Synthesis and Optimization of Chitosan Nanoparticles Loaded with l-Ascorbic Acid and Thymoquinone
Abstract
1. Introduction
2. Materials and Methods
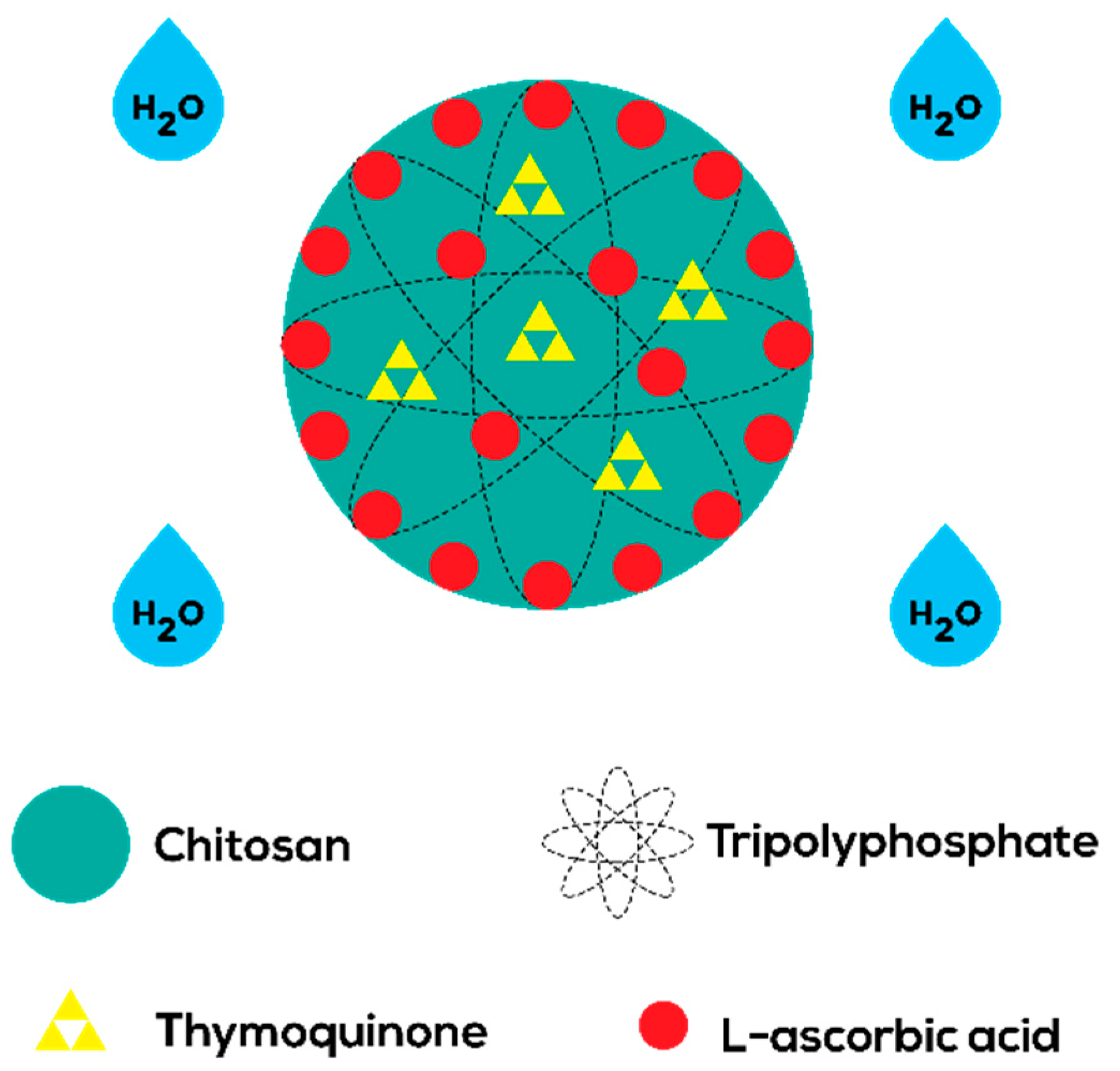
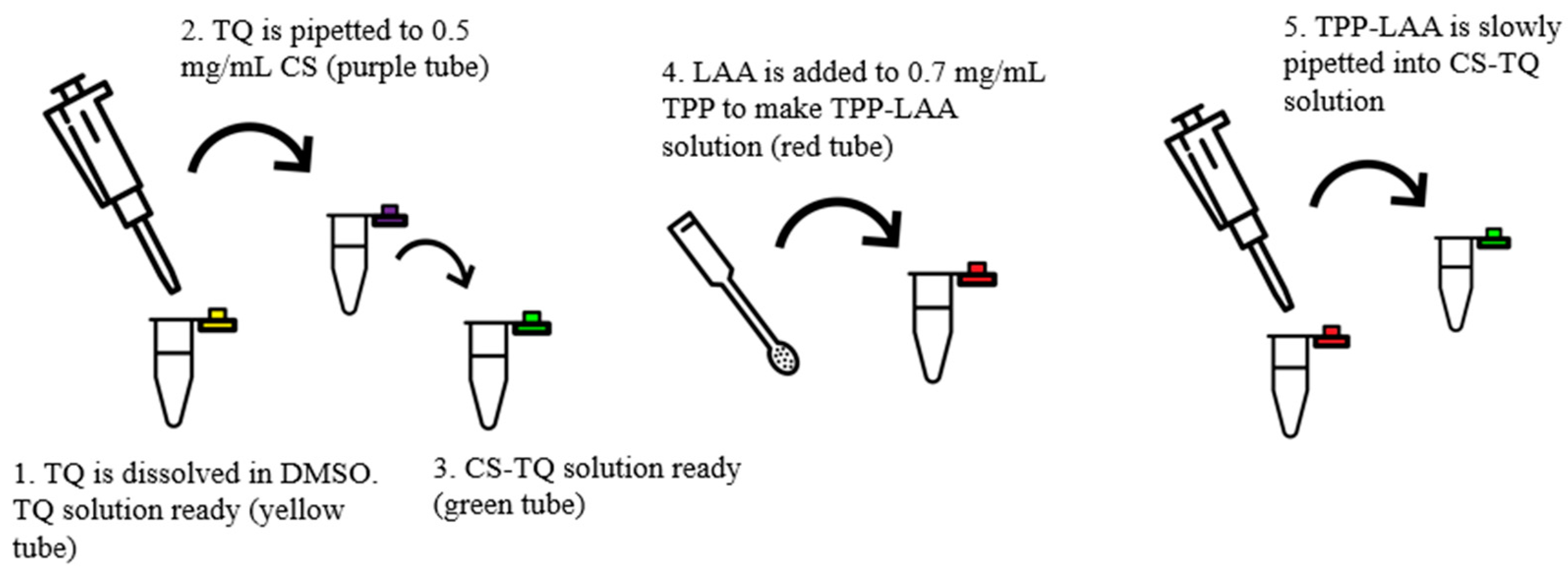
2.1. Preparation of CNP Samples
2.2. Physicochemical Characterization of CNP Samples
2.2.1. Fourier-Transform Infrared Spectroscopy (FTIR)
2.2.2. Particle Size Distribution (PSD)
2.2.3. Encapsulation Efficiency (EE)
2.2.4. Field-Emission Scanning Electron Microscopy (FESEM)
3. Results and Discussion
3.1. Determination of Free Functional Groups in Nanoparticle Samples via FTIR
3.2. Particle Size Distribution (PSD) Study by Dynamic Light Scattering (DLS)
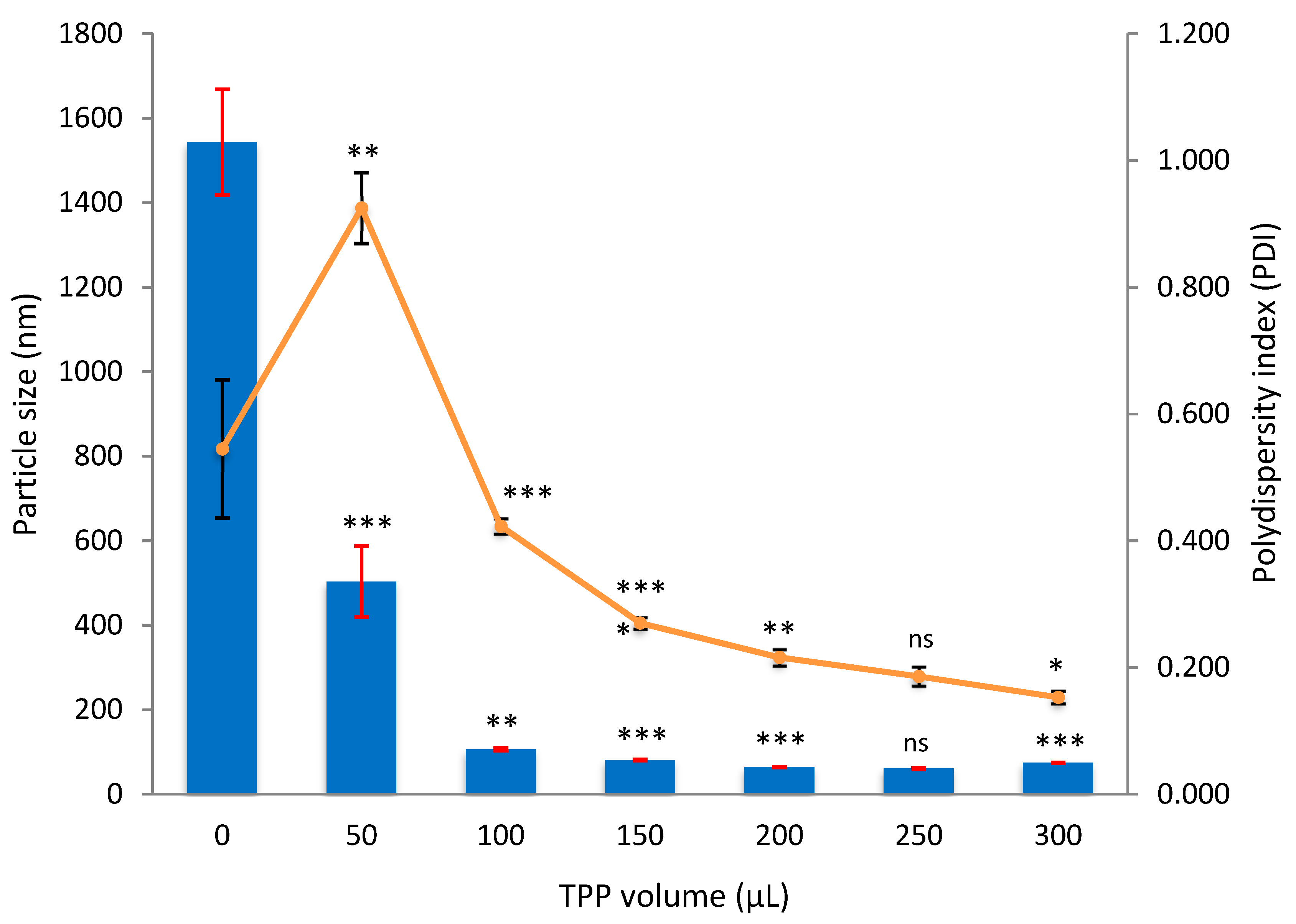
3.2.1. Empty CNP
3.2.2. Single-Loaded CNP
3.2.3. Dual-Loaded CNP
3.2.4. Comparison in Particle Size and Dispersity Between Empty, Single-Loaded and Dual-Loaded CNP
3.3. Encapsulation Efficiency (EE) Study of Nanoparticle Samples
3.3.1. Single-Loaded CNP
3.3.2. Dual-Loaded CNP
3.3.3. Comparison in EE between Single-Loaded and Dual-Loaded CNP
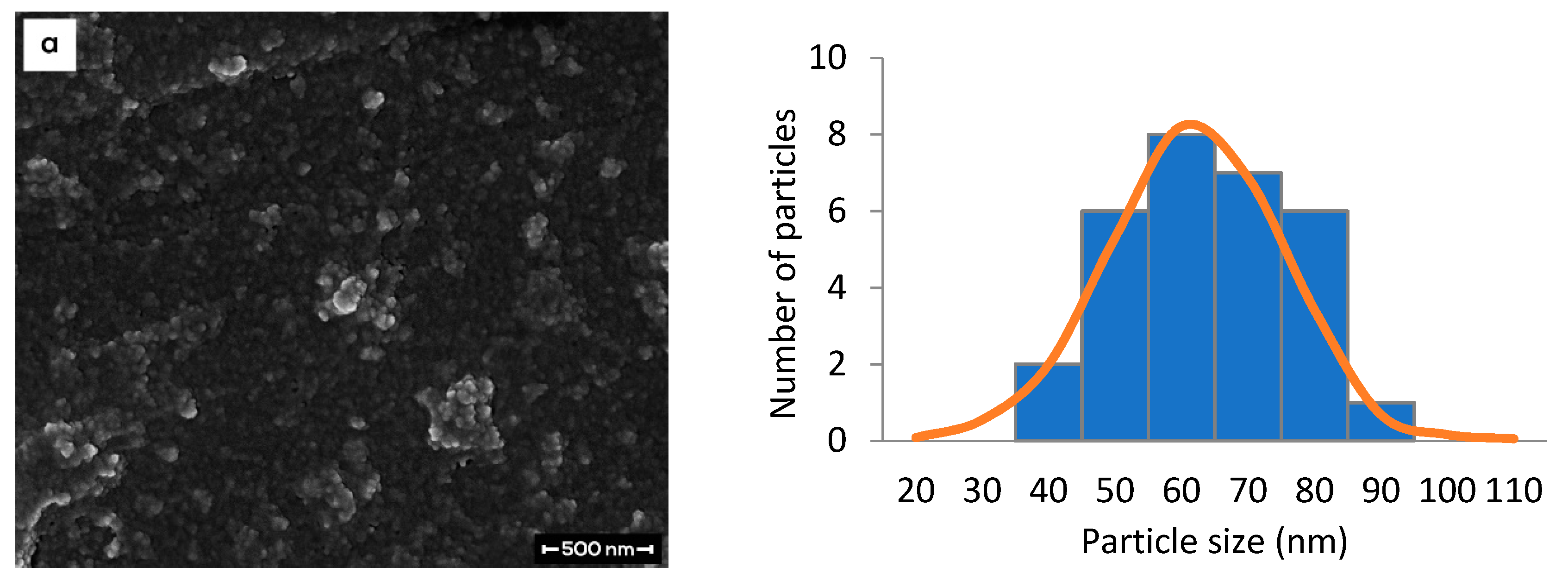
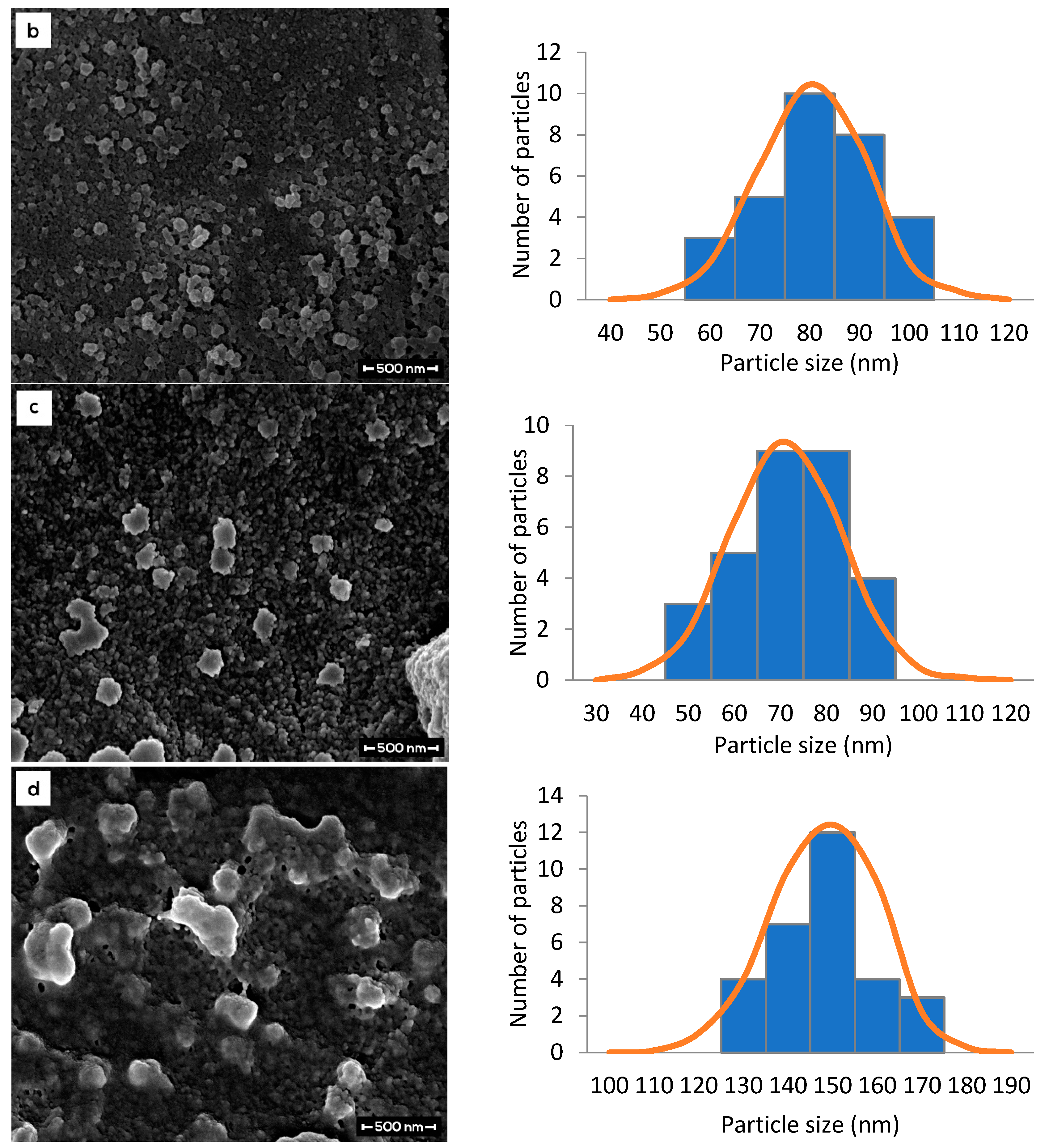
3.4. Morphological Study of Nanoparticle Samples by FESEM
3.5. Formulation Selection of Single- and Dual-Loaded CNP
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chandra, R.K. 1990 McCollum Award Lecture. Nutrition and immunity: Lessons from the past and new insights into the future. Am. J. Clin. Nutr. 1991, 53, 1087–1101. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Sun, Y.; Zhu, J.; Li, W.; Zhang, A.; Kuang, T.; Xie, J.; Yang, Z. Delivery of nanoparticles for treatment of brain tumor. Curr. Drug Metab. 2016, 17, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Neves, A.R.; Martins, S.; Segundo, M.A.; Reis, S. Nanoscale delivery of resveratrol towards enhancement of supplements and nutraceuticals. Nutrients 2016, 8, 131. [Google Scholar] [CrossRef] [PubMed]
- Paik, S.-Y.-R.; Kim, J.-S.; Shin, S.; Ko, S. Characterization, quantification, and determination of the toxicity of iron oxide nanoparticles to the bone marrow cells. Int. J. Mol. Sci. 2015, 16, 22243–22257. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Kim, J.; Park, Y.I.; Lee, N.; Hyeon, T. Recent development of inorganic nanoparticles for biomedical Imaging. ACS Cent. Sci. 2018, 4, 324–336. [Google Scholar] [CrossRef] [PubMed]
- Madureira, A.R.; Pereira, A.; Pintado, M. Current state on the development of nanoparticles for use against bacterial gastrointestinal pathogens. Focus on chitosan nanoparticles loaded with phenolic compounds. Carbohydr. Polym. 2015, 130, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Ahad, A.; Al-Saleh, A.A.; Akhtar, N.; Al-Mohizea, A.M.; Al-Jenoobi, F.I. Transdermal delivery of antidiabetic drugs: Formulation and delivery strategies. Drug Discov. Today 2015, 20, 1217–1227. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.; Sharma, A.R.; Nam, J.S.; Doss, G.P.C.; Lee, S.S.; Chakraborty, C. Nanoparticle based insulin delivery system: The next generation efficient therapy for Type 1 diabetes. J. Nanobiotechnol. 2015, 13, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.Y.; Al-Salami, H.; Dass, C.R. Potential of insulin nanoparticle formulations for oral delivery and diabetes treatment. J. Control. Release 2017, 264, 247–275. [Google Scholar] [CrossRef] [PubMed]
- Nayak, D.; Minz, A.P.; Ashe, S.; Rauta, P.R.; Kumari, M.; Chopra, P.; Nayak, B. Synergistic combination of antioxidants, silver nanoparticles and chitosan in a nanoparticle based formulation: Characterization and cytotoxic effect on MCF-7 breast cancer cell lines. J. Colloid Interface Sci. 2016, 470, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, B.; Hojjat-farsangi, M.; Mohammadi, H.; Anvari, E. Nanoparticles and targeted drug delivery in cancer therapy. Immunol. Lett. 2017, 190, 64–83. [Google Scholar] [CrossRef] [PubMed]
- Grobmyer, S.R.; Zhou, G.; Gutwein, L.G.; Iwakuma, N.; Sharma, P.; Hochwald, S.N. Nanoparticle delivery for metastatic breast cancer. Nanomedicine 2012, 8, S21–S30. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, V.; Reddy, K.R.; Sakthivel, S.; Swetha, C. Optimization and charaterization of repaglinide biodegradable polymeric nanoparticle loaded transdermal patchs: In vitro and in vivo studies. Colloids Surf. B Biointerfaces 2013, 111, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.S.; Song, M.Y.; Park, J.D.; Song, K.S.; Ryu, H.R.; Chung, Y.H.; Chang, H.K.; Lee, J.H.; Oh, K.H.; Kelman, B.J.; et al. Subchronic oral toxicity of silver nanoparticles. Part. Fibre Toxicol. 2010, 7. [Google Scholar] [CrossRef] [PubMed]
- Panyala, N.R.; Peña-Méndez, E.M.; Havel, J. Silver or silver nanoparticles: A hazardous threat to the environment and human health? J. Appl. Biomed. 2008, 6, 117–129. [Google Scholar]
- Falugi, C.; Aluigi, M.G.; Chiantore, M.C.; Privitera, D.; Ramoino, P.; Gatti, M.A.; Fabrizi, A.; Pinsino, A.; Matranga, V. Toxicity of metal oxide nanoparticles in immune cells of the sea urchin. Mar. Environ. Res. 2012, 76, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Desai, P.; Patlolla, R.R.; Singh, M. Interaction of nanoparticles and cell-penetrating peptides with skin for transdermal drug delivery. Mol. Membr. Biol. 2010, 27, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Kuen, C.; Fakurazi, S.; Othman, S.; Masarudin, M. Increased loading, efficacy and sustained release of silibinin, a poorly soluble drug using hydrophobically-modified chitosan nanoparticles for enhanced delivery of anticancer drug delivery systems. Nanomaterials 2017, 7, 379. [Google Scholar] [CrossRef] [PubMed]
- Kyzas, G.Z.; Bikiaris, D.N.; Lambropoulou, D.A. Effect of humic acid on pharmaceuticals adsorption using sulfonic acid grafted chitosan. J. Mol. Liq. 2017, 230, 1–5. [Google Scholar] [CrossRef]
- Kyzas, G.Z.; Bikiaris, D.N.; Mitropoulos, A.C. Chitosan adsorbents for dye removal: A review. Polym. Int. 2017, 66, 1800–1811. [Google Scholar] [CrossRef]
- Kyzas, G.Z.; Bikiaris, D.N. Recent modifications of chitosan for adsorption applications: A critical and systematic review. Mar. Drugs 2015, 13, 312–337. [Google Scholar] [CrossRef] [PubMed]
- Chabib, L.; Martien, R.; Ismail, H. Formulation of nanocurcumin using low viscosity chitosan polymer and its cellular uptake study into T47D cells. Indones. J. Pharm. 2012, 23, 27–35. [Google Scholar]
- Nguyen, T.V.; Nguyen, T.T.H.; Wang, S.L.; Vo, T.P.K.; Nguyen, A.D. Preparation of chitosan nanoparticles by TPP ionic gelation combined with spray drying, and the antibacterial activity of chitosan nanoparticles and a chitosan nanoparticle–amoxicillin complex. Res. Chem. Intermed. 2017, 43, 3527–3537. [Google Scholar] [CrossRef]
- Sreekumar, S.; Goycoolea, F.M.; Moerschbacher, B.M.; Rivera-Rodriguez, G.R. Parameters influencing the size of chitosan-TPP nano- and microparticles. Sci. Rep. 2018, 8, 4695. [Google Scholar] [CrossRef] [PubMed]
- Koukaras, E.N.; Papadimitriou, S.A.; Bikiaris, D.N.; Froudakis, G.E. Insight on the formation of chitosan nanoparticles through ionotropic gelation with tripolyphosphate. Mol. Pharm. 2012, 9, 2856–2862. [Google Scholar] [CrossRef] [PubMed]
- Al-Kassas, R.; Wen, J.; Cheng, A.E.M.; Kim, A.M.J.; Liu, S.S.M.; Yu, J. Transdermal delivery of propranolol hydrochloride through chitosan nanoparticles dispersed in mucoadhesive gel. Carbohydr. Polym. 2016, 153, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Yan, W.; Xu, Z.; Ni, H. Formation mechanism of monodisperse, low molecular weight chitosan nanoparticles by ionic gelation technique. Colloids Surf. B Biointerfaces 2012, 90, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Uchechi, O.; Ogbonna, J.D.N.; Attama, A.A. Nanoparticles for Dermal and Transdermal Drug Delivery; IntechOpen: London, UK, 2014; ISBN 978-953-51-1628-8. [Google Scholar]
- Tian, X.L.; Tian, D.F.; Wang, Z.Y.; Mo, F.K. Synthesis and evaluation of chitosan-vitamin C complex. Indian J. Pharm. Sci. 2009, 71, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Alishahi, A.; Mirvaghefi, A.; Tehrani, M.R.; Farahmand, H.; Shojaosadati, S.A.; Dorkoosh, F.A.; Elsabee, M.Z. Shelf life and delivery enhancement of vitamin C using chitosan nanoparticles. Food Chem. 2011, 126, 935–940. [Google Scholar] [CrossRef]
- Alam, S.; Khan, Z.I.; Mustafa, G.; Kumar, M.; Islam, F.; Bhatnagar, A.; Ahmad, F.J. Development and evaluation of thymoquinone-encapsulated chitosan nanoparticles for nose-to-brain targeting: A pharmacoscintigraphic study. Int. J. Nanomed. 2012, 7, 5705–5718. [Google Scholar] [CrossRef] [PubMed]
- Soni, P.; Kaur, J.; Tikoo, K. Dual drug-loaded paclitaxel-thymoquinone nanoparticles for effective breast cancer therapy. J. Nanopart. Res. 2015, 17. [Google Scholar] [CrossRef]
- Ullah, I.; Badshah, H.; Naseer, M.I.; Lee, H.Y.; Kim, M.O. Thymoquinone and vitamin C attenuates pentylenetetrazole-induced seizures via activation of GABA B1 receptor in adult rats cortex and hippocampus. Neuromol. Med. 2014. [Google Scholar] [CrossRef]
- Pritchard, J.R.; Bruno, P.M.; Gilbert, L.a; Capron, K.L.; Lauffenburger, D.a; Hemann, M.T. Defining principles of combination drug mechanisms of action. Proc. Natl. Acad. Sci. USA 2012, 110. [Google Scholar] [CrossRef] [PubMed]
- Naderinezhad, S.; Amoabediny, G.; Haghiralsadat, F. Co-delivery of hydrophilic and hydrophobic anticancer drugs using biocompatible pH-sensitive lipid-based nano-carriers for multidrug-resistant cancers. RSC Adv. 2017, 7, 30008–30019. [Google Scholar] [CrossRef]
- Kaur, G.; Mehta, S.K.; Kumar, S.; Bhanjana, G.; Dilbaghi, N. Coencapsulation of hydrophobic and hydrophilic antituberculosis drugs in synergistic Brij 96 microemulsions: A biophysical characterization. J. Pharm. Sci. 2015, 104, 2203–2212. [Google Scholar] [CrossRef] [PubMed]
- Esposito, E.; Cervellati, F.; Menegatti, E.; Nastruzzi, C.; Coretesi, R. Spray dried eudgragit microparticles as encapsulation devices for vitamin C. Int. J. Pharm. 2002, 242, 329–334. [Google Scholar] [CrossRef]
- Kassab, R.B.; El-Hennamy, R.E. The role of thymoquinone as a potent antioxidant in ameliorating the neurotoxic effect of sodium arsenate in female rat. Egypt. J. Basic Appl. Sci. 2017, 4, 160–167. [Google Scholar] [CrossRef]
- Masarudin, M.J. Chitosan Nanoparticles as a Delivery Vehicle for [14C]-Doxorubicin and the Formaldehyde Releasing Prodrug AN-250. Ph.D. Thesis, La Trobe University, Bundoora, Victoria, Australia, 2012. digitalTheses-20130424-100341. [Google Scholar]
- Masarudin, M.J.; Cutts, S.M.; Evison, B.J.; Phillips, D.R.; Pigram, P.J. Factors determining the stability, size distribution, and cellular accumulation of small, monodisperse chitosan nanoparticles as candidate vectors for anticancer drug delivery: Application to the passive encapsulation of [14C]-doxorubicin. Nanotechnol. Sci. Appl. 2015, 8, 67–80. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wan, A.; Shi, Y.; Zhang, Y.; Chen, Y. Experimental and mathematical studies on the drug release properties of aspirin loaded chitosan nanoparticles. BioMed Res. Int. 2014. [Google Scholar] [CrossRef] [PubMed]
- Subri, N.N.S.; Cormack, P.A.G.; Siti Nurul, S.N.A.; Abdullah, L.C.; Daik, R. Synthesis of poly(acrylonitrile-co-divinylbenzene-co-vinylbenzyl chloride)-derived hypercrosslinked polymer microspheres and a preliminary evaluation of their potential for the solid-phase capture of pharmaceuticals. J. Appl. Polym. Sci. 2018, 135, 45677. [Google Scholar] [CrossRef]
- Stetefeld, J.; McKenna, S.A.; Patel, T.R. Dynamic light scattering: A practical guide and applications in biomedical sciences. Biophys. Rev. 2016, 8, 409–427. [Google Scholar] [CrossRef] [PubMed]
- Berg, R.W. Investigation of l (+)-ascorbic acid with raman spectroscopy in visible and UV light. Appl. Spectrosc. Rev. 2015, 50, 193–239. [Google Scholar] [CrossRef]
- Ong, Y.S.; Yazan, L.S.; Ng, W.K.; Noordin, M.M.; Sapuan, S.; Foo, J.B.; Tor, Y.S. Acute and subacute toxicity profiles of thymoquinone-loaded nanostructured lipid carrier in BALB/c mice. Int. J. Nanomed. 2016, 5905–5915. [Google Scholar] [CrossRef] [PubMed]
- Goyal, S.N.; Prajapati, C.P.; Gore, P.R.; Patil, C.R.; Mahajan, U.B.; Sharma, C.; Talla, S.P.; Ojha, S.K. Therapeutic potential and pharmaceutical development of thymoquinone: A multitargeted molecule of natural origin. Front. Pharmacol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Khlibsuwan, R.; Pongjanyakul, T. Particle agglomeration of chitosan–magnesium aluminum silicate nanocomposites for direct compression tablets. Int. J. Pharm. 2018, 535, 410–419. [Google Scholar] [CrossRef] [PubMed]
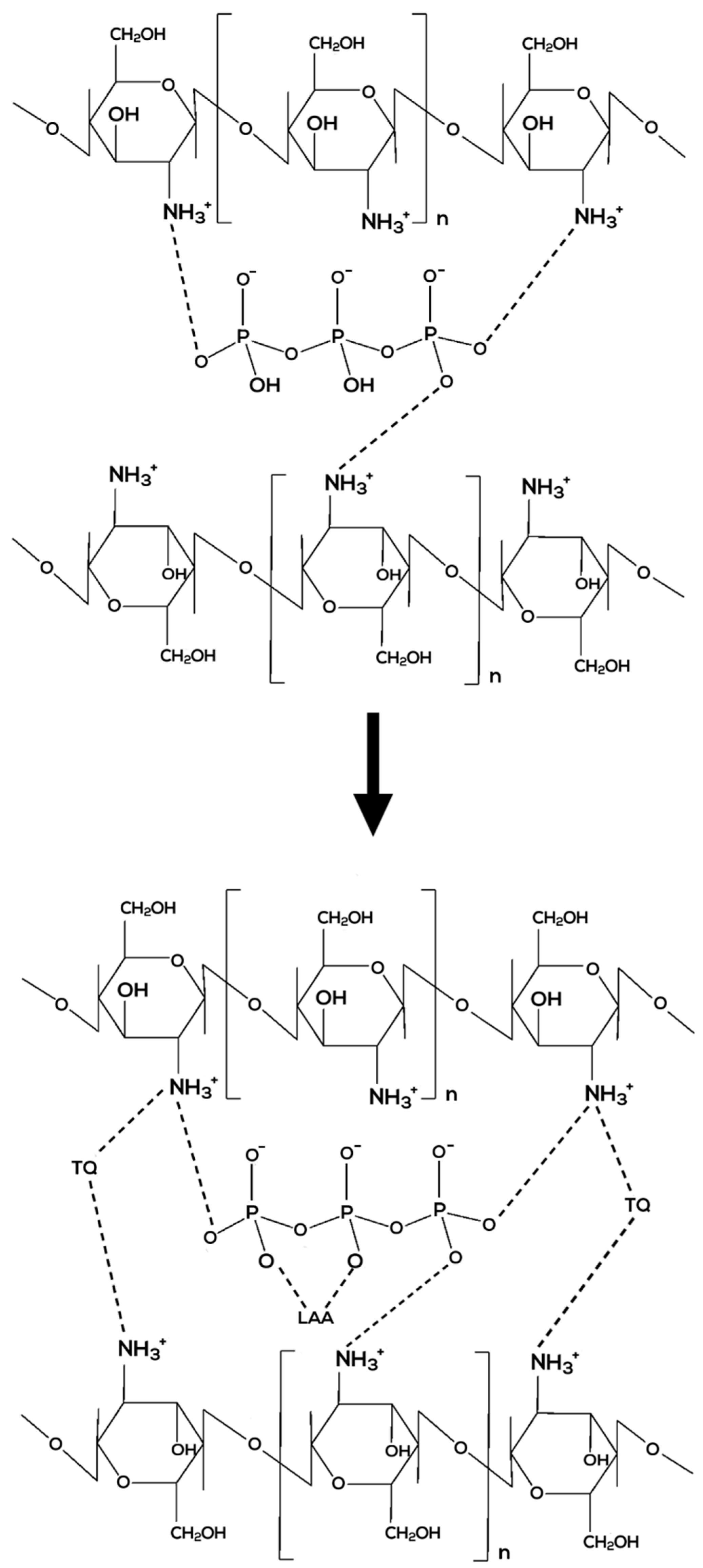


| % Transmittance | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Peak | a | b | c | d | e | f | g | h | i |
| Functional Group | NH2 Amine I, OH Stretch | CH Stretch | C=O Stretch | CN–C=O Amide II carbonyl | C=C Stretch, NH Deformation | CO Stretch | CO Stretch | CN Amide III Stretch | P=O Stretch |
| Wavenumber (cm−1) | 3352 | 2897 | 1761 | 1641 | 1532 | 1391 | 1321 | 1237 | 1007 |
| CNP | 38.7 | 74.6 | None | 42.2 | 56.3 | 84.2 | None | 56.8 | 13.1 |
| CNP-LAA | 42.2 | 75.0 | 90.3 | 47.4 | 60.0 | 72.9 | 78.3 | 62.0 | 8.1 |
| CNP-TQ | 33.9 | 75.2 | None | 47.2 | 58.0 | 86.0 | None | 61.1 | 8.1 |
| CNP-LAA-TQ | 38.4 | 73.4 | 88.1 | 49.4 | 64.4 | 74.7 | 76.2 | 67.5 | 8.2 |
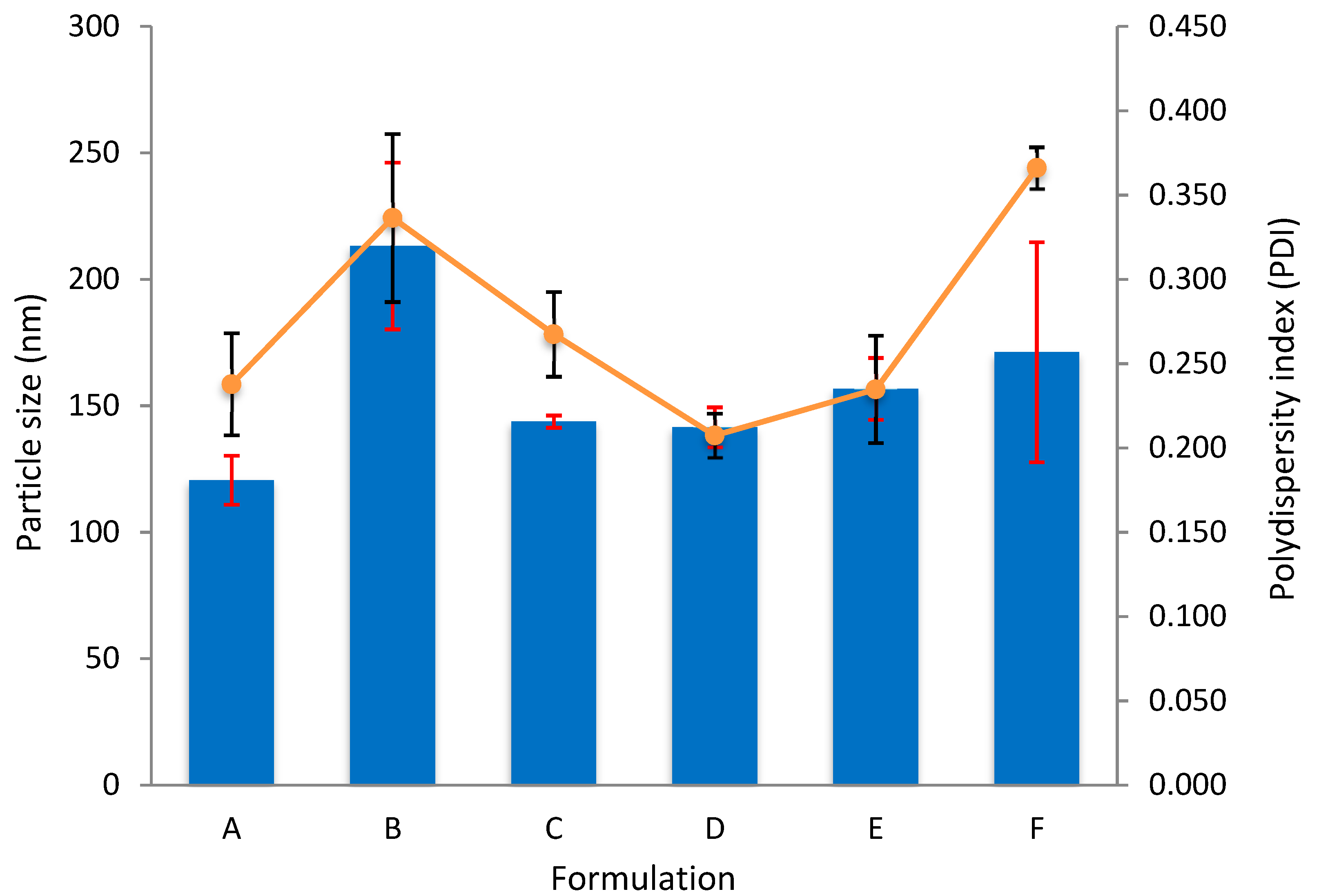
| Formulation | Concentration (μM) | Particle Size (nm ± sd) | Polydispersity Index (±sd) | |
|---|---|---|---|---|
| LAA | TQ | |||
| A | 160 | 100 | 120.6 ± 9.7 | 0.238 ± 0.030 |
| B | 235 | 100 | 213.2 ± 33.0 | 0.336 ± 0.050 |
| C | 310 | 100 | 143.7 ± 2.4 | 0.267 ± 0.025 |
| D | 160 | 150 | 141.5 ± 7.8 | 0.207 ± 0.013 |
| E | 235 | 150 | 156.6 ± 12.2 | 0.235 ± 0.032 |
| F | 310 | 150 | 171.2 ± 43.5 | 0.366 ± 0.012 |
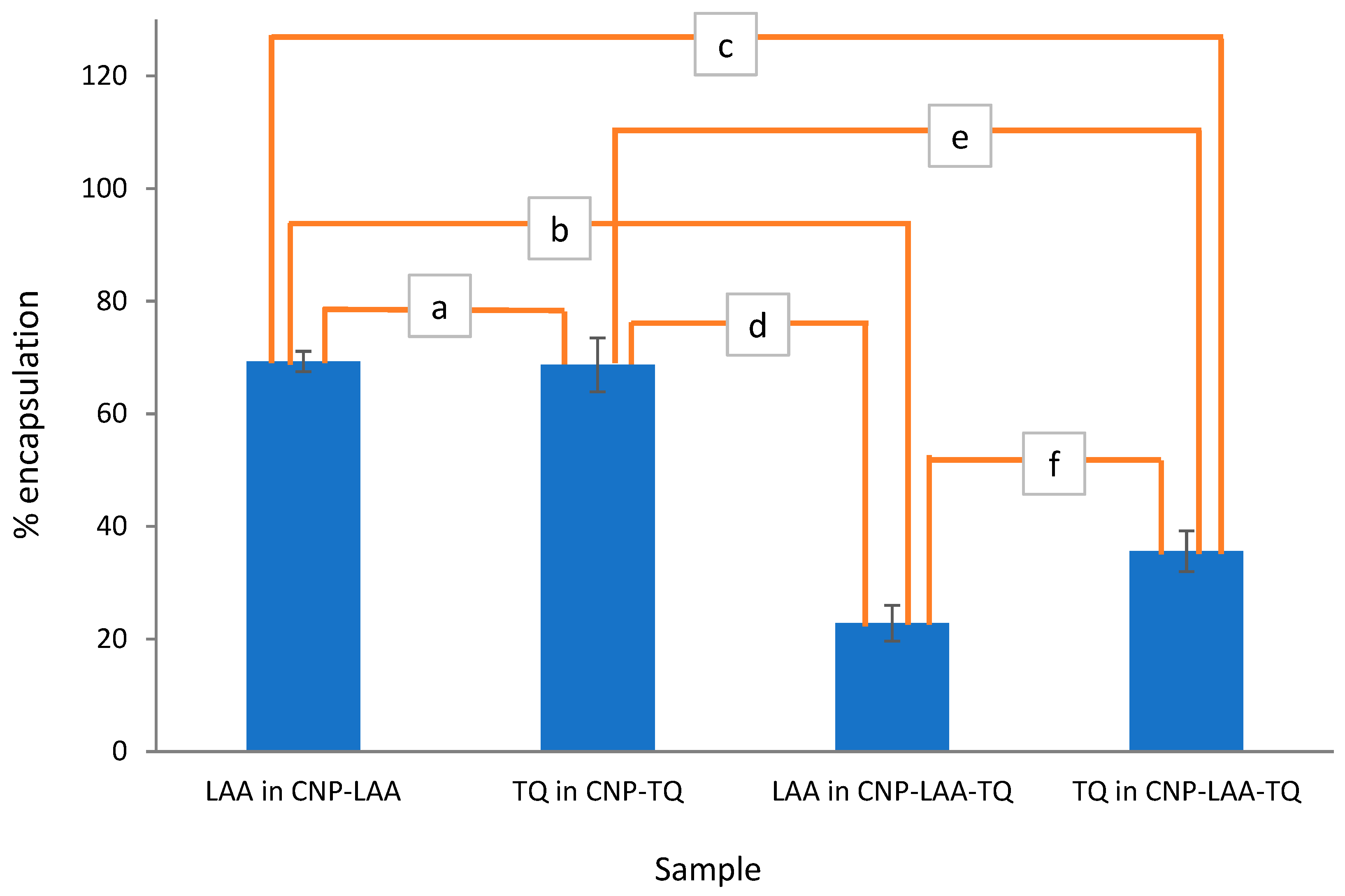
| Sample | Initial (μM) | EE of | Loading (μM) | |||
|---|---|---|---|---|---|---|
| LAA | TQ | LAA in CNP-LAA-TQ (% ± sd) | TQ in CNP-LAA-TQ (% ± sd) | LAA | TQ | |
| A | 160 | 100 | 47.9 ± 5.7 | 17.8 ± 3.1 | 76.6 | 17.8 |
| B | 235 | 100 | 31.8 ± 2.8 | 12.8 ± 9.6 | 74.7 | 12.8 |
| C | 310 | 100 | 73.5 ± 2.4 | 13.3 ± 2.8 | 227.9 | 73.5 |
| D | 160 | 150 | 22.8 ± 3.2 | 35.6 ± 3.6 | 36.5 | 53.4 |
| E | 235 | 150 | 11.1 ± 3.6 | 30.3 ± 9.0 | 26.1 | 45.5 |
| F | 310 | 150 | 21.6 ± 4.3 | 31.6 ± 7.0 | 67.0 | 47.4 |
| Type of CNP | PSD | %EE | |
|---|---|---|---|
| Particle Size (nm) | PDI | ||
| Empty | 60.5 ± 2.1 | 0.186 ± 0.015 | *nr |
| Single-loaded LAA | 87.3 ± 3.8 | 0.266 ± 0.021 | 69.3 ± 1.8 |
| TQ | 74.7 ± 8.5 | 0.364 ± 0.097 | 68.7 ± 4.8 |
| Dual-loaded LAA | 141.5 ± 7.8 (D) | 0.207 ± 0.013 (D) | 22.8 ± 3.2 (D) |
| TQ | 35.6 ± 3.6 (D) | ||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Othman, N.; Masarudin, M.J.; Kuen, C.Y.; Dasuan, N.A.; Abdullah, L.C.; Md. Jamil, S.N.A. Synthesis and Optimization of Chitosan Nanoparticles Loaded with l-Ascorbic Acid and Thymoquinone. Nanomaterials 2018, 8, 920. https://doi.org/10.3390/nano8110920
Othman N, Masarudin MJ, Kuen CY, Dasuan NA, Abdullah LC, Md. Jamil SNA. Synthesis and Optimization of Chitosan Nanoparticles Loaded with l-Ascorbic Acid and Thymoquinone. Nanomaterials. 2018; 8(11):920. https://doi.org/10.3390/nano8110920
Chicago/Turabian StyleOthman, Nurhanisah, Mas Jaffri Masarudin, Cha Yee Kuen, Nurul Azira Dasuan, Luqman Chuah Abdullah, and Siti Nurul Ain Md. Jamil. 2018. "Synthesis and Optimization of Chitosan Nanoparticles Loaded with l-Ascorbic Acid and Thymoquinone" Nanomaterials 8, no. 11: 920. https://doi.org/10.3390/nano8110920
APA StyleOthman, N., Masarudin, M. J., Kuen, C. Y., Dasuan, N. A., Abdullah, L. C., & Md. Jamil, S. N. A. (2018). Synthesis and Optimization of Chitosan Nanoparticles Loaded with l-Ascorbic Acid and Thymoquinone. Nanomaterials, 8(11), 920. https://doi.org/10.3390/nano8110920






