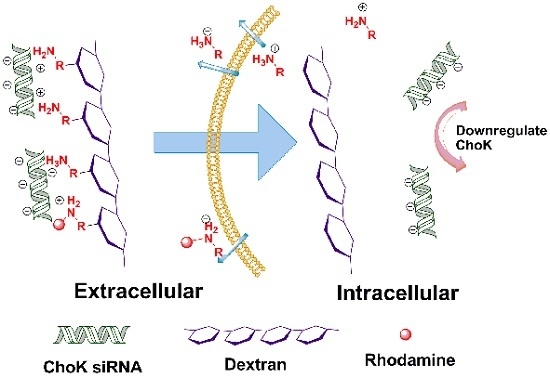
Degradable Dextran Nanopolymer as a Carrier for Choline Kinase (ChoK) siRNA Cancer Therapy
Abstract
:1. Introduction
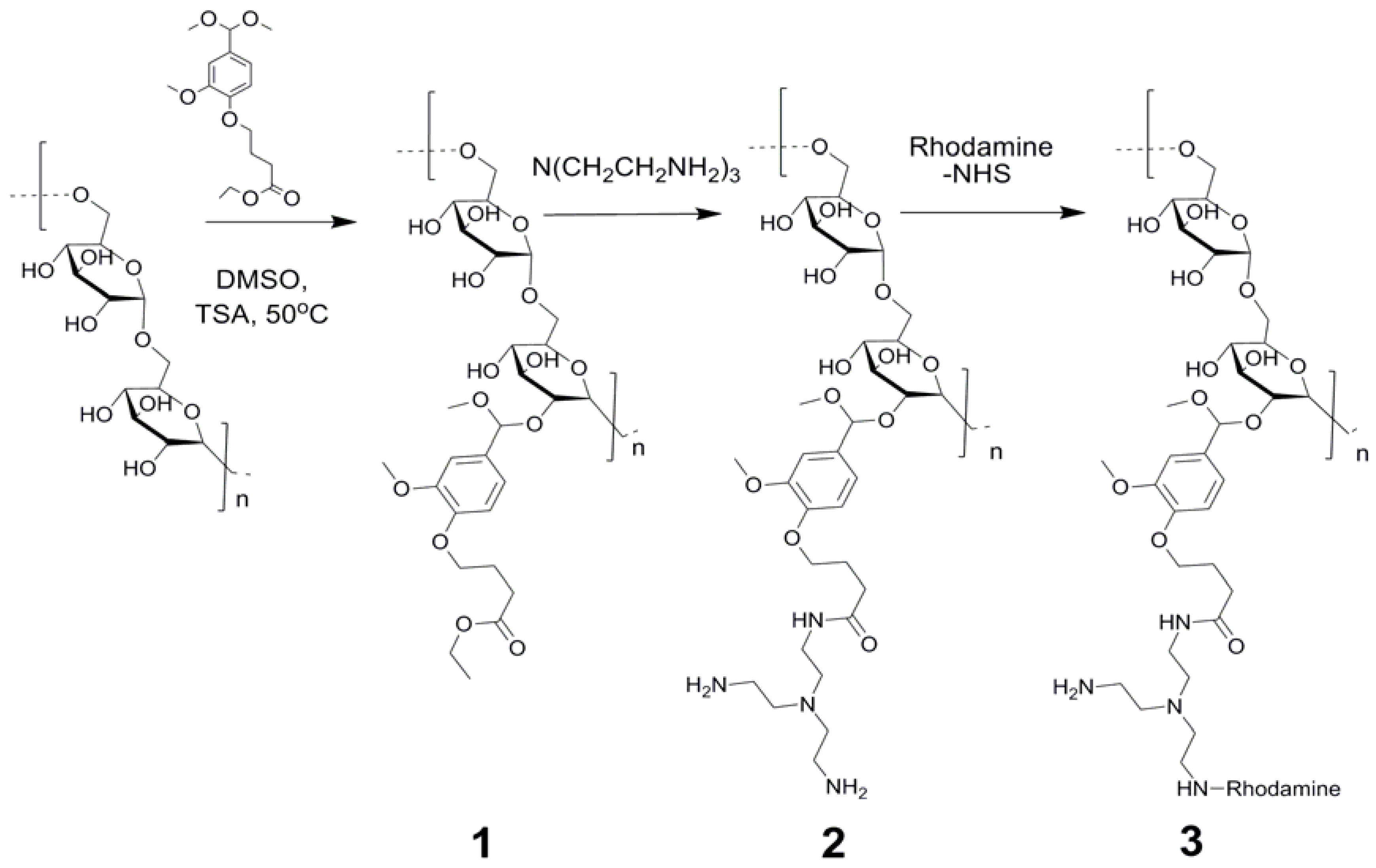
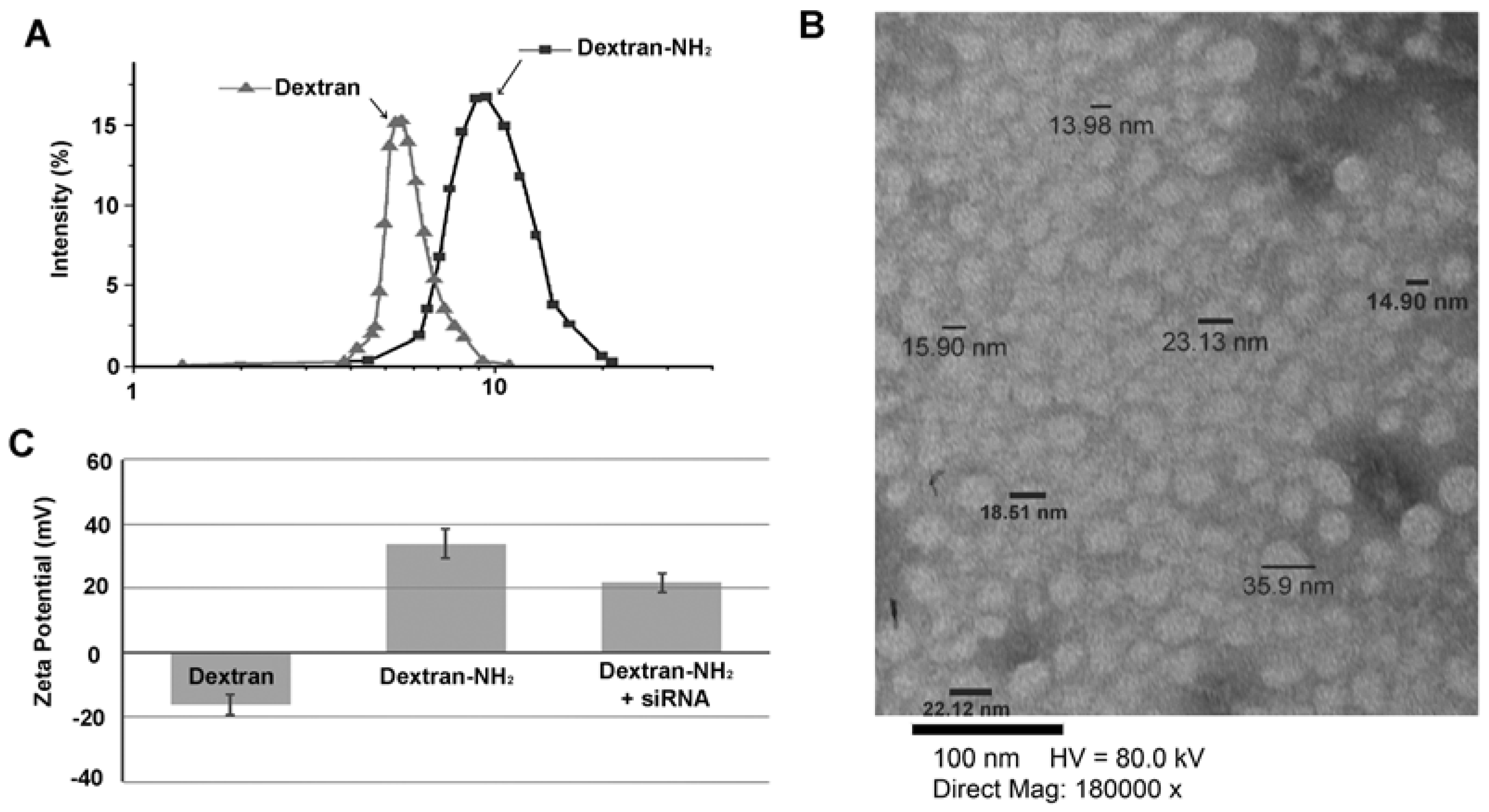
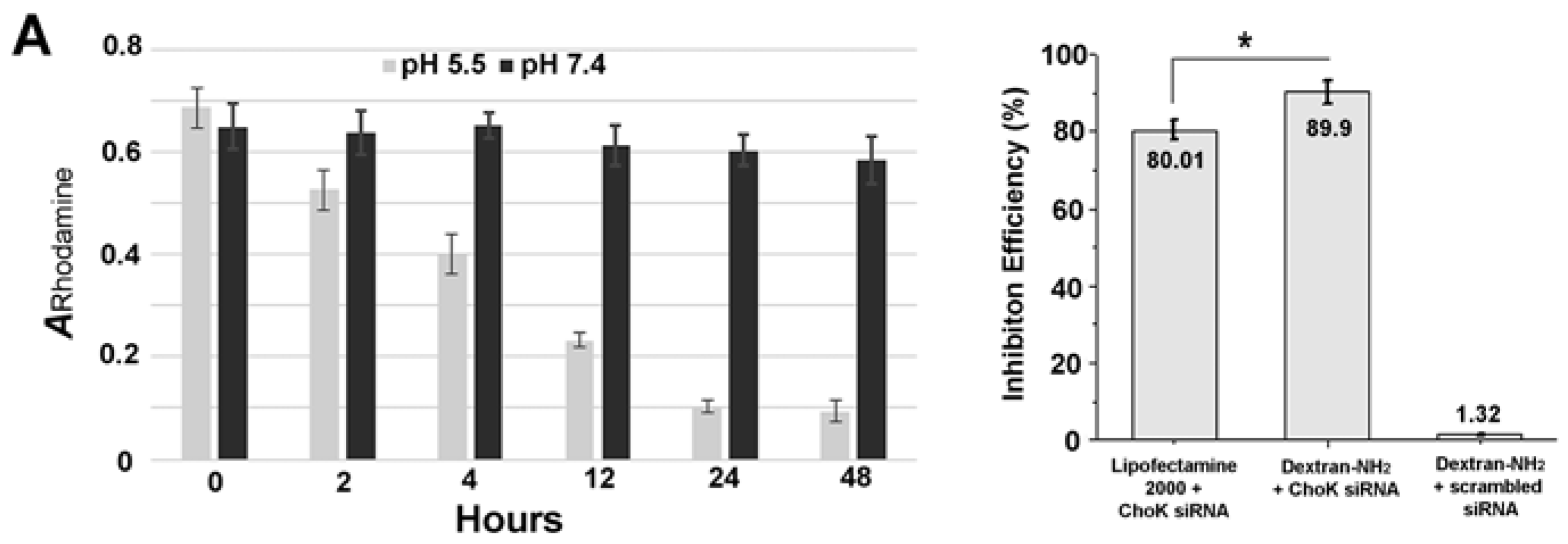
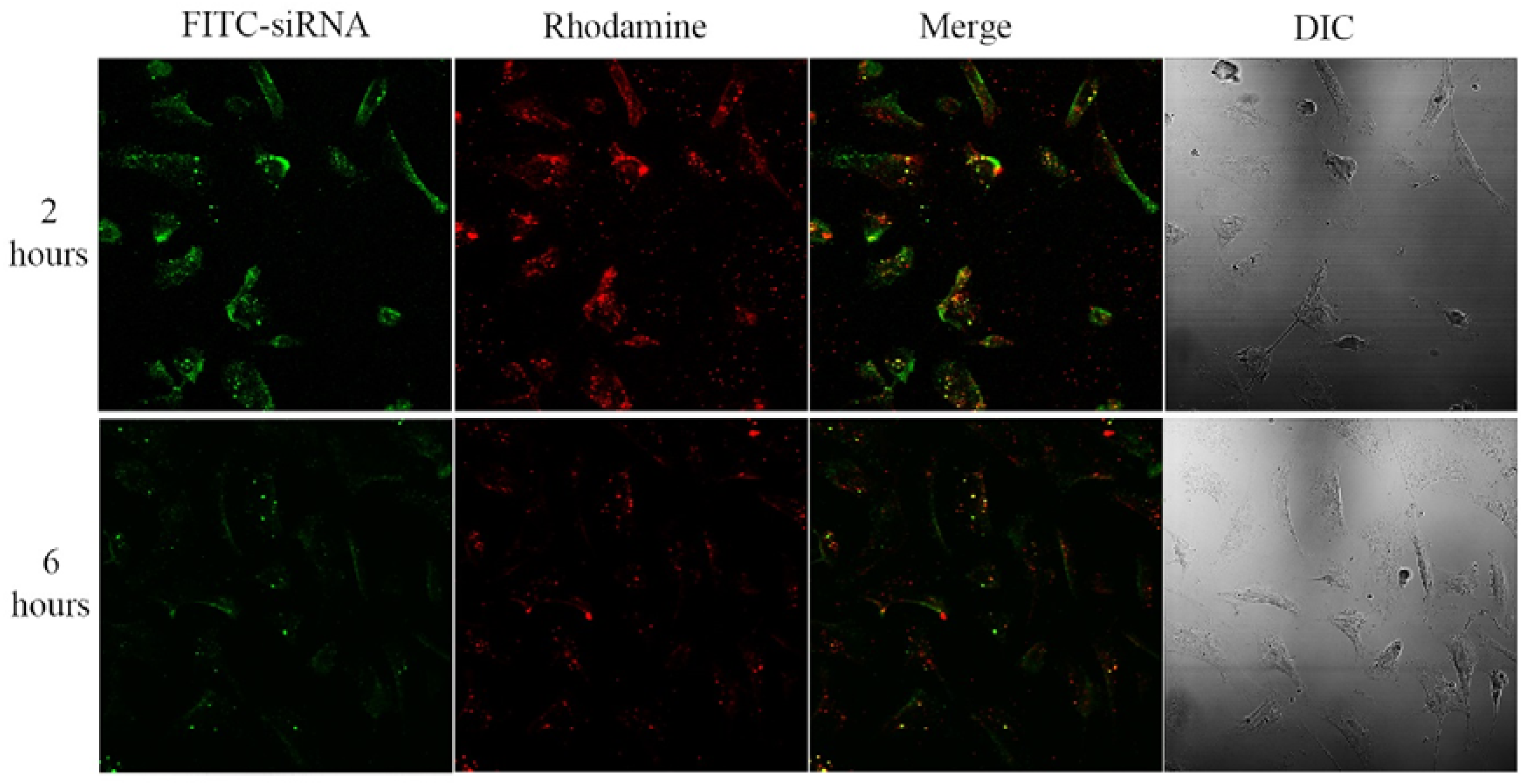
2. Results and Discussion
3. Experimental Section
3.1. Determination of Size Distribution and Zeta Potential
3.2. Colorimetric Assay of Degradation Study
3.3. Cell Culture and RNA Isolation, Quantitative Reverse Transcription-PCR (qRT-PCR)
3.4. Confocal Laser Scanning Fluorescence Microscopy
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- McManus, M.T.; Sharp, P.A. Gene silencing in mammals by small interfering RNAs. Nat. rev. Genet. 2002, 3, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Devi, G.R. siRNA-based approaches in cancer therapy. Cancer Gene Therapy 2006, 13, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Sun, T.; Ferrari, M. Nanovector delivery of sirna for cancer therapy. Cancer gene therapy 2012, 19, 367–373. [Google Scholar] [CrossRef] [PubMed]
- Moore, C.C.; Martin, E.N.; Lee, G.; Taylor, C.; Dondero, R.; Reznikov, L.L.; Dinarello, C.; Thompson, J.; Scheld, W.M. Eukaryotic translation initiation factor 5A small interference RNA-liposome complexes reduce inflammation and increase survival in murine models of severe sepsis and acute lung injury. J. Infect. Dis. 2008, 198, 1407–1414. [Google Scholar] [CrossRef] [PubMed]
- Bouzakri, K.; Zachrisson, A.; Al-Khalili, L.; Zhang, B.B.; Koistinen, H.A.; Krook, A.; Zierath, J.R. siRNA-based gene silencing reveals specialized roles of IRS-1/Akt2 and IRS-2/Akt1 in glucose and lipid metabolism in human skeletal muscle. Cell Metab. 2006, 4, 89–96. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Mao, Q.; Paulson, H.L.; Davidson, B.L. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 2002, 20, 1006–1010. [Google Scholar] [CrossRef] [PubMed]
- Singer, O.; Marr, R.A.; Rockenstein, E.; Crews, L.; Coufal, N.G.; Gage, F.H.; Verma, I.M.; Masliah, E. Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat. Neurosci. 2005, 8, 1343–1349. [Google Scholar] [CrossRef] [PubMed]
- Dykxhoorn, D.M.; Novina, C.D.; Sharp, P.A. Killing the messenger: Short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003, 4, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.J.; Kim, S.W. Efficient siRNA delivery with non-viral polymeric vehicles. Pharm. Res. 2009, 26, 657–666. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Huang, L. Nonviral methods for siRNA delivery. Mol. Pharm. 2009, 6, 651–658. [Google Scholar] [CrossRef] [PubMed]
- Tiscornia, G.; Singer, O.; Ikawa, M.; Verma, I.M. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl. Acad. Sci. USA 2003, 100, 1844–1848. [Google Scholar] [CrossRef] [PubMed]
- Semple, S.C.; Akinc, A.; Chen, J.X.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.Y.; Stebbing, D.; Crosley, E.J.; Yaworski, E.; et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Spagnou, S.; Miller, A.D.; Keller, M. Lipidic carriers of siRNA: differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry 2004, 43, 13348–13356. [Google Scholar] [CrossRef] [PubMed]
- Gary, D.J.; Puri, N.; Won, Y.Y. Polymer-based sirna delivery: Perspectives on the fundamental and phenomenological distinctions from polymer-based DNA delivery. J. Control. Release 2007, 121, 64–73. [Google Scholar] [CrossRef] [PubMed]
- Kesharwani, P.; Gajbhiye, V.; Jain, N.K. A review of nanocarriers for the delivery of small interfering RNA. Biomaterials 2012, 33, 7138–7150. [Google Scholar] [CrossRef] [PubMed]
- De Fougerolles, A.; Vornlocher, H.P.; Maraganore, J.; Lieberman, J. Interfering with disease: A progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007, 6, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Frohlich, E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomedicine 2012, 7, 5577–5591. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Alcoceba, R.; Saniger, L.; Campos, J.; Nunez, M.C.; Khaless, F.; Gallo, M.A.; Espinosa, A.; Lacal, J.C. Choline kinase inhibitors as a novel approach for antiproliferative drug design. Oncogene 1997, 15, 2289–2301. [Google Scholar] [CrossRef] [PubMed]
- Glunde, K.; Bhujwalla, Z.M.; Ronen, S.M. Choline metabolism in malignant transformation. Nat. Rev. Cancer 2011, 11, 835–848. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Penet, M.F.; Wildes, F.; Takagi, T.; Chen, Z.H.; Winnard, P.T.; Artemov, D.; Bhujwalla, Z.M. Nanoplex delivery of siRNA and prodrug enzyme for multimodality image-guided molecular pathway targeted cancer therapy. ACS Nano 2010, 4, 6707–6716. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Penet, M.F.; Nimmagadda, S.; Li, C.; Banerjee, S.R.; Winnard, P.T.; Artemov, D.; Glunde, K.; Pomper, M.G.; Bhujwalla, Z.M. PSMA-targeted theranostic nanoplex for prostate cancer therapy. ACS Nano 2012, 6, 7752–7762. [Google Scholar] [CrossRef] [PubMed]
- Mehvar, R. Dextrans for targeted and sustained delivery of therapeutic and imaging agents. J. Control. Release 2000, 69, 1–25. [Google Scholar] [CrossRef]
- Nagane, K.; Jo, J.I.; Tabata, Y. Promoted adipogenesis of rat mesenchymal stem cells by transfection of small interfering RNA complexed with a cationized dextran. Tissue Eng. Part A 2010, 16, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Oh, M.H.; Park, J.Y.; Park, T.G.; Nam, Y.S. Protein-resistant, reductively dissociable polyplexes for in vivo systemic delivery and tumor-targeting of siRNA. Biomaterials 2013, 34, 2370–2379. [Google Scholar] [CrossRef] [PubMed]
- De Backer, L.; Braeckmans, K.; Stuart, M.C.; Demeester, J.; De Smedt, S.C.; Raemdonck, K. Bio-inspired pulmonary surfactant-modified nanogels: A promising siRNA delivery system. J. Control. Release 2015, 206, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Segovia, N.; Pont, M.; Oliva, N.; Ramos, V.; Borros, S.; Artzi, N. Hydrogel doped with nanoparticles for local sustained release of siRNA in breast cancer. Adv. Healthc. Mater. 2015, 4, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.N.; Cohen, J.L.; Chu, C.K.; Wich, P.R.; Kierstead, P.H.; Frechet, J.M.J. Conjugation chemistry through acetals toward a dextran-based delivery system for controlled release of siRNA. J. Am. Chem. Soc. 2012, 134, 15840–15848. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.A.; Vanhecke, D.; Michen, B.; Blank, F.; Gehr, P.; Petri-Fink, A.; Rothen-Rutishauser, B. Different endocytotic uptake mechanisms for nanoparticles in epithelial cells and macrophages. Beilstein J. Nanotechol. 2014, 5, 1625–1636. [Google Scholar] [CrossRef] [PubMed]
- McMahon, H.T.; Boucrot, E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2011, 12, 517–533. [Google Scholar] [CrossRef] [PubMed]
- Rothen-Rutishauser, B.; Kuhn, D.A.; Ali, Z.; Gasser, M.; Amin, F.; Parak, W.J.; Vanhecke, D.; Fink, A.; Gehr, P.; Brandenberger, C. Quantification of gold nanoparticle cell uptake under controlled biological conditions and adequate resolution. Nanomedicine 2014, 9, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, T.; Varela, J.; Lynch, I.; Salvati, A.; Dawson, K.A. Effects of Transport Inhibitors on the Cellular Uptake of Carboxylated Polystyrene Nanoparticles in Different Cell Lines. PLOS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40. [Google Scholar] [CrossRef] [PubMed]
| Treatment | Relative fluorescence intensities |
|---|---|
| No inhibitor | 1 ± 0.0484 |
| Cytochalasin D (5 µg/mL) | 0.91 ± 0.0678 |
| Chlorpromazine hydrochloride (10 µg/mL) | 0.45 ± 0.0453 |
| Methyl-β-cyclodextrin (5 mM, 6.5 mg/mL) | 0.57 ± 0.0261 |
| Nocodazole (20 µM) | 0.52 ± 0.0148 |
| Low temperature (4 °C) | 0.29 ± 0.0650 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Krishnamachary, B.; Bhujwalla, Z.M. Degradable Dextran Nanopolymer as a Carrier for Choline Kinase (ChoK) siRNA Cancer Therapy. Nanomaterials 2016, 6, 34. https://doi.org/10.3390/nano6020034
Chen Z, Krishnamachary B, Bhujwalla ZM. Degradable Dextran Nanopolymer as a Carrier for Choline Kinase (ChoK) siRNA Cancer Therapy. Nanomaterials. 2016; 6(2):34. https://doi.org/10.3390/nano6020034
Chicago/Turabian StyleChen, Zhihang, Balaji Krishnamachary, and Zaver M. Bhujwalla. 2016. "Degradable Dextran Nanopolymer as a Carrier for Choline Kinase (ChoK) siRNA Cancer Therapy" Nanomaterials 6, no. 2: 34. https://doi.org/10.3390/nano6020034
APA StyleChen, Z., Krishnamachary, B., & Bhujwalla, Z. M. (2016). Degradable Dextran Nanopolymer as a Carrier for Choline Kinase (ChoK) siRNA Cancer Therapy. Nanomaterials, 6(2), 34. https://doi.org/10.3390/nano6020034