Selective Labeling of Proteins on Living Cell Membranes Using Fluorescent Nanodiamond Probes
Abstract
:1. Introduction
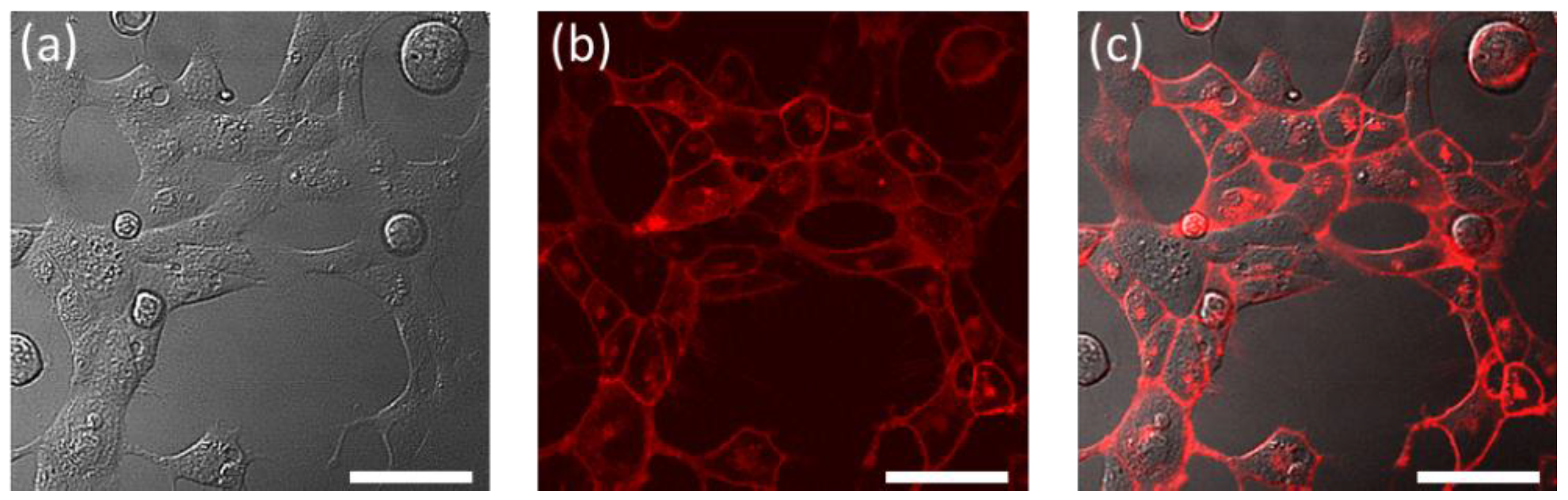
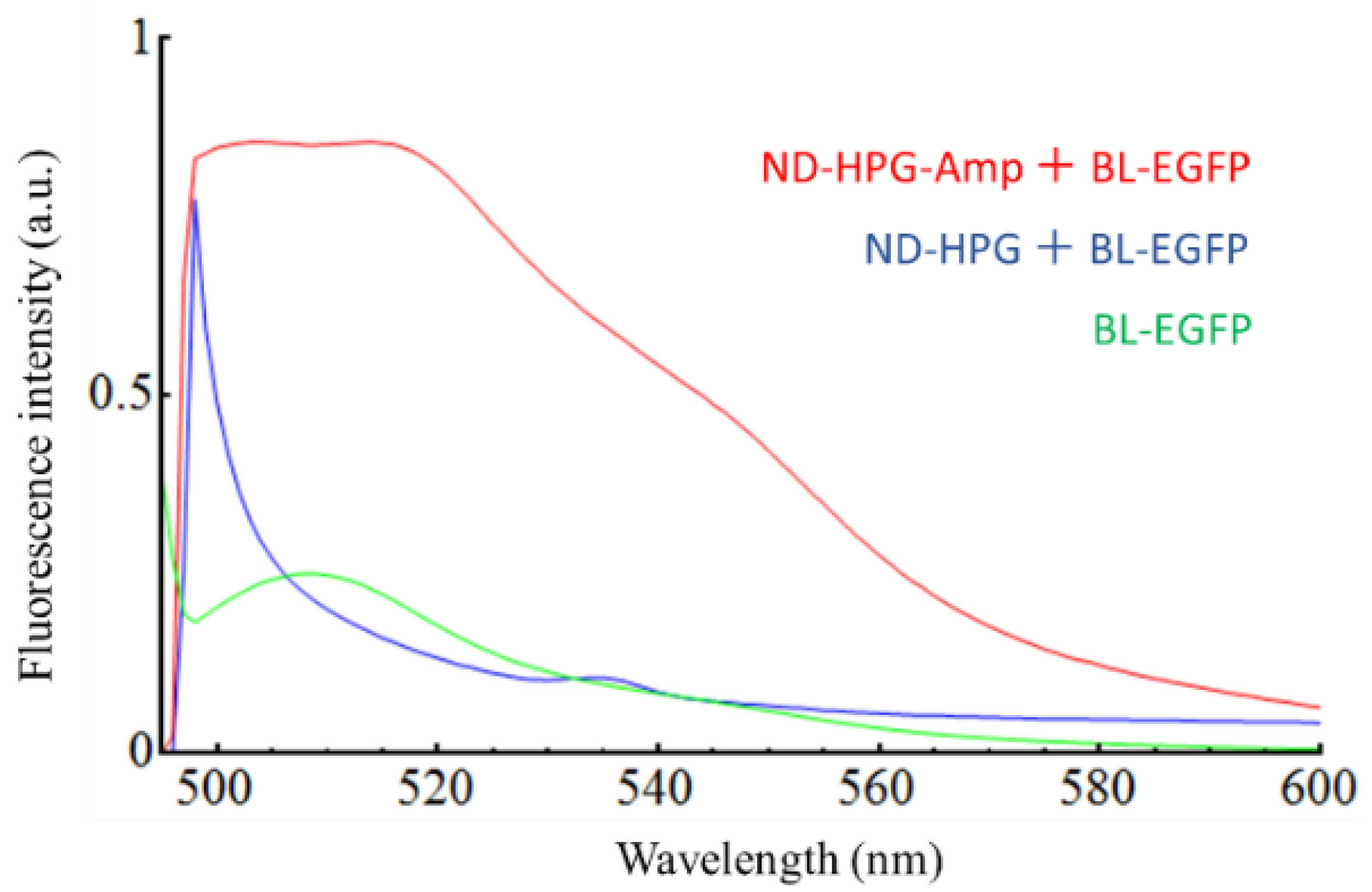
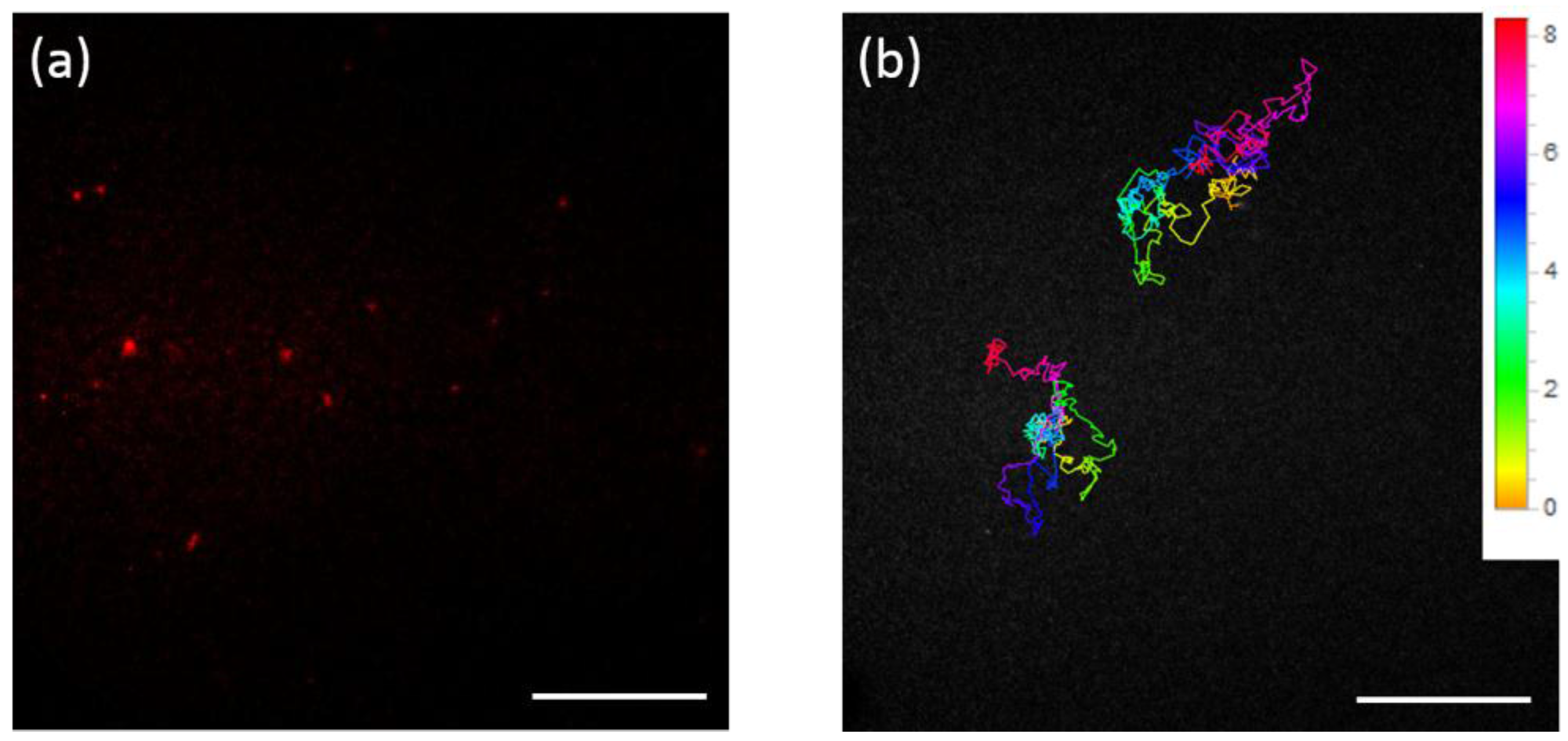
2. Results and Discussion
3. Experimental Section
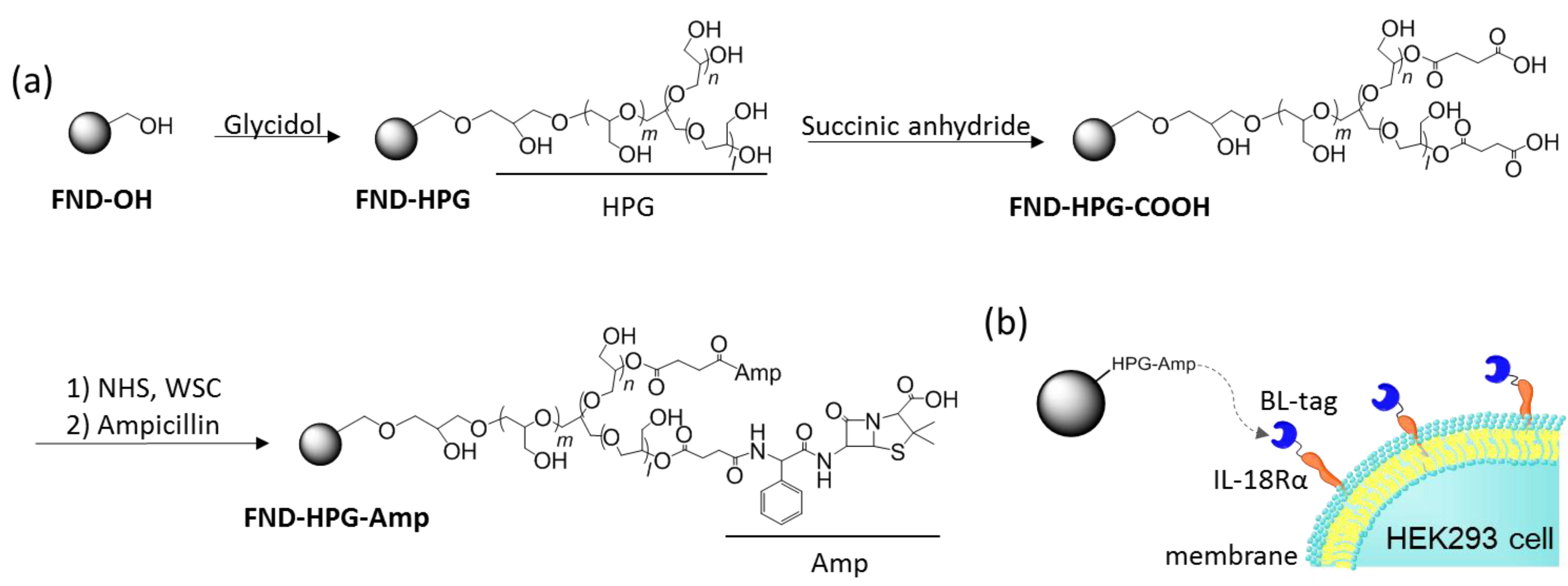
3.1. Preparation and Surface Modification of FNDs
3.1.1. Preparation of FNDs
3.1.2. FND-OH
3.1.3. FND-HPG
3.1.4. FND-HPG-COOH
3.1.5. FND-HPG-Amp
3.2. Cell Preparation
3.3. Microscopy for Single Fluorescent Molecule Tracking of FND-Labeled Proteins
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hiramoto-Yamaki, N.; Tanaka, K.A.K.; Suzuki, K.G.N.; Hirosawa, K.M.; Miyahara, M.S.H.; Kalay, Z.; Tanaka, K.; Kasai, R.S.; Kusumi, A.; Fujiwara, T.K. Ultrafast Diffusion of a Fluorescent Cholesterol Analog in Compartmentalized Plasma Membranes. Traffic 2014, 15, 583–612. [Google Scholar]
- Shibata, A.C.E.; Chen, L.H.; Nagai, R.; Ishidate, F.; Chadda, R.; Miwa, Y.; Naruse, K.; Shirai, Y.M.; Fujiwara, T.K.; Kusumi, A. Rac1 recruitment to the archipelago structure of the focal adhesion through the fluid membrane as revealed by single-molecule analysis. Cytoskeleton 2013, 70, 161–177. [Google Scholar]
- Weigel, A.V.; Simon, B.; Tamkun, M.M.; Krapf, D. Ergodic and nonergodic processes coexist in the plasma membrane as observed by single-molecule tracking. Proc. Natl. Acad. Sci. USA 2011, 108, 6438–6443. [Google Scholar]
- Fujiwara, T.; Ritchie, K.; Murakoshi, H.; Jacobson, K.; Kusumi, A. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 2002, 157, 1071–1081. [Google Scholar]
- Suzuki, K.G.N.; Fujiwara, T.K.; Sanematsu, F.; Iino, R.; Edidin, M.; Kusumi, A. GPI-anchored receptor clusters transiently recruit Lyn and Gα for temporary cluster immobilization and Lyn activation: Single-molecule tracking study 1. J. Cell Biol. 2007, 177, 717–730. [Google Scholar]
- Kasai, R.S.; Suzuki, K.G.N.; Prossnitz, E.R.; Koyama-Honda, I.; Nakada, C.; Fujiwara, T.K.; Kusumi, A. Full characterization of GPCR monomer-dimer dynamic equilibrium by single molecule imaging. J. Cell Biol. 2011, 192, 463–480. [Google Scholar]
- Yildiz, A.; Forkey, J.N.; McKinney, S.A.; Ha, T.; Goldman, Y.E.; Selvin, P.R. Myosin V walks hand-over-hand: Single fluorophore imaging with 1.5-nm localization. Science 2003, 300, 2061–2065. [Google Scholar]
- Axelrod, D. Total internal reflection fluorescence microscopy in cell biology. Traffic 2001, 2, 764–774. [Google Scholar]
- Toomre, D.; Manstein, D.J. Lighting up the cell surface with evanescent wave microscopy. Trends Cell Biol. 2001, 11, 298–303. [Google Scholar]
- Fernández-Suárez, M.; Ting, A.Y. Fluorescent probes for super-resolution imaging in living cells. Nat. Rev. Mol. Cell Biol. 2008, 9, 929–943. [Google Scholar]
- Dahan, M.; Lévi, S.; Luccardini, C.; Rostaing, P.; Riveau, B.; Triller, A. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science 2003, 302, 442–445. [Google Scholar]
- Komatsuzaki, A.; Ohyanagi, T.; Tsukasaki, Y.; Miyanaga, Y.; Ueda, M.; Jin, T. Compact Halo-Ligand-Conjugated Quantum Dots for Multicolored Single-Molecule Imaging of Overcrowding GPCR Proteins on Cell Membranes. Small 2015, 11, 1396–1401. [Google Scholar]
- Stefani, F.D.; Hoogenboom, J.P.; Barkai, E. Beyond quantum jumps: Blinking nanoscale light emitters. Phys. Today 2009, 62, 34–39. [Google Scholar]
- Cichos, F.; von Borczyskowski, C.; Orrit, M. Power-law intermittency of single emitters. Curr. Opin. Colloid Interface Sci. 2007, 12, 272–284. [Google Scholar]
- Hardman, R. A Toxicologic Review of Quantum Dots: Toxicity Depends on Physicochemical and Environmental Factors. Environ. Health Perspect. 2006, 114, 165–172. [Google Scholar]
- Gaebel, T.; Popa, I.; Gruber, A.; Domhan, M.; Jelezko, F.; Wrachtrup, J. Stable single-photon source in the near infrared. New J. Phys. 2004, 6. [Google Scholar] [CrossRef]
- Yu, S.-J.; Kang, M.-W.; Chang, H.-C.; Chen, K.-M.; Yu, Y.-C. Bright fluorescent nanodiamonds: No photobleaching and low cytotoxicity. J. Am. Chem. Soc. 2005, 127. [Google Scholar] [CrossRef]
- Fu, C.-C.; Lee, H.-Y.; Chen, K.; Lim, T.-S.; Wu, H.-Y.; Lin, P.-K.; Wei, P.-K.; Tsao, P.-H.; Chang, H.-C.; Fann, W. Characterization and application of single fluorescent nanodiamonds as cellular biomarkers. Proc. Natl. Acad. Sci. USA 2007, 104. [Google Scholar] [CrossRef]
- Chang, Y.-R.; Lee, H.-Y.; Chen, K.; Chang, C.-C.; Tsai, D.-S.; Fu, C.-C.; Lim, T.-S.; Tzeng, Y.-K.; Fang, C.-Y.; Han, C.-C.; et al. Mass production and dynamic imaging of fluorescent nanodiamonds. Nat. Nanotechnol. 2008, 3, 284–288. [Google Scholar]
- Schrand, A.M.; Huang, H.; Carlson, C.; Schlager, J.J.; Sawa, E.O.; Hussain, S.M.; Dai, L. Are diamond nanoparticles cytotoxic? J. Phys. Chem. B 2007, 111, 2–7. [Google Scholar]
- Liu, K.-K.; Cheng, C.-L.; Chang, C.-C.; Chao, J.-I. Biocompatible and detectable carboxylated nanodiamond on human cell. Nanotechnology 2007, 18. [Google Scholar] [CrossRef]
- Hui, Y.Y.; Cheng, C.-L.; Chang, H.-C. Nanodiamonds for optical bioimaging. J. Phys. D 2010, 43. [Google Scholar] [CrossRef]
- Say, J.M.; Vreden, C.; Reilly, D.J.; Brown, L.J.; Rabeau, J.R.; King, N.J.C. Luminescent nanodiamonds for biomedical applications. Biophys. Rev. 2011, 3, 171–184. [Google Scholar]
- Balasubramanian, G.; Lazariev, A.; Arumugam, S.R.; Duan, D.W. Nitrogen-Vacancy color center in diamond-emerging nanoscale applications in bioimaging and biosensing. Curr. Opin. Chem. Biol. 2014, 20, 69–77. [Google Scholar]
- Sotoma, S.; Igarashi, R.; Iimura, J.; Kumiya, Y.; Tochio, H.; Harada, Y.; Shirakawa, M. Suppression of nonspecific protein-nanodiamond adsorption enabling specific targeting of nanodiamonds to biomolecules of interest. Chem. Lett. 2015, 44, 354–356. [Google Scholar]
- Kagan, V.E.; Wipf, P.; Stoyanovsky, D.; Greenberger, J.S.; Borisenko, G.; Belikova, N.A.; Yanamala, N.; Arias, A.K.S.; Tungekar, M.A.; Jiang, J.; et al. Mitochondrial targeting of electron scavenging antioxidants: Regulation of selective oxidation vs random chain reactions. Adv. Drug Deliv. Rev. 2009, 61, 1375–1385. [Google Scholar]
- Moore, L.; Chow, E.K.-H.; Osawa, E.; Bishop, J.M.; Ho, D. Diamond-lipid hybrids enhance chemotherapeutic tolerance and mediate tumor regression. Adv. Mater. 2013, 25, 3532–3541. [Google Scholar]
- Sotoma, S.; Akagi, K.; Hosokawa, S.; Igarashi, R.; Tochio, H.; Harada, Y.; Shirakawa, M. Comprehensive and quantitative analysis for controlling the physical/chemical states and particle properties of nanodiamonds for biological applications. RSC Adv. 2015, 5, 13818–13827. [Google Scholar]
- Zhao, L.; Xu, Y.-H.; Qin, H.; Abe, S.; Akasaka, T.; Chano, T.; Watari, F.; Kimura, T.; Komatsu, N.; Chen, X. Platinum on Nanodiamond: A promising prodrug conjugated with stealth polyglycerol, targeting peptide and acid-responsive antitumor drug. Adv. Funct. Mater. 2014, 24, 5348–5357. [Google Scholar]
- Mizukami, S.; Watanabe, S.; Hori, Y.; Kikuchi, K. Covalent protein labeling based on noncatalytic β-Lactamase and a designed FRET substrate. J. Am. Chem. Soc. 2009, 131, 5016–5017. [Google Scholar]
- Watanabe, S.; Mizukami, S.; Hori, Y.; Kikuchi, K. Multicolor protein labeling in living cells using mutant β-lactamase-tag technology. Bioconjug. Chem. 2010, 21, 2320–2326. [Google Scholar]
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar]
- Kusumi, A.; Suzuki, K.G.N.; Kasai, R.S.; Ritchie, K.; Fujiwara, T.K. Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem. Sci. 2011, 36, 604–615. [Google Scholar]
- Gelles, J.; Schnapp, B.J.; Sheetz, M.P. Tracking kinesin-driven movements with nanometre-scale precision. Nature 1988, 331, 450–453. [Google Scholar]
- Toba, S.; Watanabe, T.M.; Yamaguchi-Okimoto, L.; Toyoshima, Y.Y.; Higuchi, H. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc. Natl. Acad. Sci. USA 2006, 103, 5741–5745. [Google Scholar]
- Gruber, A.; Dräbenstedt, C.; Tietz, L.; Fleury, J.; Wrachtrup, J.; Borczyskowski, C. Scanning Confocal Optical Microscopy and Magnetic Resonance on Single Defect Centers. Science 1997, 276, 2012–2014. [Google Scholar]
- Schirhagl, R.; Chang, K.; Loretz, M.; Degen, C.L. Nitrogen-vacancy centers in diamond: Nanoscale sensors for physics and biology. Annu. Rev. Phys. Chem. 2014, 65, 83–105. [Google Scholar]
- Huo, Q.; Worden, J.G. Monofunctional gold nanoparticles: Synthesis and applications. J. Nanopart. Res. 2007, 9, 1013–1025. [Google Scholar]
- Ackerson, C.J.; Sykes, M.T.; Kornberg, R.D. Defined DNA/nanoparticle conjugates. Proc. Natl. Acad. Sci. USA 2005, 102, 13383–13385. [Google Scholar]
- Sung, K.-M.; Mosley, D.W.; Peelle, B.R.; Zhang, S.; Jacobson, J.M. Synthesis of monofunctionalized gold nanoparticles by fmoc solid-phase reactions. J. Am. Chem. Soc. 2004, 126, 5064–5065. [Google Scholar] [PubMed]
- Chung, P.; Perevedentseva, E.; Tu, J.; Chang, C.; Cheng, C. Spectroscopic study of bio-functionalized nanodiamonds. Diam. Relat. Mater. 2006, 15, 622–625. [Google Scholar]
- Krüger, A.; Liang, Y.; Jarre, G.; Stegk, J. Surface functionalisation of detonation diamond suitable for biological applications. J. Mater. Chem. 2006, 16, 2322–2328. [Google Scholar]
- Zhao, L.; Takimoto, T.; Ito, M.; Kitagawa, N.; Kimura, T.; Komatsu, N. Chromatographic separation of highly soluble diamond nanoparticles prepared by polyglycerol grafting. Angew. Chem. Int. Ed. Engl. 2011, 50, 1388–1392. [Google Scholar] [PubMed]
- Boudou, J.-P.; David, M.-O.; Joshi, V.; Eidi, H.; Curmi, P.A. Hyperbranched polyglycerol modified fluorescent nanodiamond for biomedical research. Diam. Relat. Mater. 2013, 38, 131–138. [Google Scholar]
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sotoma, S.; Iimura, J.; Igarashi, R.; Hirosawa, K.M.; Ohnishi, H.; Mizukami, S.; Kikuchi, K.; Fujiwara, T.K.; Shirakawa, M.; Tochio, H. Selective Labeling of Proteins on Living Cell Membranes Using Fluorescent Nanodiamond Probes. Nanomaterials 2016, 6, 56. https://doi.org/10.3390/nano6040056
Sotoma S, Iimura J, Igarashi R, Hirosawa KM, Ohnishi H, Mizukami S, Kikuchi K, Fujiwara TK, Shirakawa M, Tochio H. Selective Labeling of Proteins on Living Cell Membranes Using Fluorescent Nanodiamond Probes. Nanomaterials. 2016; 6(4):56. https://doi.org/10.3390/nano6040056
Chicago/Turabian StyleSotoma, Shingo, Jun Iimura, Ryuji Igarashi, Koichiro M. Hirosawa, Hidenori Ohnishi, Shin Mizukami, Kazuya Kikuchi, Takahiro K. Fujiwara, Masahiro Shirakawa, and Hidehito Tochio. 2016. "Selective Labeling of Proteins on Living Cell Membranes Using Fluorescent Nanodiamond Probes" Nanomaterials 6, no. 4: 56. https://doi.org/10.3390/nano6040056
APA StyleSotoma, S., Iimura, J., Igarashi, R., Hirosawa, K. M., Ohnishi, H., Mizukami, S., Kikuchi, K., Fujiwara, T. K., Shirakawa, M., & Tochio, H. (2016). Selective Labeling of Proteins on Living Cell Membranes Using Fluorescent Nanodiamond Probes. Nanomaterials, 6(4), 56. https://doi.org/10.3390/nano6040056




