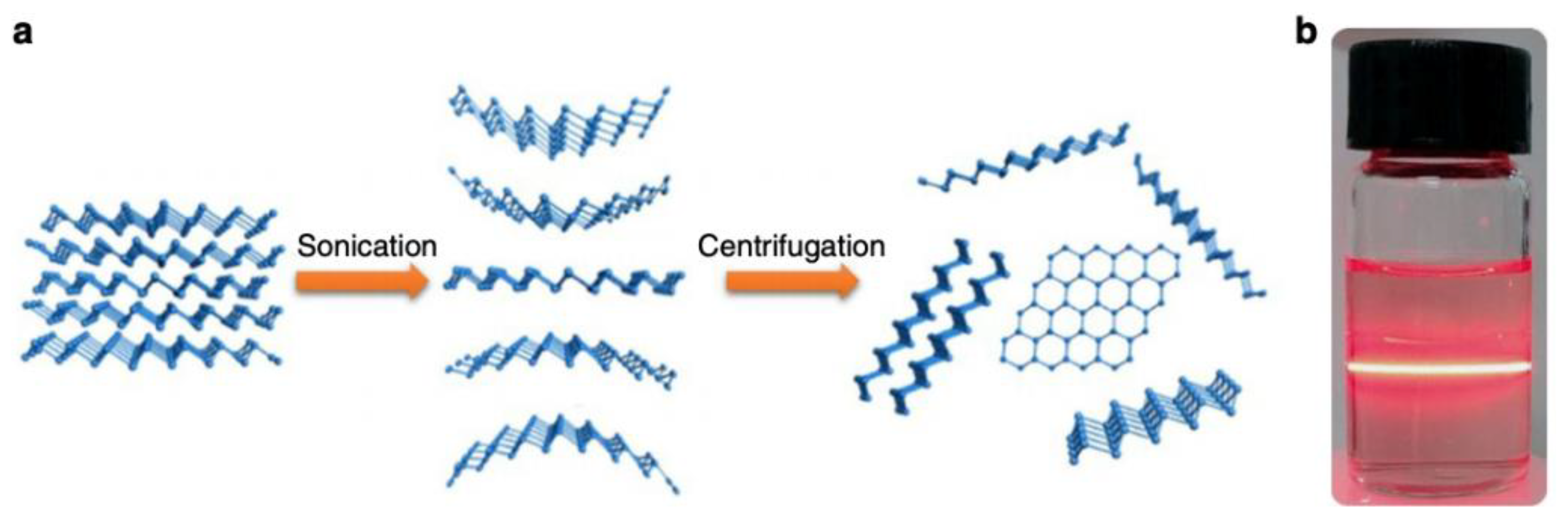
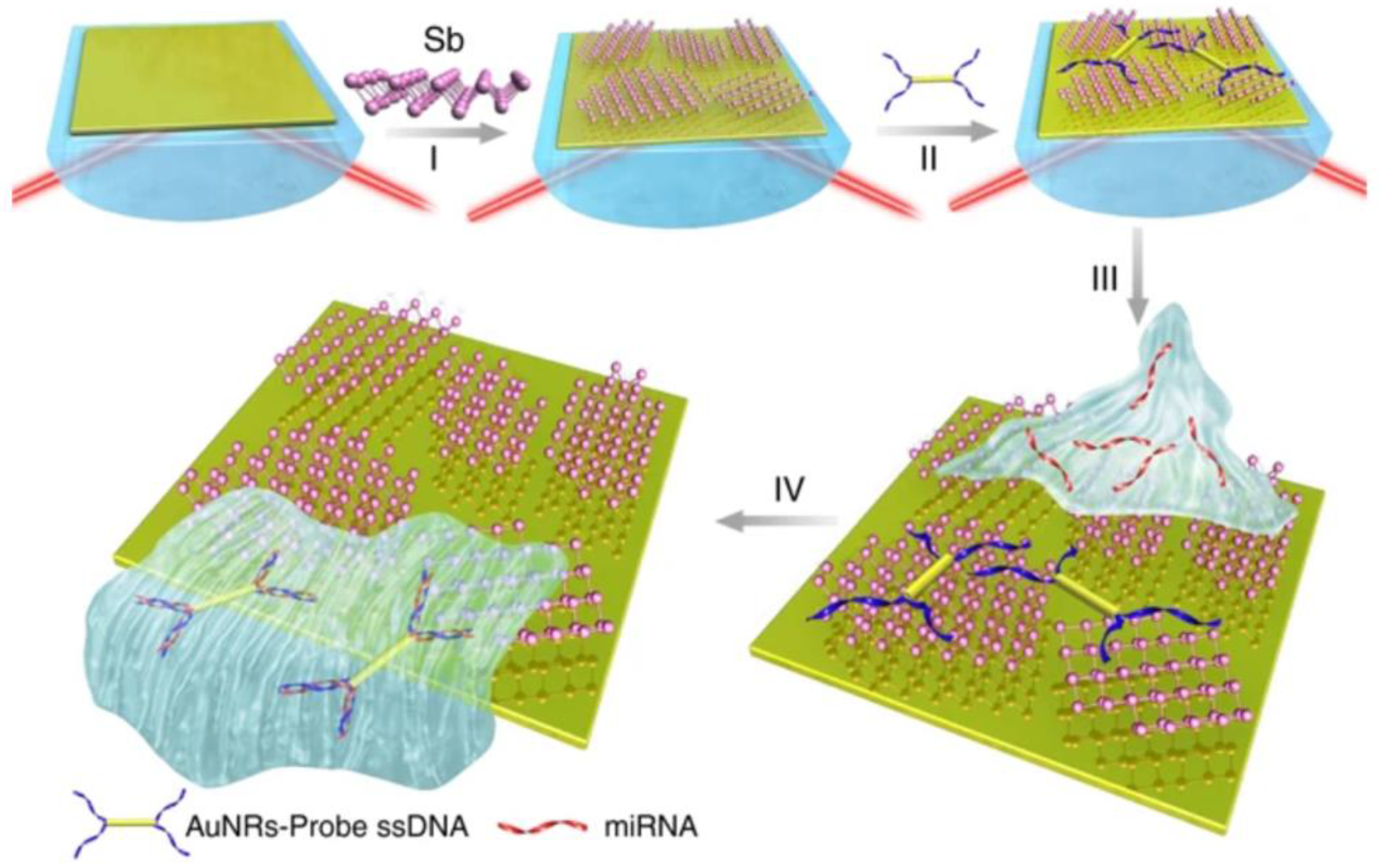
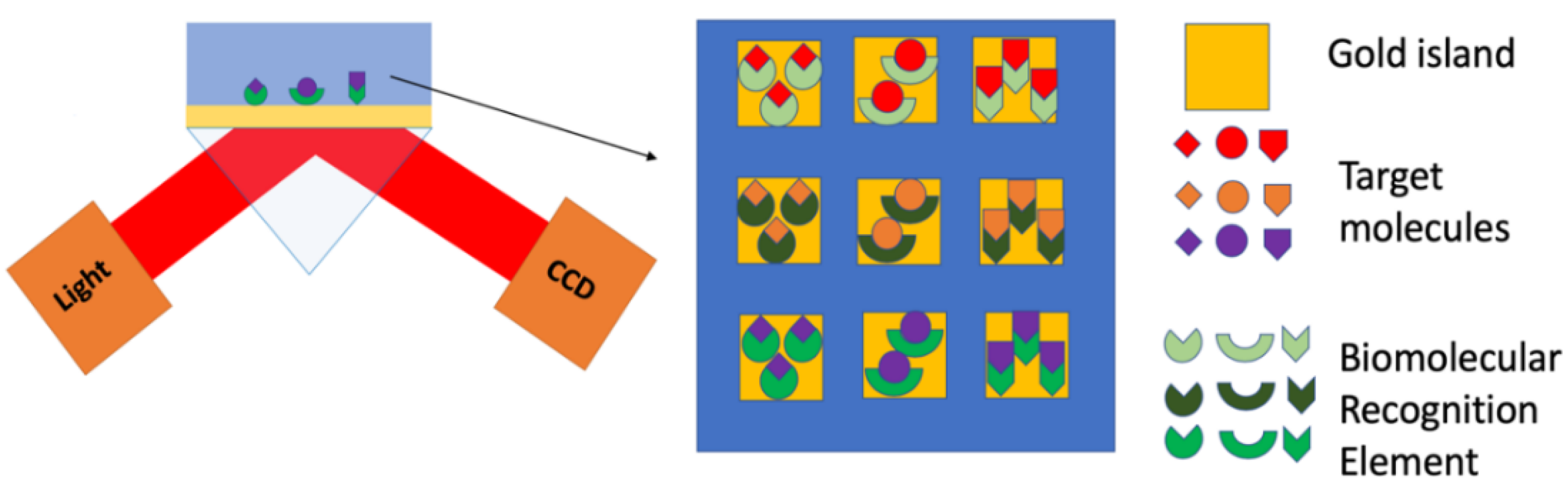
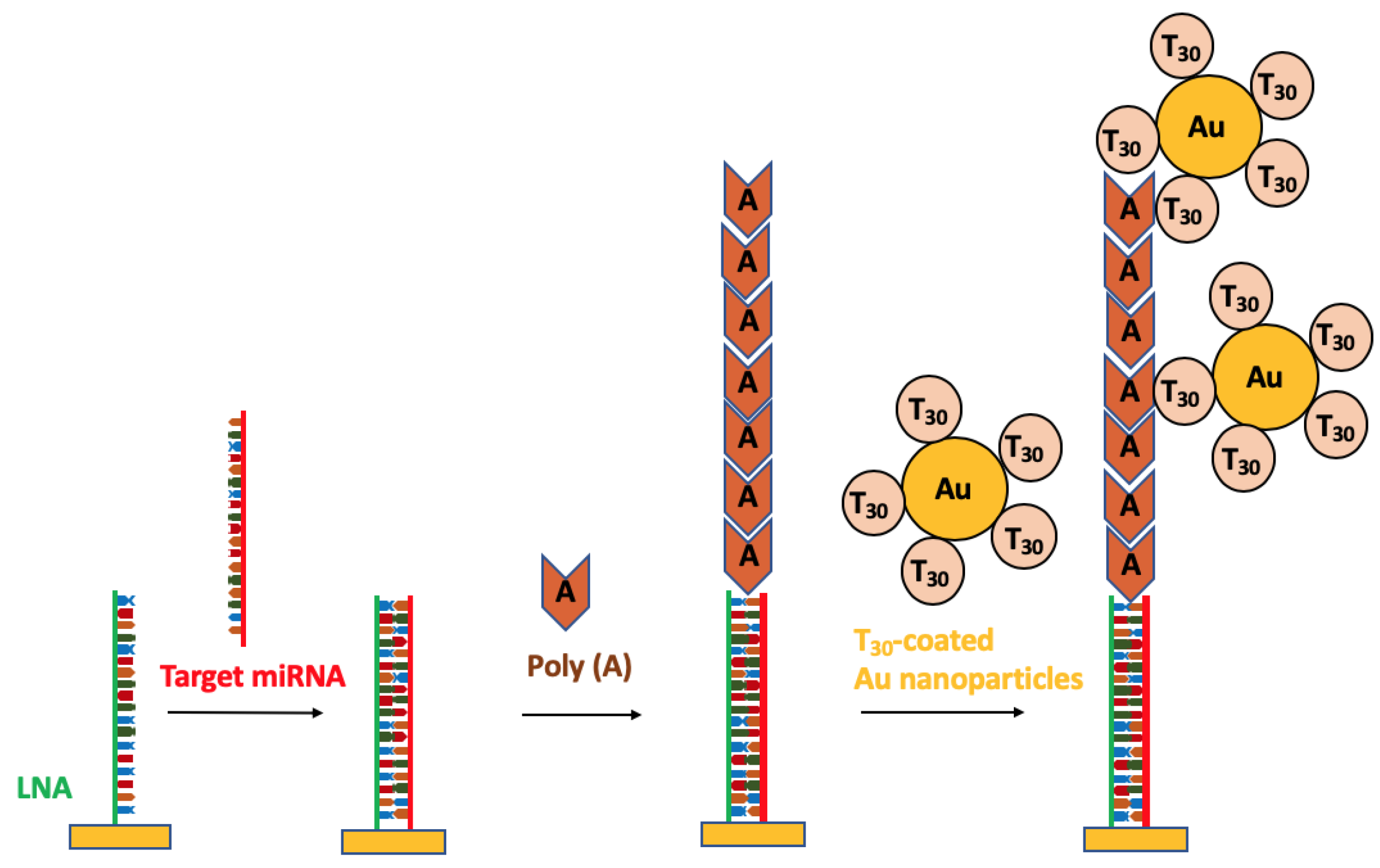
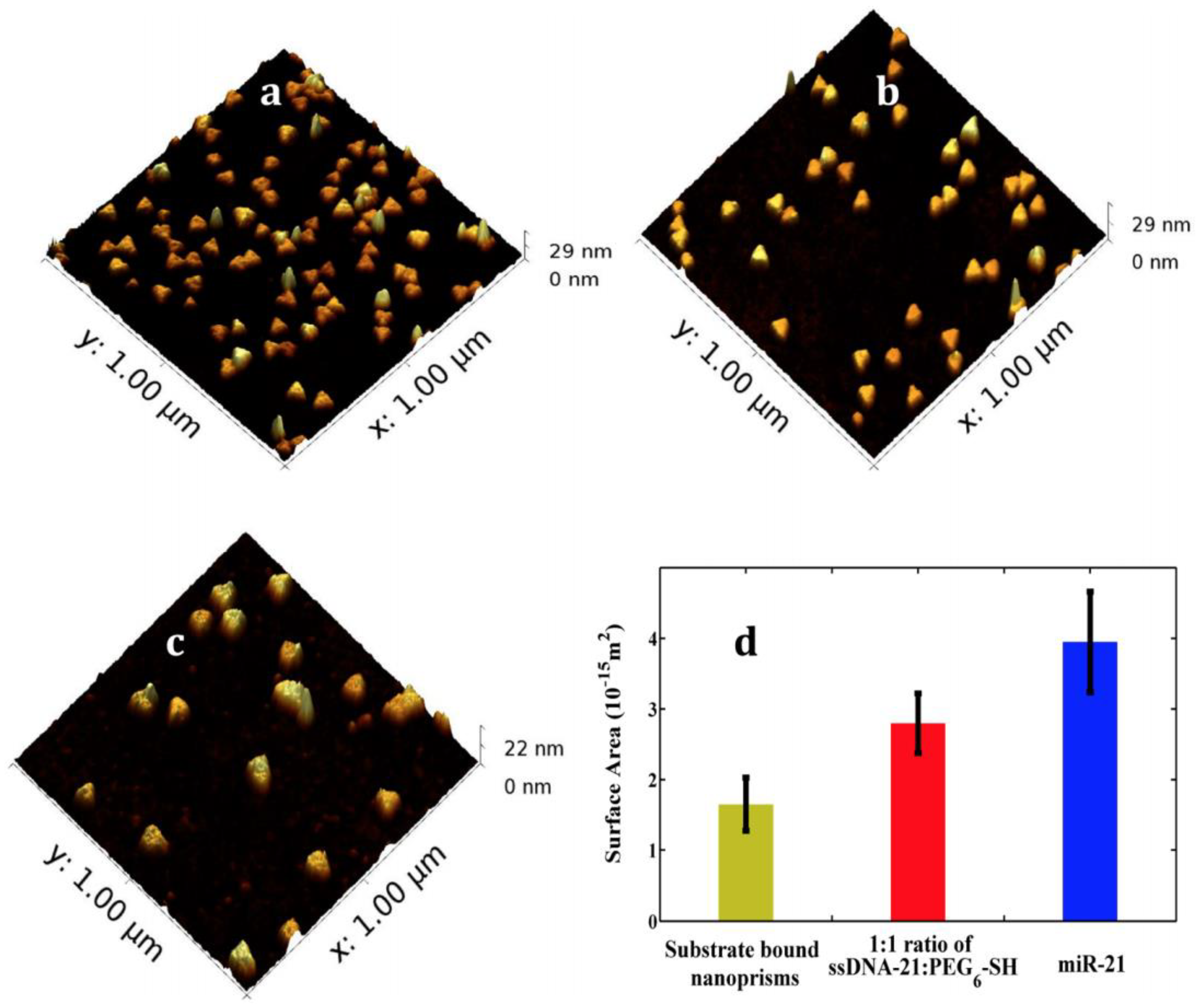
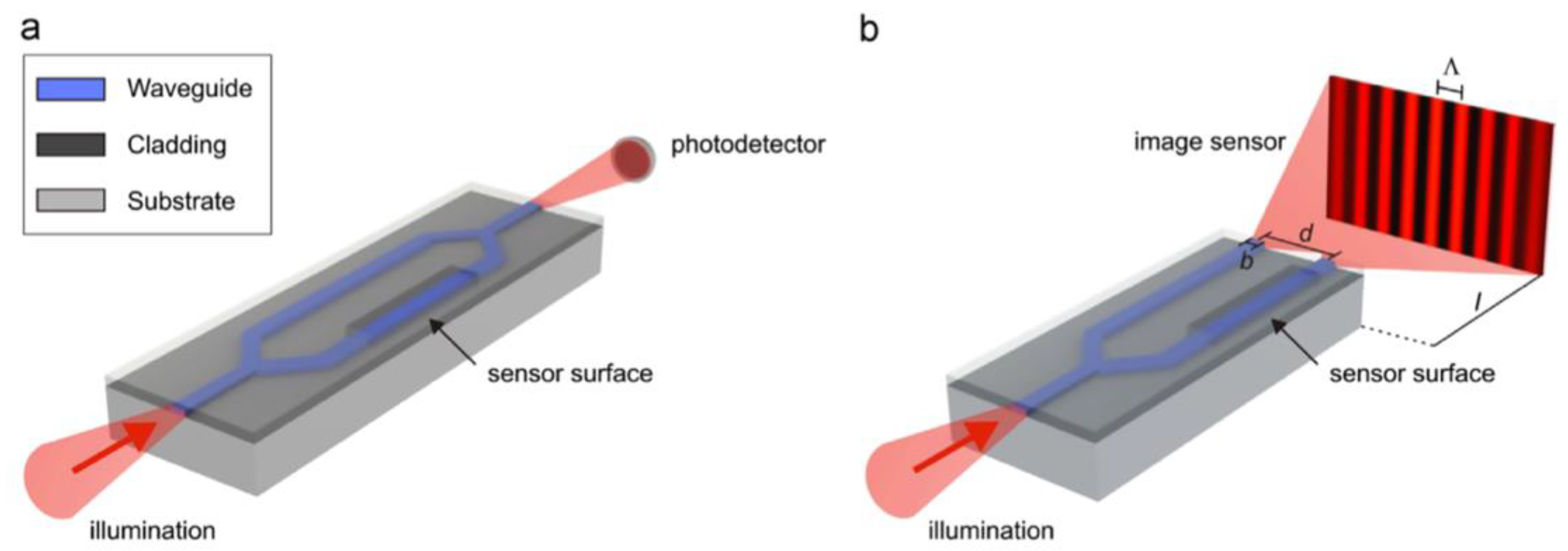
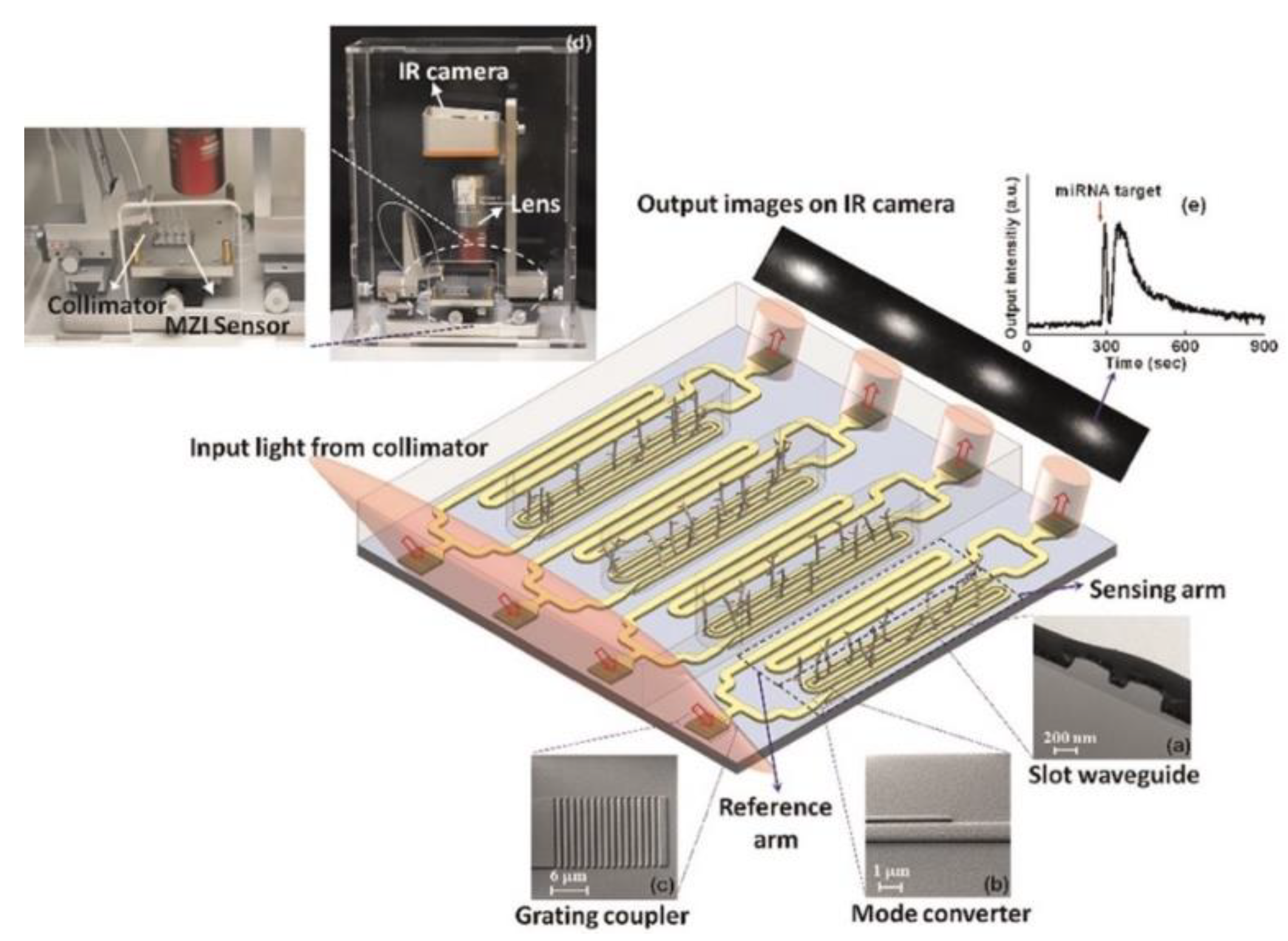
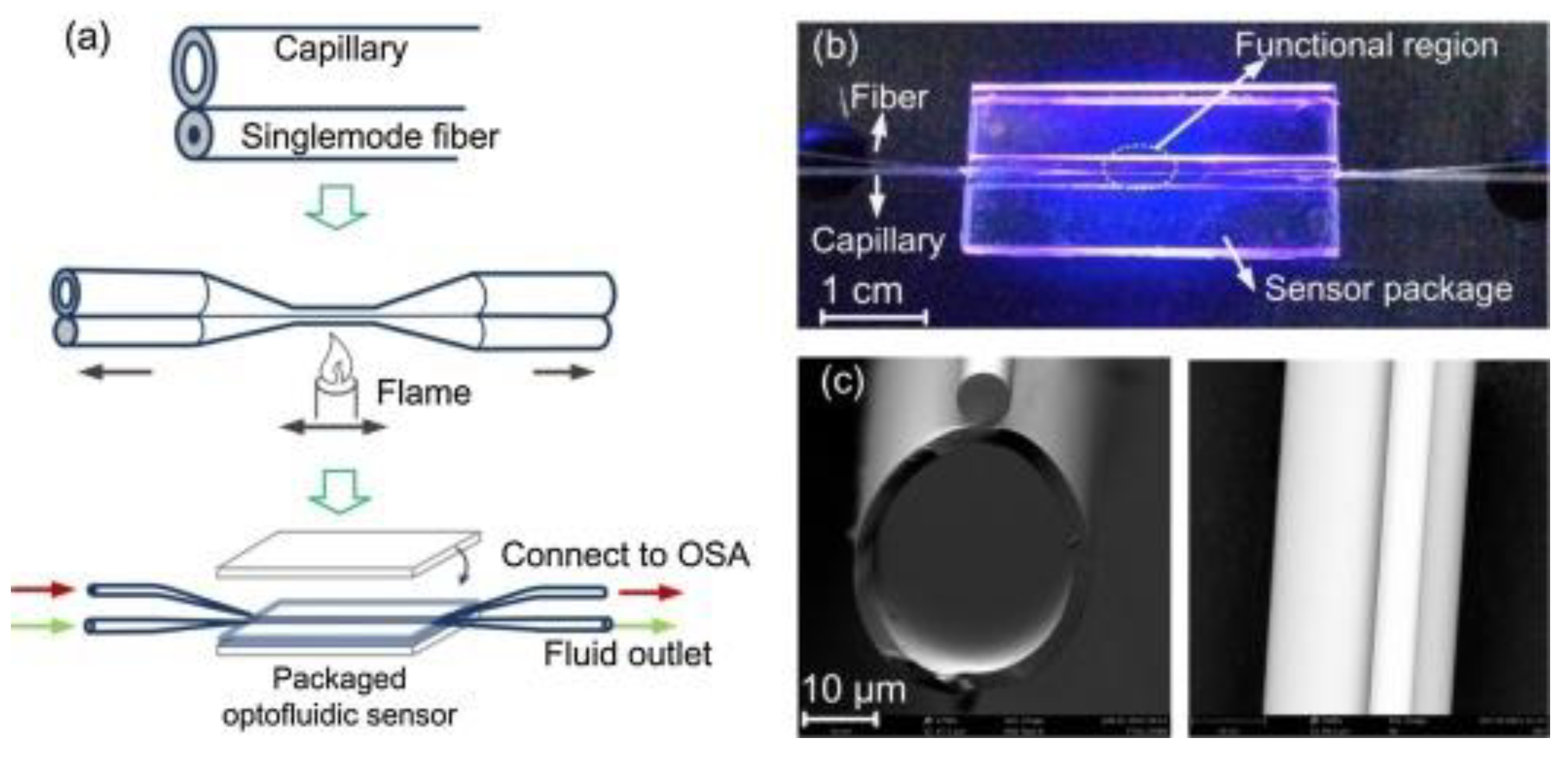
| miRNA Type | Sample Source | Method | LOD | Sample Volume | Reference |
|---|---|---|---|---|---|
| miR-16, miR-23b, and miR-122b | mouse liver tissue | SPRi | 10 fM | 500 µL | Fang 2006 [77] |
| miR-133b, miR-21, miR-24-1, and let-7c | U87 MG cells | microrings | 150 fM | 75 µL. | Qavi 2010 [73] |
| miR-16, miR-21, miR-24-1, and miR-26a | total mouse brain RNA | microrings | 10 pM | 35 μL | Qavi 2011 [78] |
| miR-21, and let-7a | clinical urine samples | MZI | 1 nM | 70 μL (PBS) 200 mL (urine) | Liu 2015 [31] |
| miR-let7a | synthetic RNA | MZI | 202 pM | -- | Liang 2017 [79] |
| miR-122 | mouse liver tissue | SPR | 2 pM | 440 μL | Sipova 2010 [71] |
| miR-21 and miR-10b | human plasma | LSPR | 23–35 fM | 50 µL | Joshi 2014 [62] |
| miR-10b | human plasma | LSPR | aM | 2.5 mL | Joshi 2015 [63] |
| miR-15a | Human plasma | SPRi | 0.56 fM | Hu 2017 [75] | |
| miR-93 | synthetic RNA | SPR | 10 pM | 25 μl | Schmieder 2016 [25] |
| miR-21and miR-155 | synthetic RNA | LSPR | 10 aM | 5 μL | Xue 2019 [44] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, M.; Slaughter, G. Label-Free MicroRNA Optical Biosensors. Nanomaterials 2019, 9, 1573. https://doi.org/10.3390/nano9111573
Lai M, Slaughter G. Label-Free MicroRNA Optical Biosensors. Nanomaterials. 2019; 9(11):1573. https://doi.org/10.3390/nano9111573
Chicago/Turabian StyleLai, Meimei, and Gymama Slaughter. 2019. "Label-Free MicroRNA Optical Biosensors" Nanomaterials 9, no. 11: 1573. https://doi.org/10.3390/nano9111573
APA StyleLai, M., & Slaughter, G. (2019). Label-Free MicroRNA Optical Biosensors. Nanomaterials, 9(11), 1573. https://doi.org/10.3390/nano9111573






