Heteromer Nanostars by Spontaneous Self-Assembly
Abstract
:1. Introduction
2. Materials and Methods
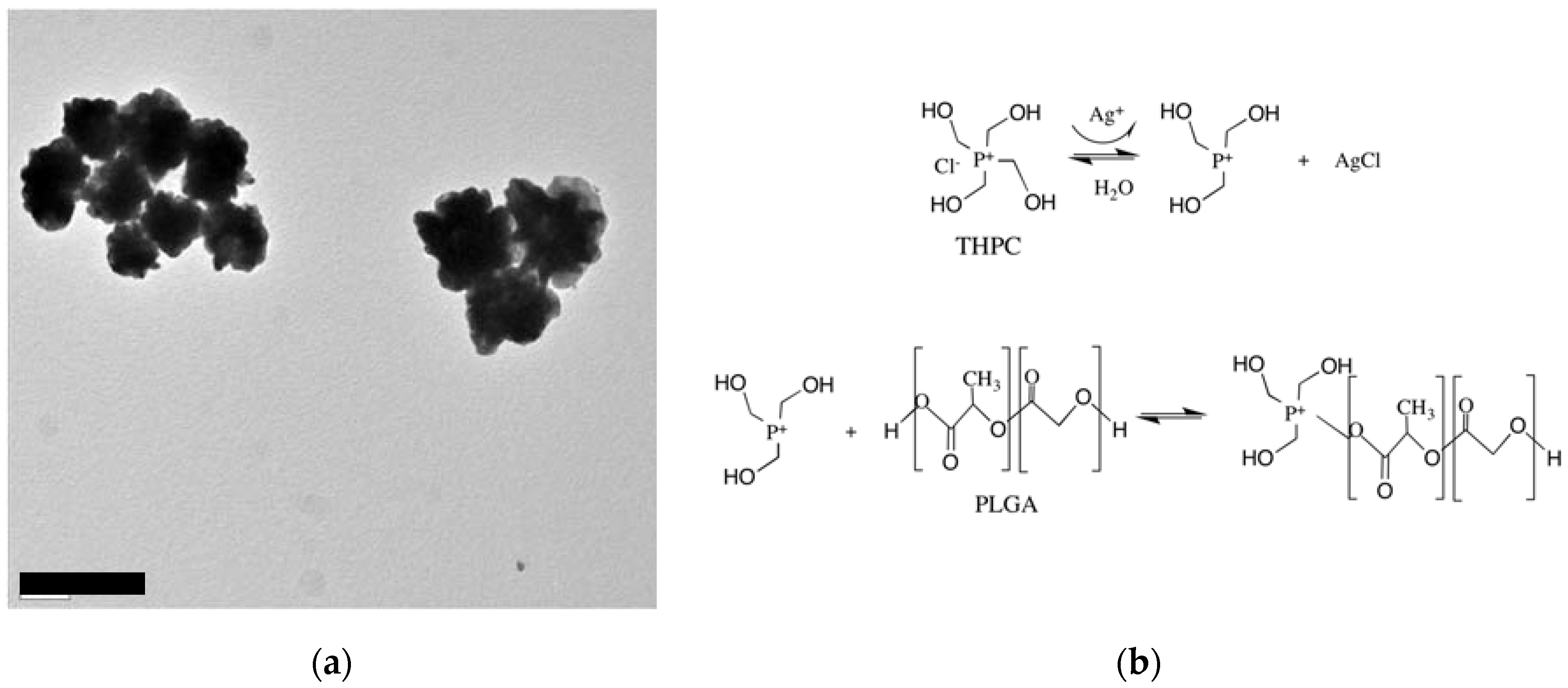
2.1. Synthesis of PLGA-THPC Heteromer Nanostars
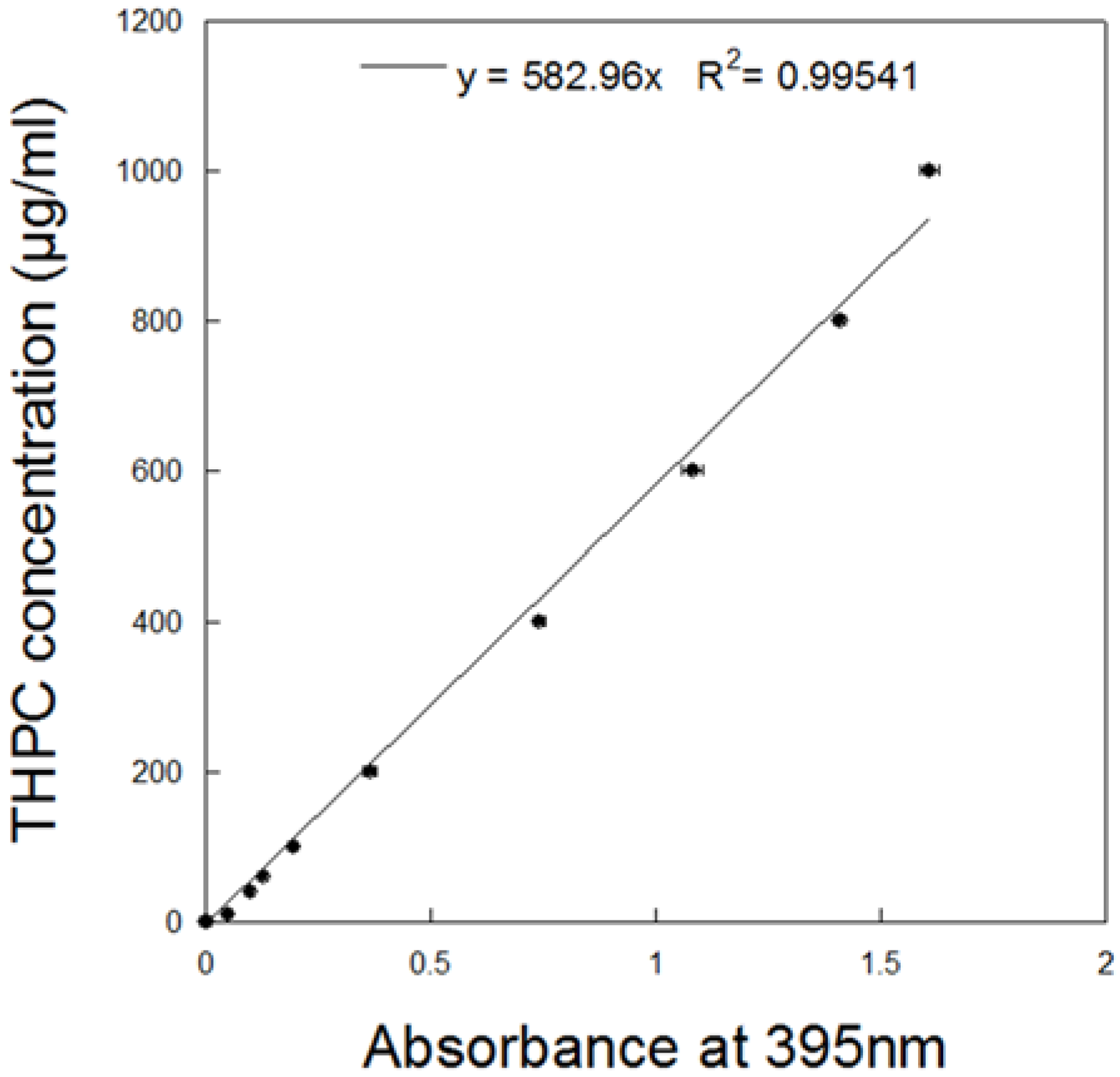
2.2. Measurement of THPC in Nanostars
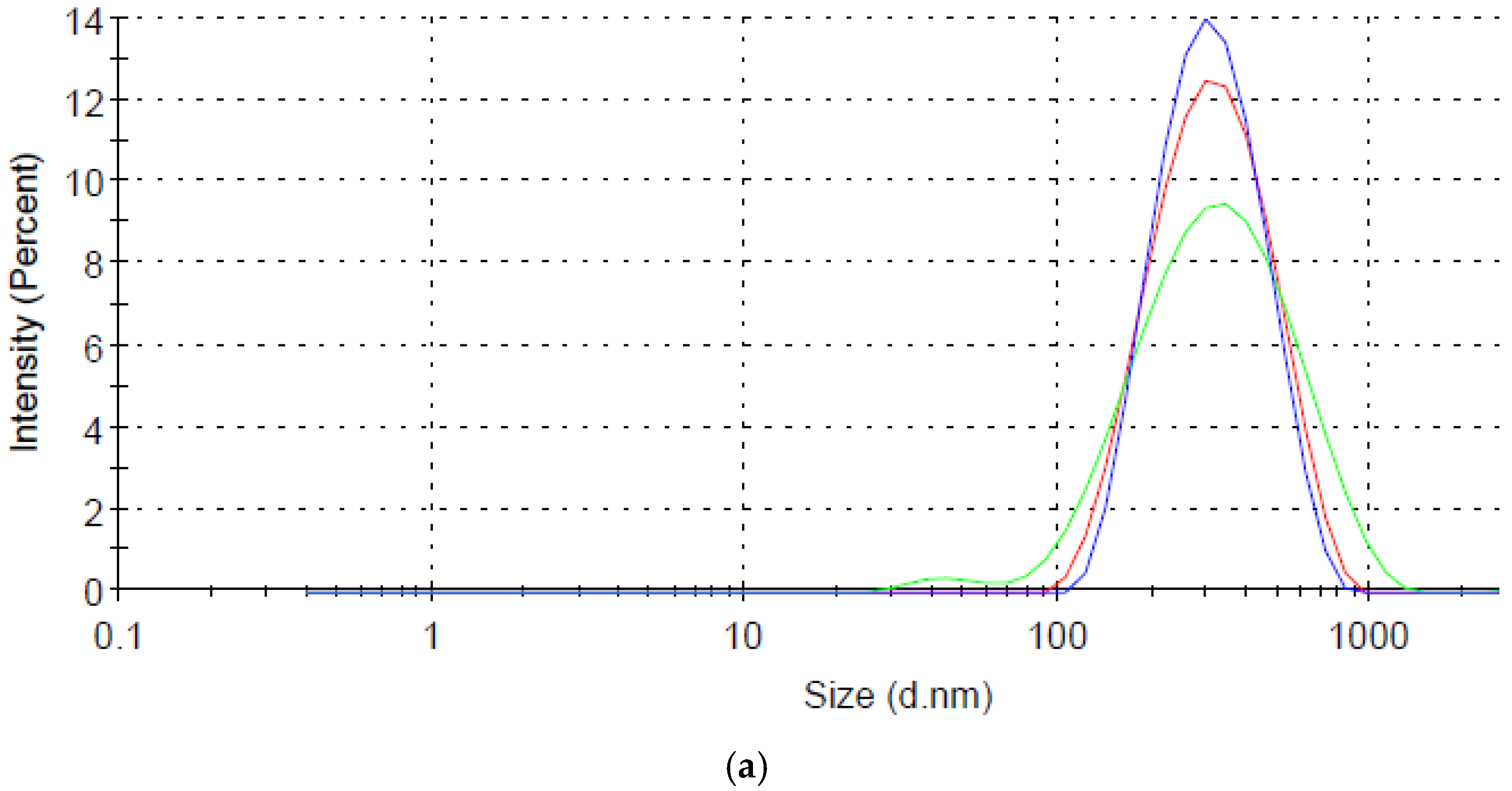
2.3. Characterization of Nanostars
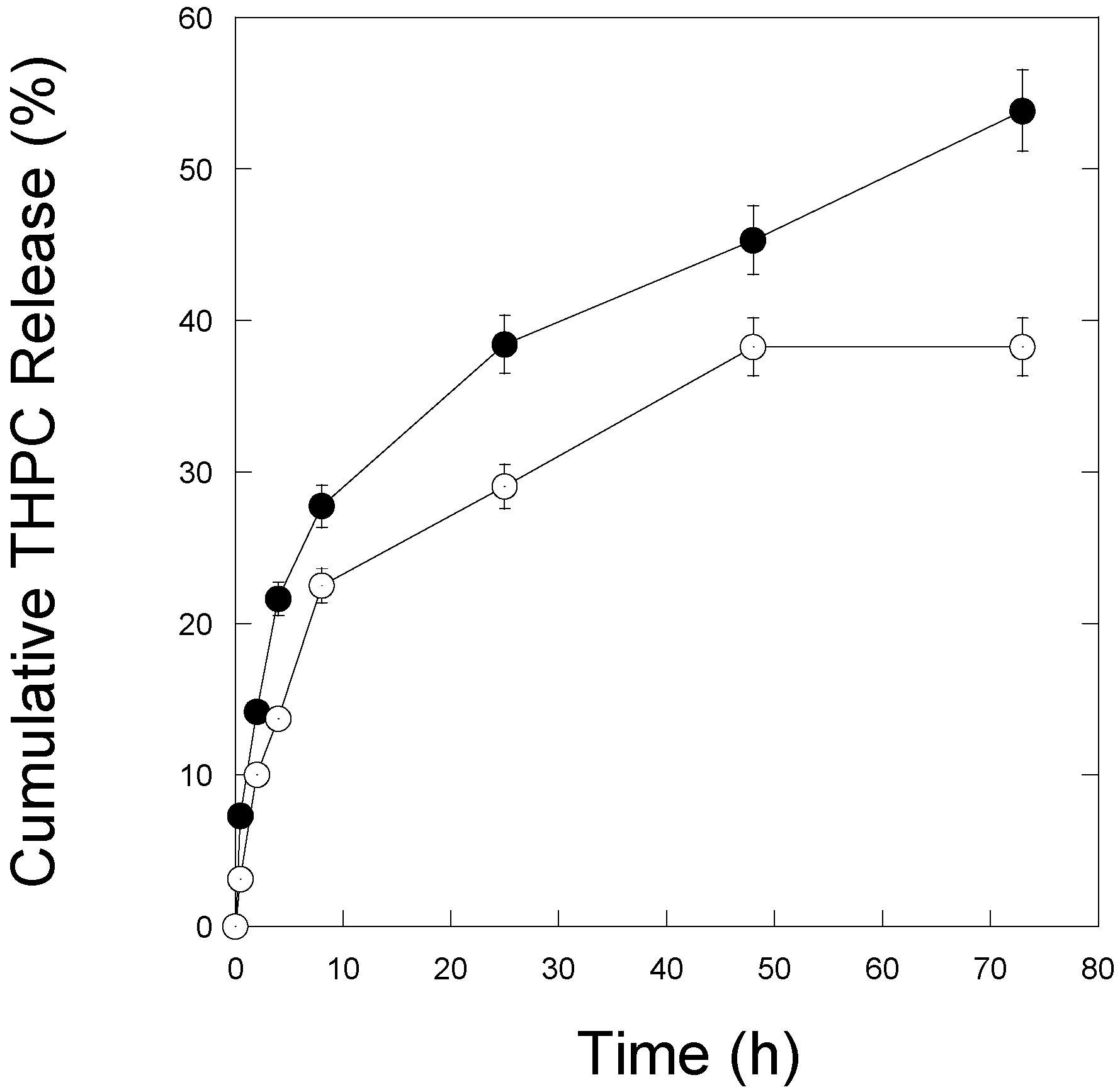
2.4. Quantification of THPC Release
2.5. Intracellular Uptake
2.6. In Vitro Cytotoxicity of Heteromer Nanostars in MDA-MB-231 Breast Cancer Cells
2.7. Statistical Analysis
3. Results and Discussion
3.1. Synthesis of Heteromer Nanostars
3.2. THPC Release
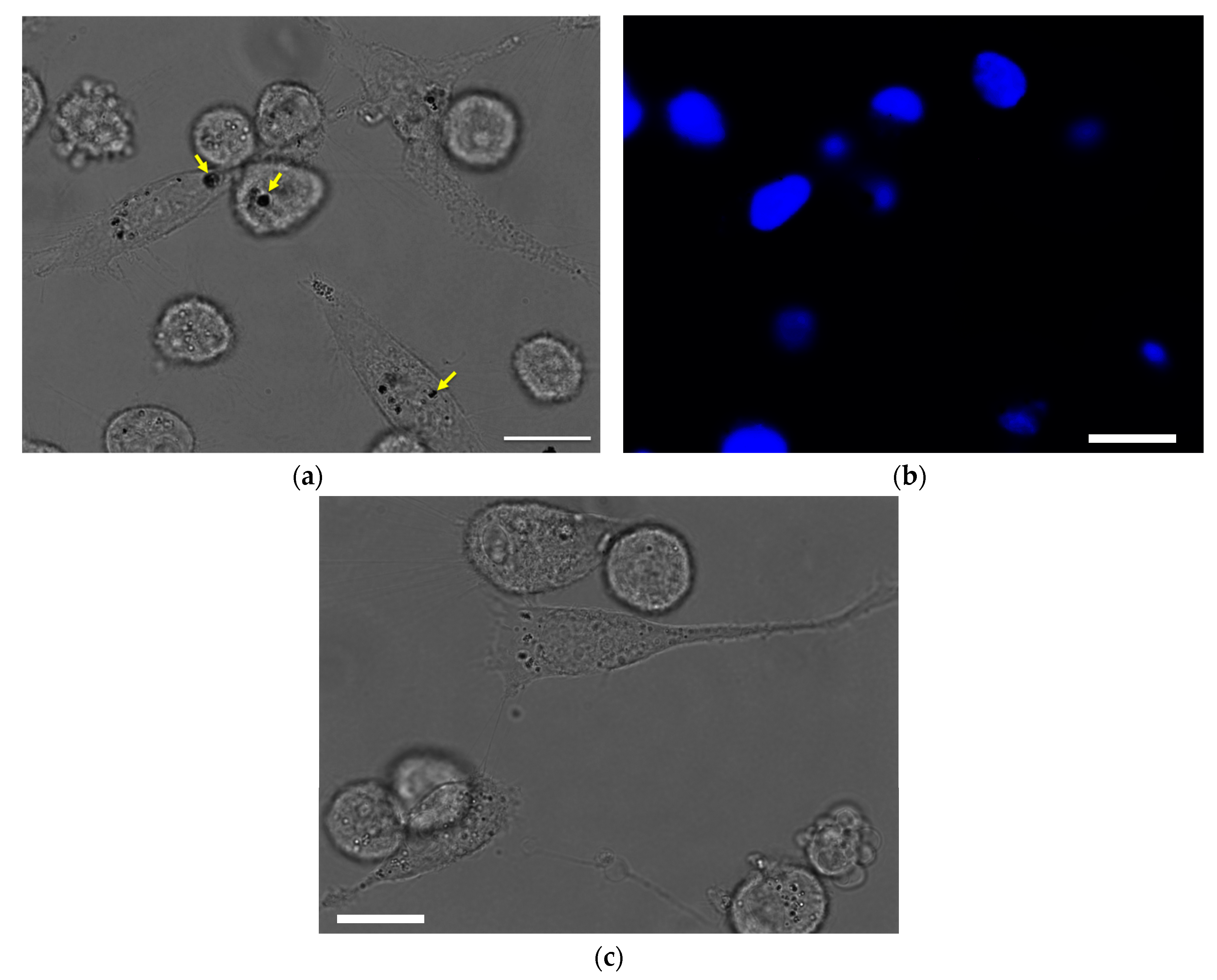
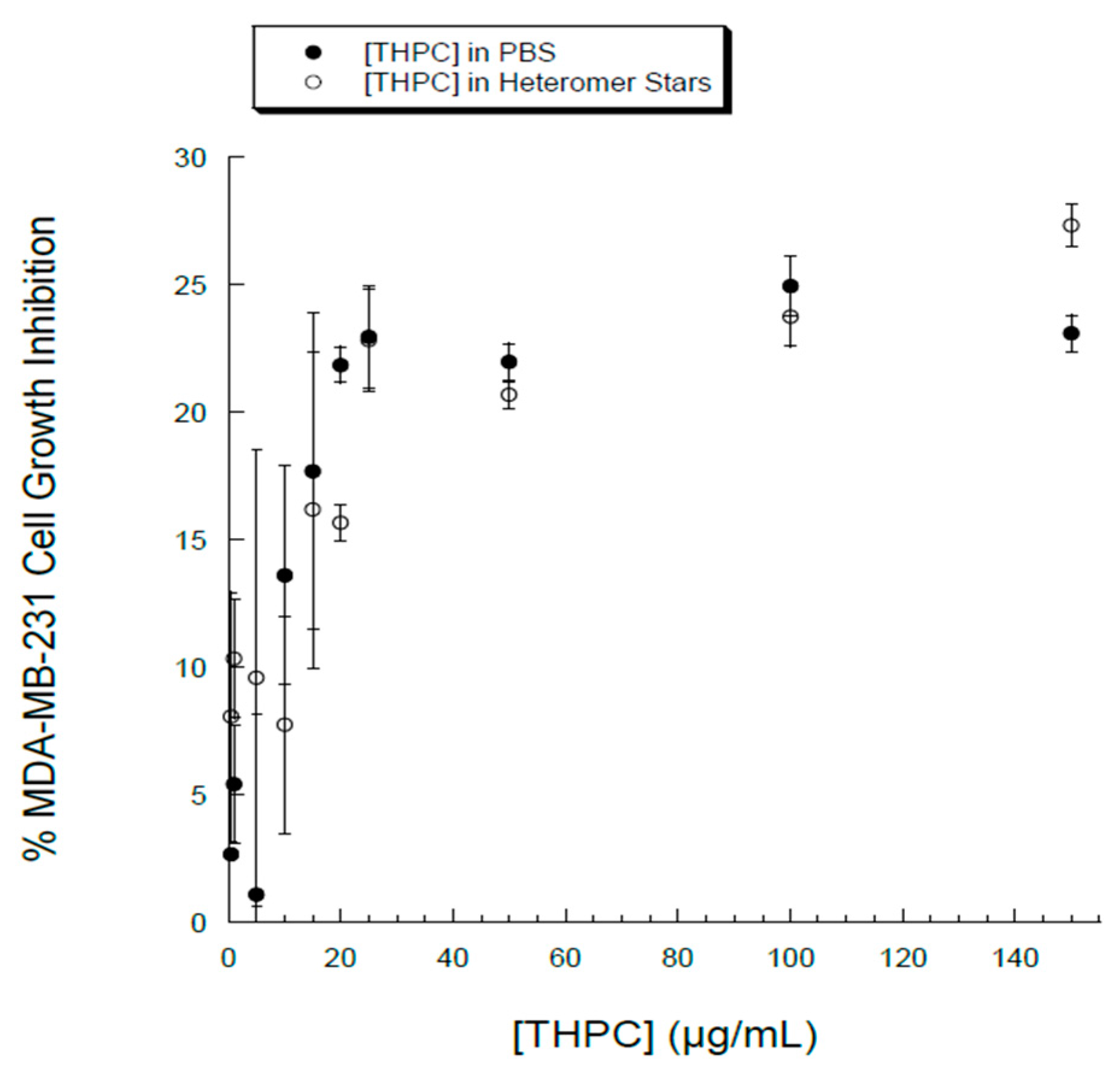
3.3. Cellular Uptake and Cytotoxicity in Breast Cancer Cells
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Gratton, S.E.A.; Ropp, P.A.; Pohlhaus, P.D.; Luft, J.C.; Madden, V.J.; Napier, M.E.; DeSimone, J.M. The effect of particle design on cellular internalization pathways. Proc. Natl. Acad. Sci. USA 2008, 105, 11613–11618. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.L.; Herlihy, K.P.; Napier, M.E.; DeSimone, J.M. PRINT: A Novel Platform Toward Shape and Size Specific Nanoparticle Theranostics. Acc. Chem. Res. 2011, 44, 990–998. [Google Scholar] [CrossRef] [PubMed]
- Champion, J.A.; Katare, Y.K.; Mitragotri, S. Making polymeric micro- and nanoparticles of complex shapes. Proc. Natl. Acad. Sci. USA 2007, 104, 11901–11904. [Google Scholar] [CrossRef] [PubMed]
- Mitragotri, S.; Lahann, J. Physical approaches to biomaterial design. Nat. Mater. 2009, 8, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Kolhar, P.; Doshi, N.; Mitragotri, S. Polymer Nanoneedle-Mediated Intracellular Drug Delivery. Small 2011, 7, 2094–2100. [Google Scholar] [CrossRef] [PubMed]
- Bhaskar, S.; Pollock, K.M.; Yoshida, M.; Lahann, J. Towards designer microparticles: Simultaneous control of anisotropv, shape and size. Small 2010, 6, 404–411. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Dalhaimer, P.; Cai, S.; Tsai, R.; Tewari, M.; Minko, T.; Discher, D.E. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat. Nanotechnol. 2007, 2, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Discher, D.E. Hydrolytic degradation of poly(ethylene oxide)-block-polycaprolactone worm micelles. J. Am. Chem. Soc. 2005, 127, 12780–12781. [Google Scholar] [CrossRef] [PubMed]
- Laemthong, T.; Kim, H.H.; Dunlap, K.; Brocker, C.; Barua, D.; Forciniti, D.; Huang, Y.-W.; Barua, S. Bioresponsive polymer coated drug nanorods for breast cancer treatment. Nanotechnology 2017, 28, 045601. [Google Scholar] [CrossRef] [PubMed]
- Barua, S.; Yoo, J.-W.; Kolhar, P.; Wakankar, A.; Gokarn, Y.R.; Mitragotri, S. Particle shape enhances specificity of antibody-displaying nanoparticles. Proc. Natl. Acad. Sci. USA 2013, 110, 3270–3275. [Google Scholar] [CrossRef] [PubMed]
- Champion, J.; Mitragotri, S. Shape Induced Inhibition of Phagocytosis of Polymer Particles. Pharm. Res. 2009, 26, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Champion, J.A.; Mitragotri, S. Role of target geometry in phagocytosis. Proc. Natl. Acad. Sci. USA 2006, 103, 4930–4934. [Google Scholar] [CrossRef] [PubMed]
- Devarajan, P.V.; Jindal, A.B.; Patil, R.R.; Mulla, F.; Gaikwad, R.V.; Samad, A. Particle shape: A new design parameter for passive targeting in splenotropic drug delivery. J. Pharm.Sci. 2010, 99, 2576–2581. [Google Scholar] [CrossRef] [PubMed]
- Danko, M.; Libiszowski, J.; Biela, T.; Wolszczak, M.; Duda, A. Molecular dynamics of star-shaped poly(l-lactide)s in Tetrahydrofuran as Solvent Monitored by Fluorescence Spectroscopy. J. Polym. Sci. Part A Polym. Chem. 2005, 43, 4586–4599. [Google Scholar] [CrossRef]
- Elkins, C.L.; Viswanathan, K.; Long, T.E. Synthesis and characterization of star-shaped poly(ethylene-co-propylene) polymers bearing terminal self-complementary multiple hydrogen-bonding sites. Macromolecules 2006, 39, 3132–3139. [Google Scholar] [CrossRef]
- Pitsikalis, M.; Hadjichristidis, N. Model mono-, di-, and tri-ω-functionalized three-arm star polybutadienes. Synthesis and association in dilute solutions by membrane osmometry and static light scattering. Macromolecules 1995, 28, 3904–3910. [Google Scholar] [CrossRef]
- Nanjwade, B.K.; Bechra, H.M.; Derkar, G.K.; Manvi, F.V.; Nanjwade, V.K. Dendrimers: Emerging polymers for drug-delivery systems. Eur. J. Pharm. Sci. 2009, 38, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Torchilin, V.P. Micellar Nanocarriers: Pharmaceutical Perspectives. Pharm. Res. 2006, 24, 1. [Google Scholar] [CrossRef] [PubMed]
- Grayson, S.M.; Fréchet, J.M.J. Convergent dendrons and dendrimers: From synthesis to applications. Chem. Rev. 2001, 101, 3819–3867. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.-L.; van Cuong, N.; Hsieh, M.-F. Endocytosis Pathways of the Folate Tethered Star-Shaped PEG-PCL Micelles in Cancer Cell Lines. Polymers 2014, 6, 634–650. [Google Scholar] [CrossRef]
- Makadia, H.K.; Siegel, S.J. Poly Lactic-co-Glycolic Acid (PLGA) as Biodegradable Controlled Drug Delivery Carrier. Polymers 2011, 3, 1377–1397. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.I.; Kim, J.C.; Kim, Y.H.; Tae, G.; Lee, S.Y.; Kim, K.; Kwon, I.C. The effect of surface functionalization of PLGA nanoparticles by heparin- or chitosan-conjugated Pluronic on tumor targeting. J. Control. Release 2010, 143, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Thomasin, C.; Nam-Trân, H.; Merkle, H.P.; Gander, B. Drug microencapsulation by PLA/PLGA coacervation in the light of thermodynamics. 1. Overview and theoretical considerations. J. Pharm. Sci. 1998, 87, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Murakami, H.; Kobayashi, M.; Takeuchi, H.; Kawashima, Y. Preparation of poly(DL-lactide-co-glycolide) nanoparticles by modified spontaneous emulsification solvent diffusion method. Int. J. Pharm. 1999, 187, 143–152. [Google Scholar] [CrossRef]
- Yang, Y.-Y.; Chung, T.-S.; Ng, N.P. Morphology, drug distribution, and in vitro release profiles of biodegradable polymeric microspheres containing protein fabricated by double-emulsion solvent extraction/evaporation method. Biomaterials 2001, 22, 231–241. [Google Scholar] [CrossRef]
- Betancourt, T.; Brown, B.; Brannon-Peppas, L. Doxorubicin-loaded PLGA nanoparticles by nanoprecipitation: Preparation, characterization and in vitro evaluation. Nanomedicine 2007, 2, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Kocbek, P.; Obermajer, N.; Cegnar, M.; Kos, J.; Kristl, J. Targeting cancer cells using PLGA nanoparticles surface modified with monoclonal antibody. J. Control. Release 2007, 120, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, V.; Ankola, D.D.; Gupta, S.C.; Schneider, M.; Lehr, C.M.; Kumar, M.R. PLGA Nanoparticles Stabilized with Cationic Surfactant: Safety Studies and Application in Oral Delivery of Paclitaxel to Treat Chemical-Induced Breast Cancer in Rat. Pharm. Res. 2009, 26, 2495–2503. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.C.; Dawson, M.; Lai, S.K.; Wang, Y.-Y.; Suk, J.S.; Yang, M.; Zeitlin, P.; Boyle, M.P.; Fu, J.; Hanes, J. Biodegradable polymer nanoparticles that rapidly penetrate the human mucus barrier. Proc. Natl. Acad. Sci. USA 2009, 106, 19268–19273. [Google Scholar] [CrossRef] [PubMed]
- Kumari, A.; Yadav, S.K.; Yadav, S.C. Biodegradable polymeric nanoparticles based drug delivery systems. Colloids Surf. B Biointerfaces 2010, 75, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.; Lampe, K.J.; Heilshorn, S.C. Tetrakis(hydroxymethyl) Phosphonium Chloride as a Covalent Cross-Linking Agent for Cell Encapsulation within Protein-Based Hydrogels. Biomacromolecules 2012, 13, 3912–3916. [Google Scholar] [CrossRef] [PubMed]
- Gulka, C.P.; Wong, A.C.; Wright, D.W. Spontaneous Self-Assembly and Disassembly of Colloidal Gold Nanoparticles Induced by Tetrakis(hydroxymethyl) Phosphonium Chloride. Chem. Commun. (Camb. Engl.) 2016, 52, 1266–1269. [Google Scholar] [CrossRef] [PubMed]
- Hueso, J.L.; Sebastian, V.; Mayoral, A.; Uson, L.; Arruebo, M.; Santamaria, J. Beyond gold: Rediscovering tetrakis-(hydroxymethyl)-phosphonium chloride (THPC) as an effective agent for the synthesis of ultra-small noble metal nanoparticles and Pt-containing nanoalloys. RSC Adv. 2013, 3, 10427–10433. [Google Scholar] [CrossRef]
- Homan, K.A.; Chen, J.; Schiano, A.; Mohamed, M.; Willets, K.A.; Murugesan, S.; Stevenson, K.J.; Emelianov, S. Silver-Polymer Composite Stars: Synthesis and Applications. Adv. Funct. Mater. 2011, 21, 1673–1680. [Google Scholar] [CrossRef] [PubMed]
- Stevanovic, M.; Uskokovic, D. Poly(lactide-co-glycolide)-based Micro and Nanoparticles for the Controlled Drug Delivery of Vitamins. Curr. Nanosci. 2009, 5, 1–14. [Google Scholar]
- Pasparakis, G.; Manouras, T.; Vamvakaki, M.; Argitis, P. Harnessing photochemical internalization with dual degradable nanoparticles for combinatorial photo-chemotherapy. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Duff, D.G.; Baiker, A.; Edwards, P.P. A new hydrosol of gold clusters. 1. Formation and particle size variation. Langmuir 1993, 9, 2301–2309. [Google Scholar] [CrossRef]
- Cairns, R.; Papandreou, I.; Denko, N. Overcoming Physiologic Barriers to Cancer Treatment by Molecularly Targeting the Tumor Microenvironment. Mol. Cancer Res. 2006, 4, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Nie, S. Cell-penetrating quantum dots based on multivalent and endosome-disrupting surface coatings. J. Am. Chem. Soc. 2007, 129. [Google Scholar] [CrossRef] [PubMed]
- Dominska, M.; Dykxhoorn, D.M. Breaking down the barriers: siRNA delivery and endosome escape. J. Cell Sci. 2010, 123, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Panyam, J.; Zhou, W.-Z.; Prabha, S.; Sahoo, S.K.; Labhasetwar, V. Rapid endo-lysosomal escape of poly(DL-lactide-co-glycolide) nanoparticles: implications for drug and gene delivery. FASEB J. 2002, 16, 1217–1226. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, S. (64)Cu-Labeled Phosphonium Cations as PET Radiotracers for Tumor Imaging. Bioconjugate Chem. 2011, 22, 1459–1472. [Google Scholar] [CrossRef] [PubMed]
- Ross, M.F.; Kelso, G.F.; Blaikie, F.H.; James, A.M.; Cochemé, H.M.; Filipovska, A.; da Ros, T.; Hurd, T.R.; Smith, R.A.J.; Murphy, M.P. Lipophilic triphenylphosphonium cations as tools in mitochondrial bioenergetics and free radical biology. Biochemistry (Moscow) 2005, 70, 222–230. [Google Scholar] [CrossRef]
- Sharom, F.J. The P-glycoprotein efflux pump: How does it transport drugs? J. Membr. Biol. 1997, 160, 161–175. [Google Scholar] [CrossRef] [PubMed]
- Roepe, P.D. The P-glycoprotein efflux pump: How does it transport drugs? J. Membr. Biol. 1998, 166, 71–72. [Google Scholar] [CrossRef] [PubMed]
- Kirtane, A.; Kalscheuer, S.; Panyam, J. Exploiting Nanotechnology to Overcome Tumor Drug Resistance: Challenges and Opportunities. Adv. Drug Deliv. Rev. 2013, 65. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Hazari, S.; Mehra, S.; Kaushal, D.; Moroz, K.; Dash, S. Increased Expression of P-Glycoprotein and Doxorubicin Chemoresistance of Metastatic Breast Cancer Is Regulated by miR-298. Am. J. Pathol. 2012, 180, 2490–2503. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Liang, X.-J. Overcoming drug efflux-based multidrug resistance in cancer with nanotechnology. Chin. J. Cancer 2012, 31, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Punfa, W.; Yodkeeree, S.; Pitchakarn, P.; Ampasavate, C.; Limtrakul, P. Enhancement of cellular uptake and cytotoxicity of curcumin-loaded PLGA nanoparticles by conjugation with anti-P-glycoprotein in drug resistance cancer cells. Acta Pharmacol. Sin. 2012, 33, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Omelyanenko, V.; Kopečková, P.; Gentry, C.; Kopeček, J. Targetable HPMA copolymer-adriamycin conjugates. Recognition, internalization, and subcellular fate. J. Control. Release 1998, 53, 25–37. [Google Scholar] [CrossRef]
- Gong, J.; Chen, M.; Zheng, Y.; Wang, S.; Wang, Y. Polymeric micelles drug delivery system in oncology. J. Control. Release 2012, 159, 312–323. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, W.; Gu, J.; Ren, Q.; Fan, Z.; Zhong, W.; Fang, X.; Sha, X. Enhanced antitumor efficacy by methotrexate conjugated Pluronic mixed micelles against KBv multidrug resistant cancer. Int. J. Pharm. 2013, 452, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Kang, K.W.; Chun, M.-K.; Kim, O.; Subedi, R.K.; Ahn, S.-G.; Yoon, J.-H.; Choi, H.-K. Doxorubicin-loaded solid lipid nanoparticles to overcome multidrug resistance in cancer therapy. Nanomed. Nanotechnol. Biol. Med. 2010, 6, 210–213. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, L.; Chan, H.F.; Xie, W.; Chen, S.; He, C.; Wang, Y.; Chen, M. Co-delivery of paclitaxel and tetrandrine via iRGD peptide conjugated lipid-polymer hybrid nanoparticles overcome multidrug resistance in cancer cells. Sci. Rep. 2017, 7, 46057. [Google Scholar] [CrossRef] [PubMed]
- Thierry, A.R.; Vigé, D.; Coughlin, S.S.; Belli, J.A.; Dritschilo, A.; Rahman, A. Modulation of doxorubicin resistance in multidrug-resistant cells by liposomes. FASEB J. 1993, 7, 572–579. [Google Scholar] [PubMed]
- Li, B.; Xu, H.; Li, Z.; Yao, M.; Xie, M.; Shen, H.; Shen, S.; Wang, X.; Jin, Y. Bypassing multidrug resistance in human breast cancer cells with lipid/polymer particle assemblies. Int. J. Nanomed. 2012, 7, 187–197. [Google Scholar]
- Susa, M.; Iyer, A.K.; Ryu, K.; Choy, E.; Hornicek, F.J.; Mankin, H.; Milane, L.; Amiji, M.M.; Duan, Z. Inhibition of ABCB1 (MDR1) Expression by an siRNA Nanoparticulate Delivery System to Overcome Drug Resistance in Osteosarcoma. PLoS ONE 2010, 5, e10764. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, D.; Campbell, N.R.; Das, S.; Gupta, S.; Chenna, V.; Bisht, S.; Sysa-Shah, P.; Bedja, D.; Karikari, C.; Steenbergen, C.; et al. A composite polymer nanoparticle overcomes multidrug resistance and ameliorates doxorubicin-associated cardiomyopathy. Oncotarget 2012, 3, 640–650. [Google Scholar] [CrossRef] [PubMed]

© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brocker, C.; Kim, H.; Smith, D.; Barua, S. Heteromer Nanostars by Spontaneous Self-Assembly. Nanomaterials 2017, 7, 127. https://doi.org/10.3390/nano7060127
Brocker C, Kim H, Smith D, Barua S. Heteromer Nanostars by Spontaneous Self-Assembly. Nanomaterials. 2017; 7(6):127. https://doi.org/10.3390/nano7060127
Chicago/Turabian StyleBrocker, Caitlin, Hannah Kim, Daniel Smith, and Sutapa Barua. 2017. "Heteromer Nanostars by Spontaneous Self-Assembly" Nanomaterials 7, no. 6: 127. https://doi.org/10.3390/nano7060127
APA StyleBrocker, C., Kim, H., Smith, D., & Barua, S. (2017). Heteromer Nanostars by Spontaneous Self-Assembly. Nanomaterials, 7(6), 127. https://doi.org/10.3390/nano7060127




