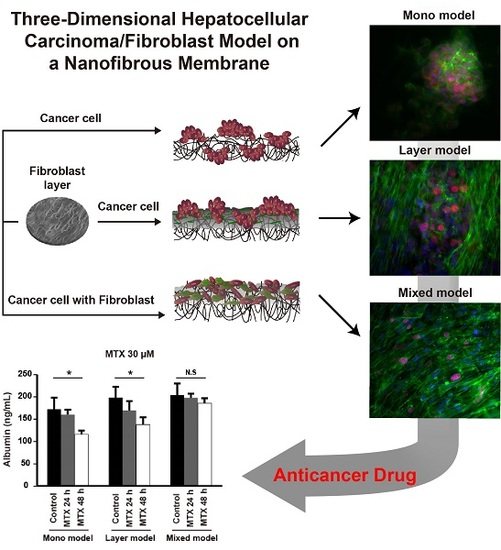
Three-Dimensional Hepatocellular Carcinoma/Fibroblast Model on a Nanofibrous Membrane Mimics Tumor Cell Phenotypic Changes and Anticancer Drug Resistance
Abstract
:1. Introduction
2. Results
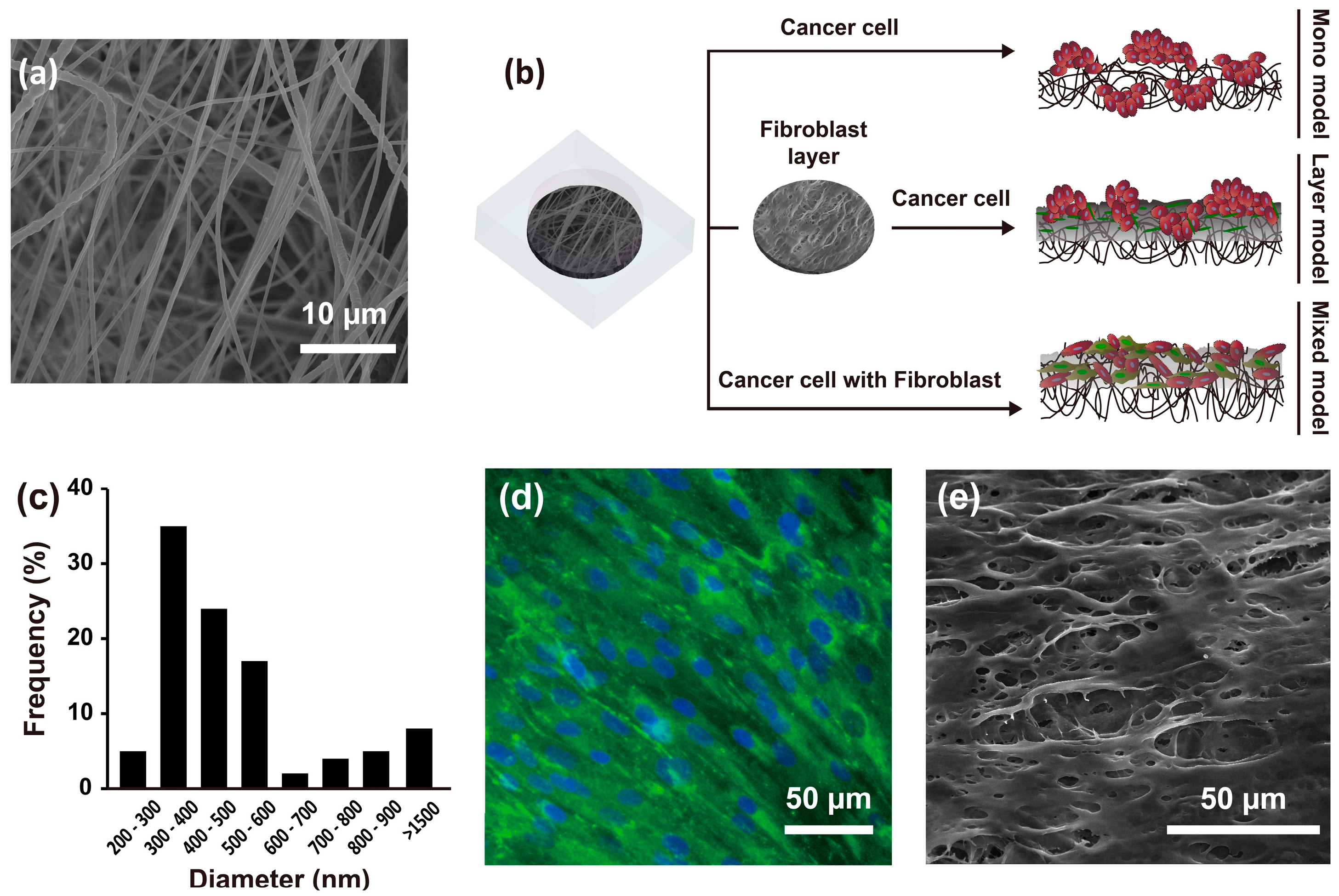
2.1. Experimental Design and Characteristics of the Nanofibrous Membrane
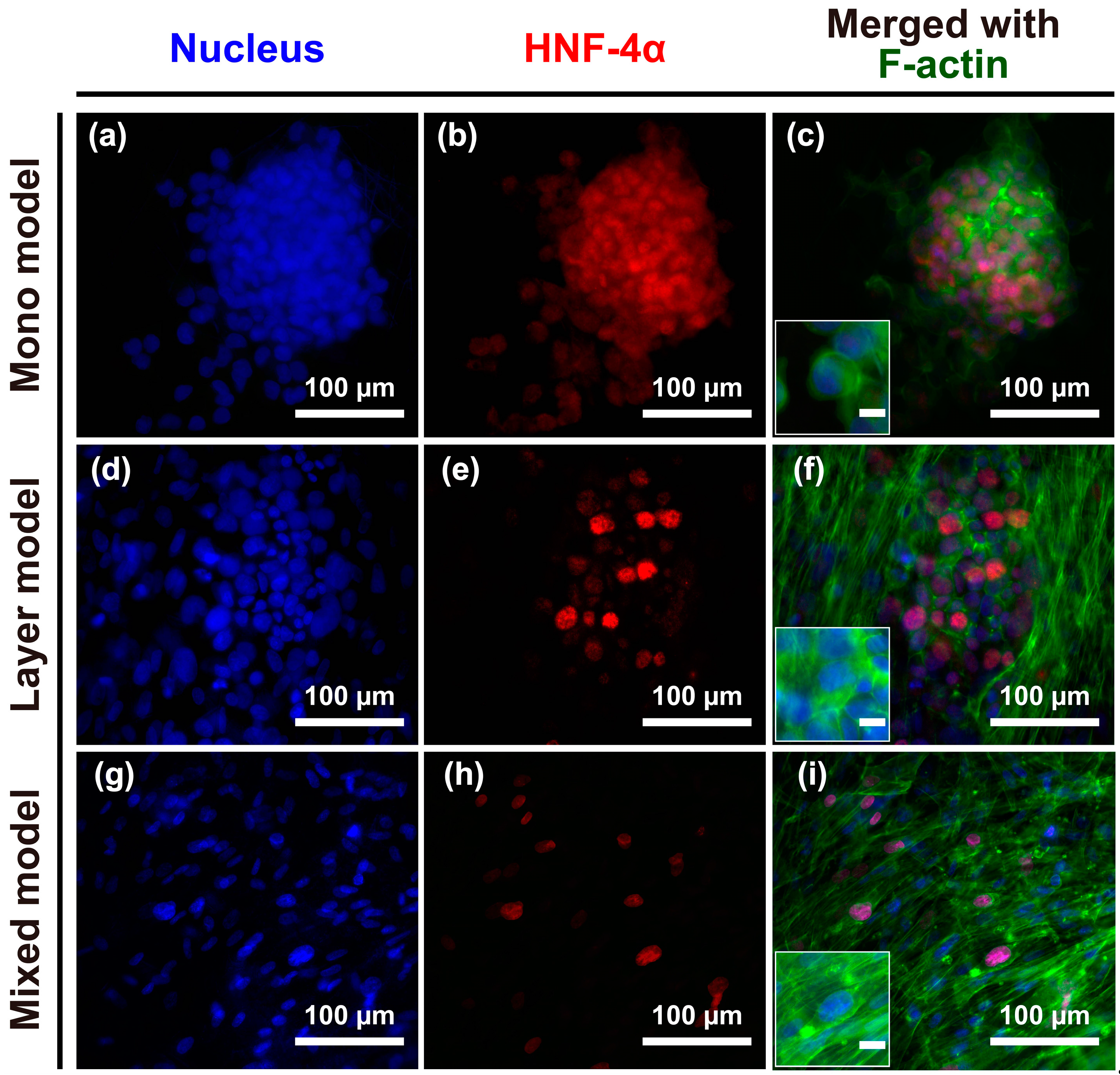
2.2. Morphological Characteristics of Hep G2 Cells and Fibroblasts
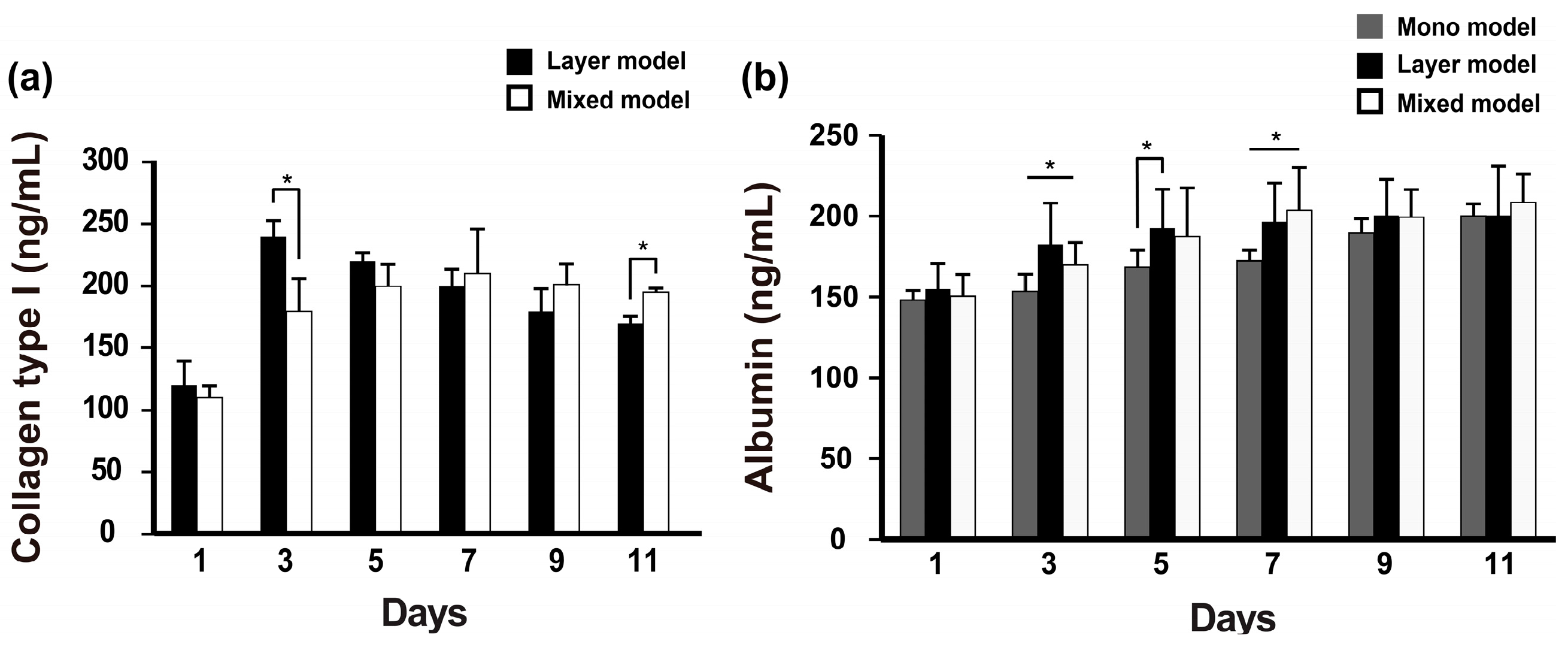
2.3. Albumin and Collagen Secretion
2.4. Cell-Cell Junctions in Hep G2 Cells
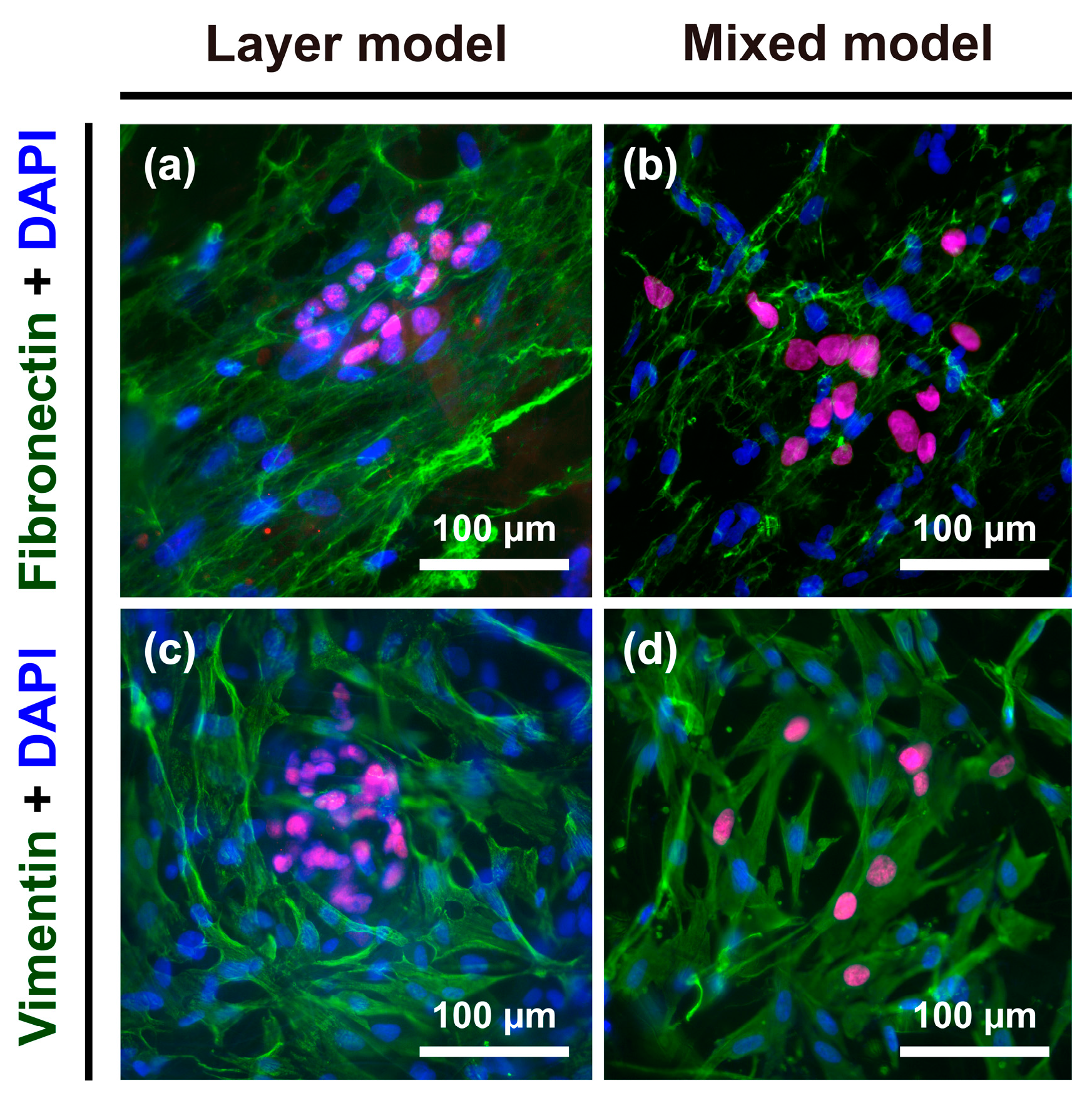
2.5. Vimentin and Fibronectin Expression
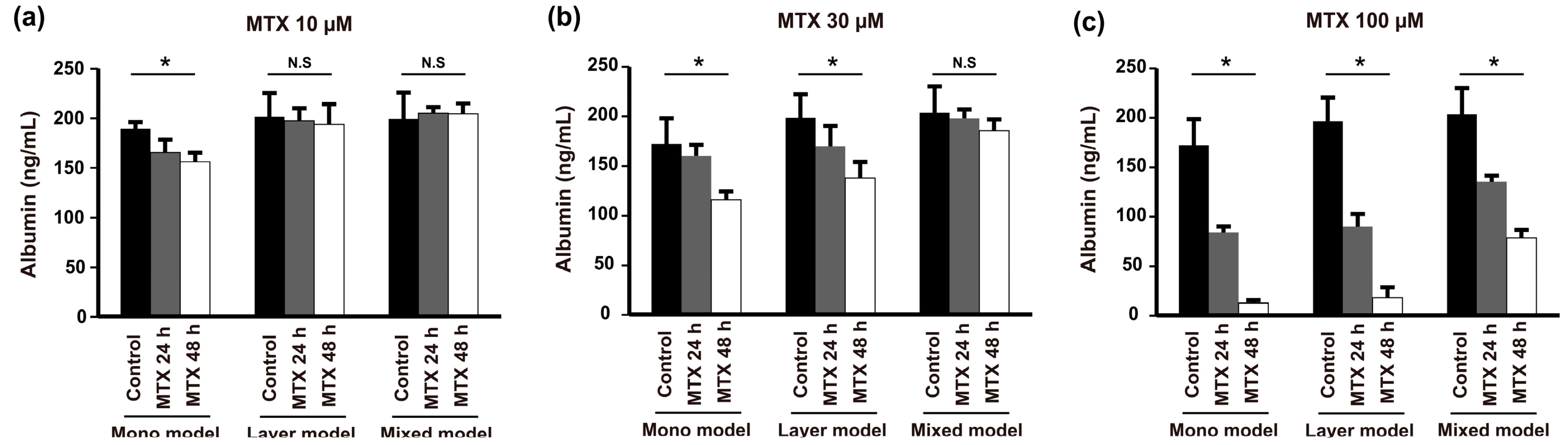
2.6. Anticancer Drug Resistance
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Electrospinning Method to Fabricate the Polycaprolactone (PCL) Nanofibrous Membrane
4.3. Characterization of the Nanofibrous Membrane
4.4. Cell Culture
4.5. Immunocytochemistry
4.6. Measurement of Albumin and Collagen Type I Levels Using Enzyme-Linked Immunosorbent Assay (ELISA) and Enzyme Immunoassay (EIA)
4.7. Methotrexate (MTX) Drug Screening
4.8. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef] [PubMed]
- Hugo, H.; Ackland, M.L.; Blick, T.; Lawrence, M.G.; Clements, J.A.; Williams, E.D.; Thompson, E.W. Epithelial—Mesenchymal and mesenchymal—Epithelial transitions in carcinoma progression. J. Cell Physiol. 2007, 213, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Zeisberg, M. Fibroblasts in cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Akhurst, R.J. Differentiation plasticity regulated by TFG-β family proteins in development and disease. Nat. Cell Biol. 2007, 9, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- Pannu, J.; Nakerakanti, S.; Smith, E.; ten Dijke, P.; Trojanowska, M. Transforming Growth Factor-β Receptor Type I-dependent Fibrogenic Gene Program Is Mediated via Activation of Smad1 and ERK1/2 Pathways. J. Biol. Chem. 2007, 282, 10405–10413. [Google Scholar] [CrossRef] [PubMed]
- Van Meeteren, L.A.; Ten Dijke, P. Regulation of endothelial cell plasticity by TFG-β. Cell Tissue Res. 2012, 347, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Ramos, C.; Becerril, C.; Montaño, M.; García-De-Alba, C.; Ramírez, R.; Checa, M.; Pardo, A.; Selman, M. FGF-1 reverts epithelial-mesenchymal transition induced by TGF-β1 through MAPK/ERK kinase pathway. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2010, 299, L222–L231. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Duan, X.; Wu, L.; Lo, P.-K.; Chen, H.; Wang, Q. Electrospun Fibrous Scaffolds Promote Breast Cancer Cell Alignment and Epithelial–Mesenchymal Transition. Langmuir 2011, 28, 2028–2034. [Google Scholar] [CrossRef] [PubMed]
- McLane, J.S.; Rivet, C.J.; Gilbert, R.J.; Ligon, L.A. A biomaterial model of tumor stromal microenvironment promotes mesenchymal morphology but not epithelial to mesenchymal transition in epithelial cells. Acta Biomater. 2014, 10, 4811–4821. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Fu, F.; Shang, L.; Cheng, Y.; Gu, Z.; Zhao, Y. Bioinspired helical microfibers from microfluidics. Adv. Mater. 2017, 29, 1605765. [Google Scholar] [CrossRef] [PubMed]
- Tamiya, S.; Liu, L.; Kaplan, H.J. Epithelial-mesenchymal transition and proliferation of retinal pigment epithelial cells initiated upon loss of cell-cell contact. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2755–2763. [Google Scholar] [CrossRef] [PubMed]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.-M.; Barbone, D.; Fennell, D.A.; Broaddus, V.C. Bcl-2 Family Proteins Contribute to Apoptotic Resistance in Lung Cancer Multicellular Spheroids. Am. J. Respir. Cell Mol. Biol. 2009, 41, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Hirschhaeuser, F.; Menne, H.; Dittfeld, C.; West, J.; Mueller-Klieser, W.; Kunz-Schughart, L.A. Multicellular tumor spheroids: An underestimated tool is catching up again. J. Biotechnol. 2010, 148, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.M.; Cukierman, E. Modeling tissue morphogenesis and cancer in 3D. Cell 2007, 130, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Yip, D.; Cho, C.H. A multicellular 3D heterospheroid model of liver tumor and stromal cells in collagen gel for anti-cancer drug testing. Biochem. Biophys. Res. Commun. 2013, 433, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, Y. Pharma firms push for sharing of cancer trial data. Science 2012, 338, 29. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ding, B.; Li, B. Biomimetic electrospun nanofibrous structures for tissue engineering. Mater. Today 2013, 16, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Nasrollahi, S.; Pathak, A. Topographic confinement of epithelial clusters induces epithelial-to-mesenchymal transition in compliant matrices. Sci. Rep. 2016, 6, 18831. [Google Scholar] [CrossRef] [PubMed]
- Onder, T.T.; Gupta, P.B.; Mani, S.A.; Yang, J.; Lander, E.S.; Weinberg, R.A. Loss of e-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008, 68, 3645–3654. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Dedhar, S.; Kalluri, R.; Thompson, E.W. The epithelial–mesenchymal transition: New insights in signaling, development, and disease. J. Cell Biol. 2006, 172, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, C.; Kong, H.J.; Hsiong, S.X.; Evangelista, M.B.; Yuen, W.; Mooney, D.J. Cancer cell angiogenic capability is regulated by 3D culture and integrin engagement. Proc. Natl. Acad. Sci. USA 2009, 106, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Dolznig, H.; Rupp, C.; Puri, C.; Haslinger, C.; Schweifer, N.; Wieser, E.; Kerjaschki, D.; Garin-Chesa, P. Modeling colon adenocarcinomas in vitro: A 3d Co-Culture System Induces Cancer-Relevant Pathways upon Tumor Cell and Stromal Fibroblast Interaction. Am. J. Pathol. 2011, 179, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Riedl, A.; Schlederer, M.; Pudelko, K.; Stadler, M.; Walter, S.; Unterleuthner, D.; Unger, C.; Kramer, N.; Hengstschläger, M.; Kenner, L.; et al. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT–mTOR–S6K signaling and drug responses. J. Cell Sci. 2017, 130, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yao, R.; Ouyang, L.; Ding, H.; Zhang, T.; Zhang, K.; Cheng, S.; Sun, W. Three-dimensional printing of hela cells for cervical tumor model in vitro. Biofabrication 2014, 6, 035001. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Mooney, D.J. Biomaterials and emerging anticancer therapeutics: Engineering the microenvironment. Nat. Rev. Cancer 2016, 16, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Inman, D.R.; Eliceiri, K.W.; Knittel, J.G.; Yan, L.; Rueden, C.T.; White, J.G.; Keely, P.J. Collagen density promotes mammary tumor initiation and progression. BMC Med. 2008, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.; Kim, J.H.; Jeong, Y.H.; Kwak, J.-Y.; Yoon, S.; Jin, S. Endothelial monolayers on collagen-coated nanofibrous membranes: Cell-cell and cell-ecm interactions. Biofabrication 2016, 8, 025008. [Google Scholar] [CrossRef] [PubMed]
- Hotaling, N.A.; Bharti, K.; Kriel, H.; Simon, C.G. Diameterj: A validated open source nanofiber diameter measurement tool. Biomaterials 2015, 61, 327–338. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, B.D.; Kang, D.; Yun, S.; Jeong, Y.H.; Kwak, J.-Y.; Yoon, S.; Jin, S. Three-Dimensional Hepatocellular Carcinoma/Fibroblast Model on a Nanofibrous Membrane Mimics Tumor Cell Phenotypic Changes and Anticancer Drug Resistance. Nanomaterials 2018, 8, 64. https://doi.org/10.3390/nano8020064
Le BD, Kang D, Yun S, Jeong YH, Kwak J-Y, Yoon S, Jin S. Three-Dimensional Hepatocellular Carcinoma/Fibroblast Model on a Nanofibrous Membrane Mimics Tumor Cell Phenotypic Changes and Anticancer Drug Resistance. Nanomaterials. 2018; 8(2):64. https://doi.org/10.3390/nano8020064
Chicago/Turabian StyleLe, Binh Duong, Donggu Kang, Seokhwan Yun, Young Hun Jeong, Jong-Young Kwak, Sik Yoon, and Songwan Jin. 2018. "Three-Dimensional Hepatocellular Carcinoma/Fibroblast Model on a Nanofibrous Membrane Mimics Tumor Cell Phenotypic Changes and Anticancer Drug Resistance" Nanomaterials 8, no. 2: 64. https://doi.org/10.3390/nano8020064
APA StyleLe, B. D., Kang, D., Yun, S., Jeong, Y. H., Kwak, J.-Y., Yoon, S., & Jin, S. (2018). Three-Dimensional Hepatocellular Carcinoma/Fibroblast Model on a Nanofibrous Membrane Mimics Tumor Cell Phenotypic Changes and Anticancer Drug Resistance. Nanomaterials, 8(2), 64. https://doi.org/10.3390/nano8020064