Comparison of Lung Inflammatory and Transcriptional Responses in Mice and Rats Following Pulmonary Exposure to a Fiber Paradigm-Compatible and Non-Compatible MWCNT
Abstract
1. Introduction
2. Materials and Methods
2.1. Multi-Walled Carbon Nanotubes
2.2. Animals and Exposure
2.3. Sample Collection
2.4. Bronchoalveolar Lavage Cell Composition
2.5. RNA Extraction and Quality Assessment
2.6. Microarray Hybridization
2.7. Transcriptomic Data Preprocessing
2.8. Functional Enrichment
3. Results
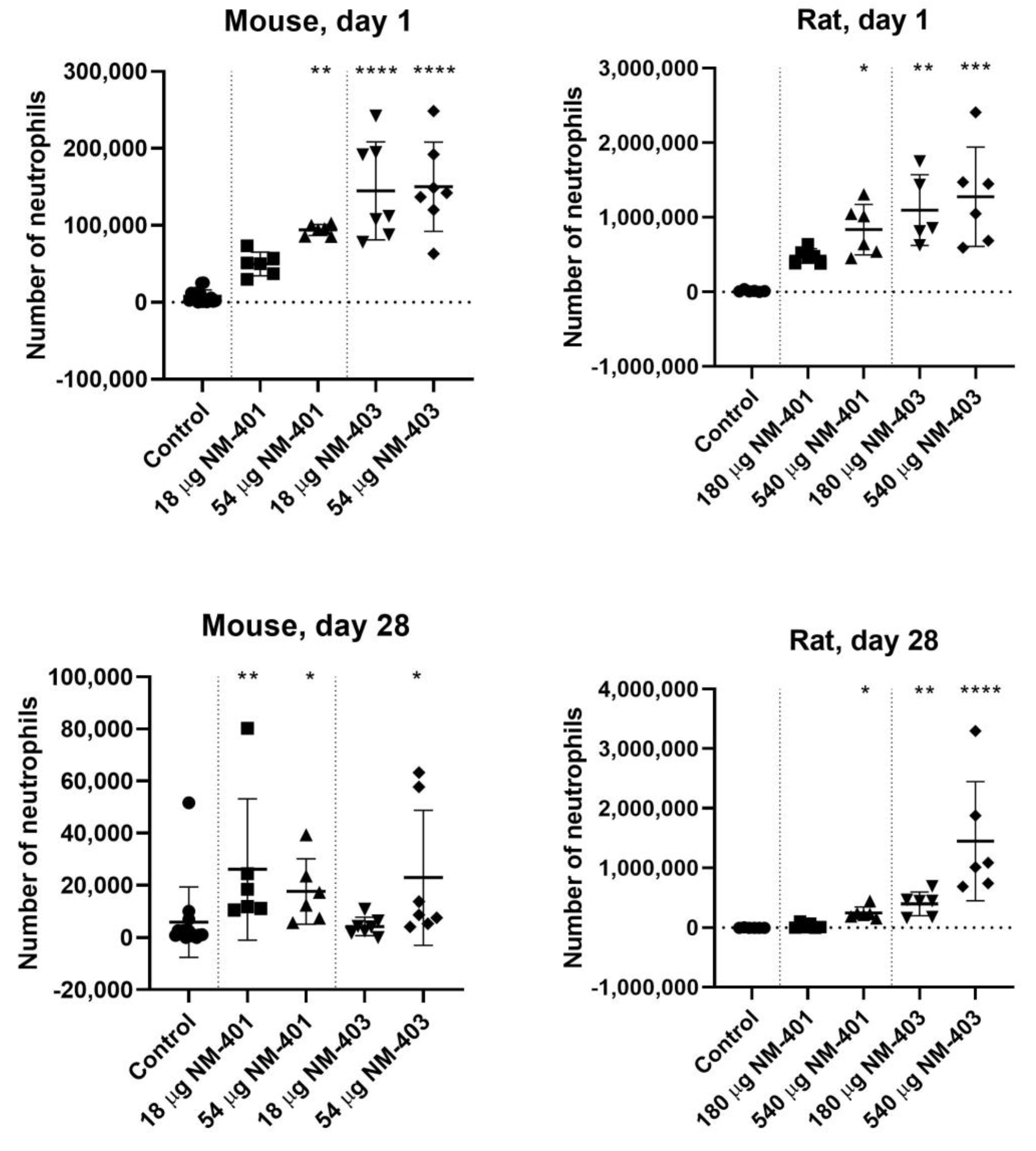
3.1. Neutrophil Influx
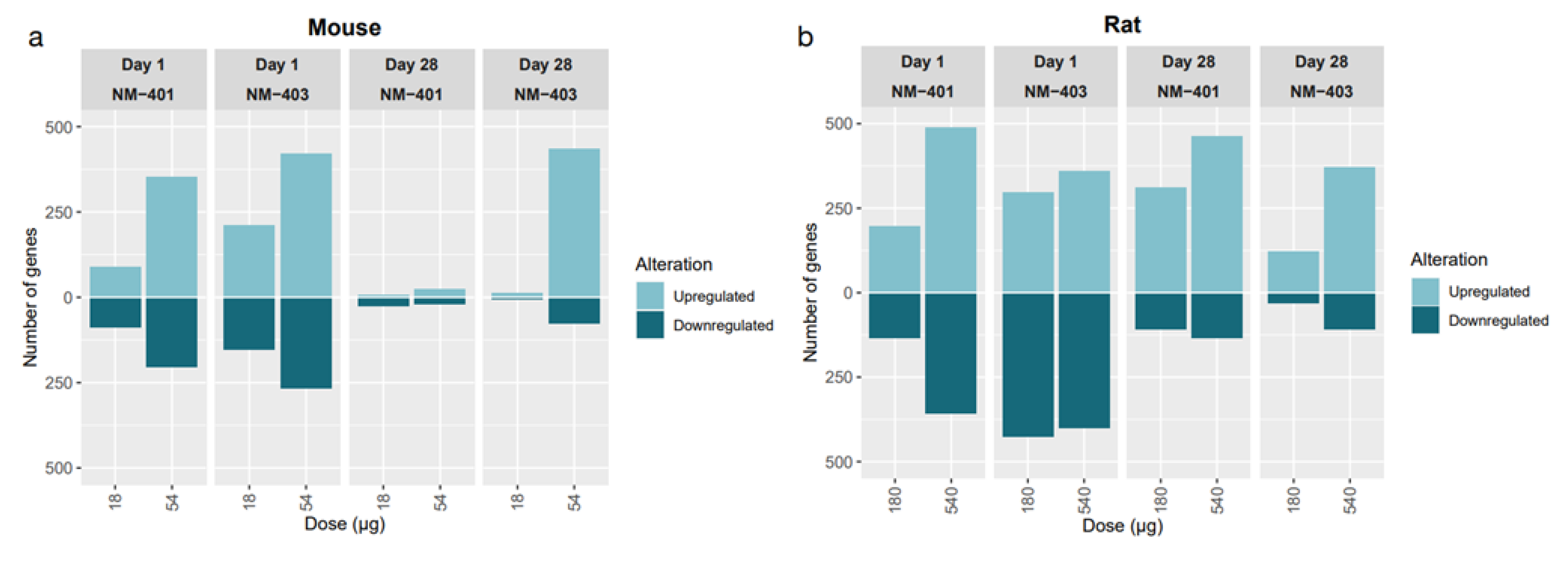
3.2. Transcriptomic Alterations
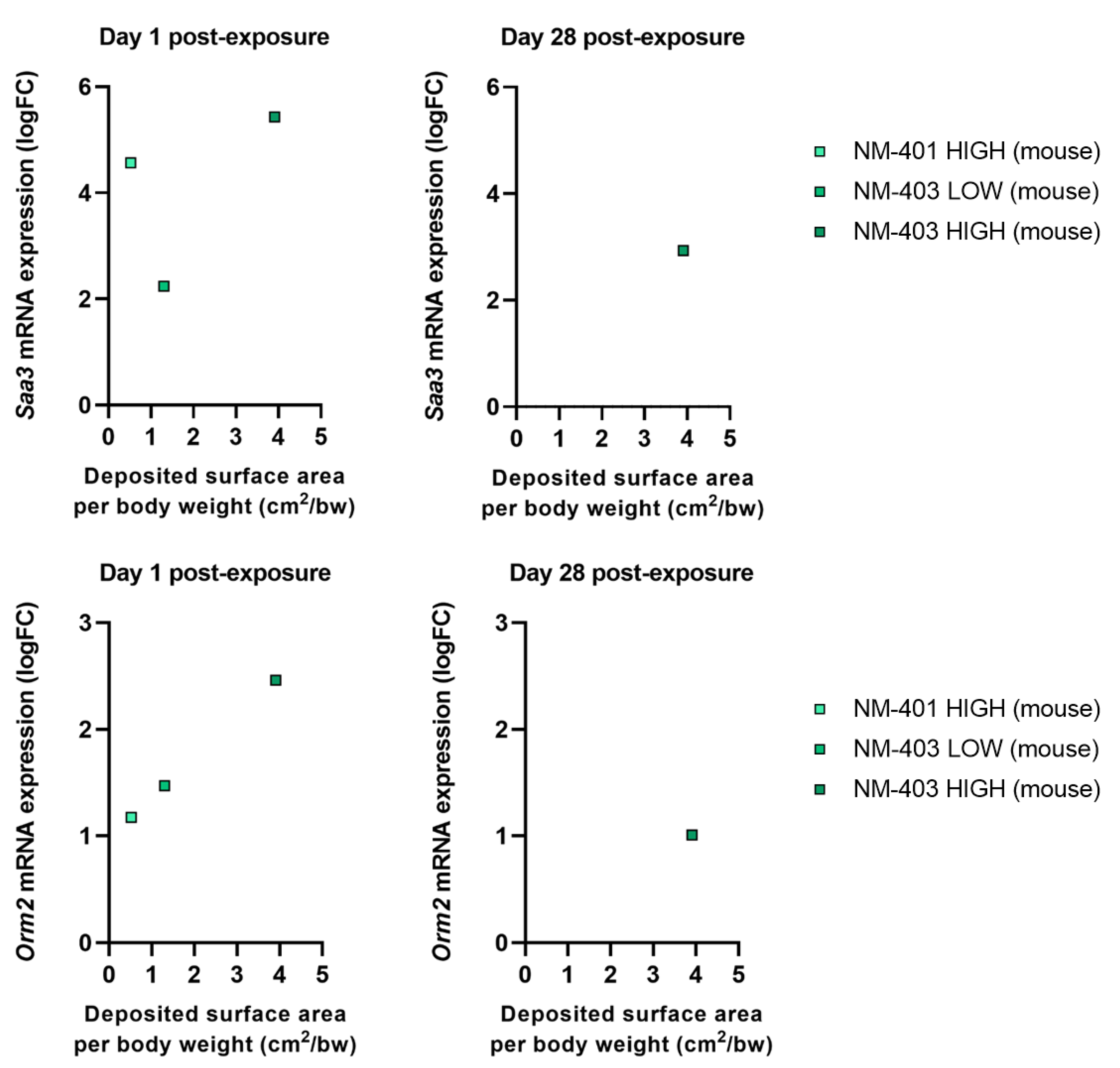
3.3. Fibrosis
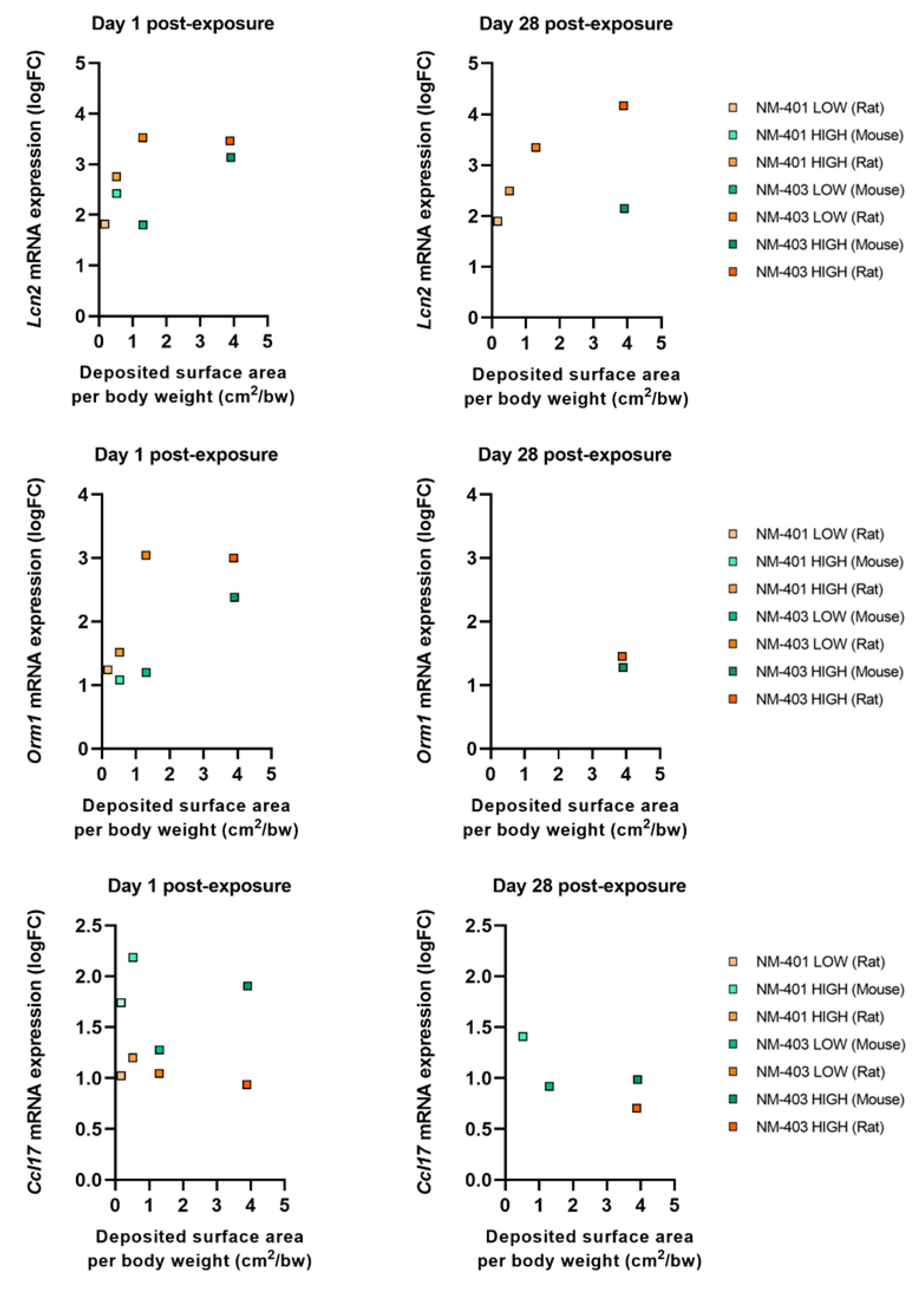
3.4. Acute Phase Response
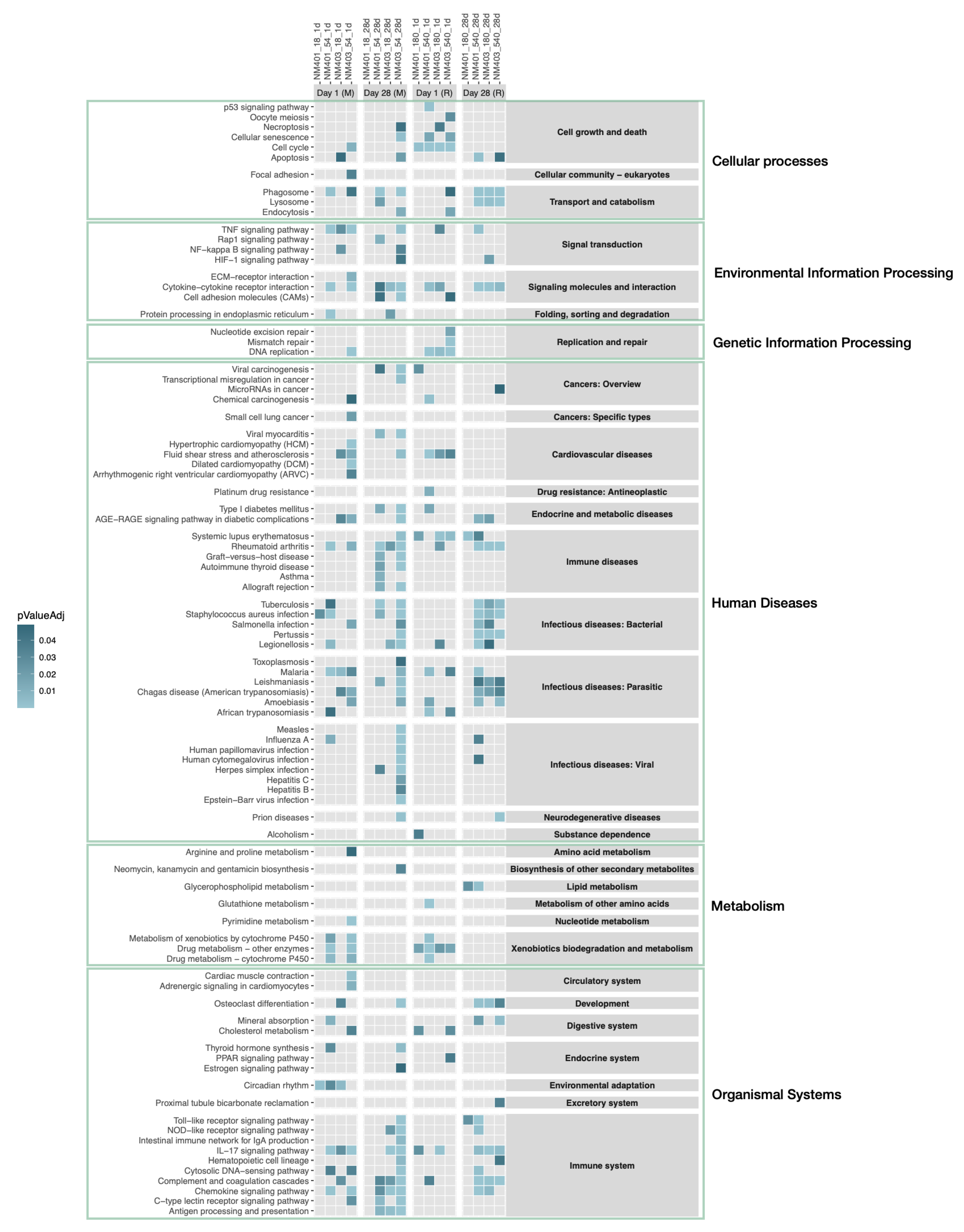
3.5. Functional Enrichment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Beg, S.; Rizwan, M.; Sheikh, A.M.; Hasnain, M.S.; Anwer, K.; Kohli, K. Advancement in carbon nanotubes: Basics, biomedical applications and toxicity. J. Pharm. Pharmacol. 2011, 63, 141–163. [Google Scholar] [CrossRef] [PubMed]
- Pauluhn, J. Subchronic 13-Week Inhalation Exposure of Rats to Multiwalled Carbon Nanotubes: Toxic Effects Are Determined by Density of Agglomerate Structures, Not Fibrillar Structures. Toxicol. Sci. 2009, 113, 226–242. [Google Scholar] [CrossRef]
- Kasai, T.; Umeda, Y.; Ohnishi, M.; Mine, T.; Kondo, H.; Takeuchi, T.; Matsumoto, M.; Fukushima, S. Lung carcinogenicity of inhaled multi-walled carbon nanotube in rats. Part. Fibre Toxicol. 2015, 13, 53. [Google Scholar] [CrossRef] [PubMed]
- Nikota, J.; Banville, A.; Goodwin, L.R.; Wu, D.; Williams, A.; Yauk, C.L.; Wallin, H.; Vogel, U.; Halappanavar, S. Stat-6 signaling pathway and not Interleukin-1 mediates multi-walled carbon nanotube-induced lung fibrosis in mice: Insights from an adverse outcome pathway framework. Part. Fibre Toxicol. 2017, 14, 37. [Google Scholar] [CrossRef] [PubMed]
- Christophersen, D.V.; Jacobsen, N.R.; Andersen, M.H.; Connell, S.P.; Barfod, K.K.; Thomsen, M.B.; Miller, M.R.; Duffin, R.; Lykkesfeldt, J.; Vogel, U.; et al. Cardiovascular health effects of oral and pulmonary exposure to multi-walled carbon nanotubes in ApoE-deficient mice. Toxicology 2016, 371, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Rahman, L.; Jacobsen, N.R.; Aziz, S.A.; Wu, D.; Williams, A.; Yauk, C.L.; White, P.; Wallin, H.; Vogel, U.; Halappanavar, S. Multi-walled carbon nanotube-induced genotoxic, inflammatory and pro-fibrotic responses in mice: Investigating the mechanisms of pulmonary carcinogenesis. Mutat. Res. Toxicol. Environ. Mutagen. 2017, 823, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, S.S.; Saber, A.T.; Mortensen, A.; Szarek, J.; Wu, D.; Williams, A.; Andersen, O.; Jacobsen, N.R.; Yauk, C.L.; Wallin, H.; et al. Changes in cholesterol homeostasis and acute phase response link pulmonary exposure to multi-walled carbon nanotubes to risk of cardiovascular disease. Toxicol. Appl. Pharmacol. 2015, 283, 210–222. [Google Scholar] [CrossRef]
- Poulsen, S.S.; Jackson, P.; Kling, K.; Knudsen, K.B.; Skaug, V.; Kyjovska, Z.O.; Thomsen, B.L.; Clausen, P.A.; Atluri, R.; Berthing, T.; et al. Multi-walled carbon nanotube physicochemical properties predict pulmonary inflammation and genotoxicity. Nanotoxicology 2016, 10, 1263–1275. [Google Scholar] [CrossRef]
- Poulsen, S.S.; Knudsen, K.B.; Jackson, P.; Weydahl, I.E.K.; Saber, A.T.; Wallin, H.; Vogel, U.; Stoeger, T. Multi-walled carbon nanotube-physicochemical properties predict the systemic acute phase response following pulmonary exposure in mice. PLoS ONE 2017, 12, e0174167. [Google Scholar] [CrossRef]
- Saleh, D.M.; Alexander, W.T.; Numano, T.; Ahmed, O.H.M.; Gunasekaran, S.; Alexander, D.B.; Abdelgied, M.; El-Gazzar, A.M.; Takase, H.; Xu, J.; et al. Comparative carcinogenicity study of a thick, straight-type and a thin, tangled-type multi-walled carbon nanotube administered by intra-tracheal instillation in the rat. Part. Fibre Toxicol. 2020, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Saleh, D.M.; Luo, S.; Ahmed, O.H.M.; Alexander, D.B.; Alexander, W.T.; Gunasekaran, S.; El-Gazzar, A.M.; Abdelgied, M.; Numano, T.; Takase, H.; et al. Assessment of the toxicity and carcinogenicity of double-walled carbon nanotubes in the rat lung after intratracheal instillation: A two-year study. Part. Fibre Toxicol. 2022, 19, 30. [Google Scholar] [CrossRef]
- Jackson, P.; Kling, K.; Jensen, K.A.; Clausen, P.A.; Madsen, A.M.; Wallin, H.; Vogel, U. Characterization of genotoxic response to 15 multiwalled carbon nanotubes with variable physicochemical properties including surface functionalizations in the FE1-Muta(TM) mouse lung epithelial cell line. Environ. Mol. Mutagen. 2014, 56, 183–203. [Google Scholar] [CrossRef]
- Danielsen, P.H.; Poulsen, S.S.; Knudsen, K.B.; Clausen, P.A.; Jensen, K.A.; Wallin, H.; Vogel, U. Physicochemical properties of 26 carbon nanotubes as predictors for pulmonary inflammation and acute phase response in mice following intratracheal lung exposure. Environ. Toxicol. Pharmacol. 2024, 107, 104413. [Google Scholar] [CrossRef]
- Hadrup, N.; Knudsen, K.B.; Carriere, M.; Mayne-L’hErmite, M.; Bobyk, L.; Allard, S.; Miserque, F.; Pibaleau, B.; Pinault, M.; Wallin, H.; et al. Safe-by-design strategies for lowering the genotoxicity and pulmonary inflammation of multiwalled carbon nanotubes: Reduction of length and the introduction of COOH groups. Environ. Toxicol. Pharmacol. 2021, 87, 103702. [Google Scholar] [CrossRef]
- Guo, C.; Wright, M.D.; Buckley, A.; Laycock, A.; Berthing, T.; Vogel, U.; Cosnier, F.; Gaté, L.; Leonard, M.O.; Smith, R. Pulmonary Toxicity of Long, Thick MWCNT and Very Long, Thin Carboxylated MWCNT Aerosols Following 28 Days Whole-Body Exposure. Toxics 2025, 13, 401. [Google Scholar] [CrossRef] [PubMed]
- Danielsen, P.H.; Poulsen, S.S.; Mortensen, A.; Berthing, T.; Gargouri, D.; Filoramo, A.; Kohonen, P.; Grafström, R.; Testard, F.; Vogel, U. Imogolite nanotube modifications impact pulmonary toxicity in mice: Implications for safe and sustainable by design (SSbD). J. Nanobiotechnol. 2025, 23, 571. [Google Scholar] [CrossRef] [PubMed]
- Halappanavar, S.; Brule, S.v.D.; Nymark, P.; Gaté, L.; Seidel, C.; Valentino, S.; Zhernovkov, V.; Danielsen, P.H.; De Vizcaya, A.; Wolff, H.; et al. Adverse outcome pathways as a tool for the design of testing strategies to support the safety assessment of emerging advanced materials at the nanoscale. Part. Fibre Toxicol. 2020, 17, 16. [Google Scholar] [CrossRef] [PubMed]
- Halappanavar, S.; Sharma, M.; Solorio-Rodriguez, S.; Wallin, H.; Vogel, U.; Sullivan, K.; Clippinger, A.J. Substance Interaction with the Pulmonary Resident Cell Membrane Components Leading to Pulmonary Fibrosis; OECD series on Adverse Outcome Pathways, No. 33; OECD Publishing: Paris, France, 2023. [Google Scholar] [CrossRef]
- Solorio-Rodriguez, S.A.; Williams, A.; Poulsen, S.S.; Knudsen, K.B.; Jensen, K.A.; Clausen, P.A.; Danielsen, P.H.; Wallin, H.; Vogel, U.; Halappanavar, S. Single-Walled vs. Multi-Walled Carbon Nanotubes: Influence of Physico-Chemical Properties on Toxicogenomics Responses in Mouse Lungs. Nanomaterials 2023, 13, 1059. [Google Scholar] [CrossRef]
- Ma-Hock, L.; Treumann, S.; Strauss, V.; Brill, S.; Luizi, F.; Mertler, M.; Wiench, K.; Gamer, A.O.; van Ravenzwaay, B.; Landsiedel, R. Inhalation Toxicity of Multiwall Carbon Nanotubes in Rats Exposed for 3 Months. Toxicol. Sci. 2009, 112, 468–481. [Google Scholar] [CrossRef]
- Poulsen, S.S.; Saber, A.T.; Williams, A.; Andersen, O.; Købler, C.; Atluri, R.; Pozzebon, M.E.; Mucelli, S.P.; Simion, M.; Rickerby, D.; et al. MWCNTs of different physicochemical properties cause similar inflammatory responses, but differences in transcriptional and histological markers of fibrosis in mouse lungs. Toxicol. Appl. Pharmacol. 2015, 284, 16–32. [Google Scholar] [CrossRef]
- Gaté, L.; Knudsen, K.B.; Seidel, C.; Berthing, T.; Chézeau, L.; Jacobsen, N.R.; Valentino, S.; Wallin, H.; Bau, S.; Wolff, H.; et al. Pulmonary toxicity of two different multi-walled carbon nanotubes in rat: Comparison between intratracheal instillation and inhalation exposure. Toxicol. Appl. Pharmacol. 2019, 375, 17–31. [Google Scholar] [CrossRef]
- Rittinghausen, S.; Hackbarth, A.; Creutzenberg, O.; Ernst, H.; Heinrich, U.; Leonhardt, A.; Schaudien, D. The carcinogenic effect of various multi-walled carbon nanotubes (MWCNTs) after intraperitoneal injection in rats. Part. Fibre Toxicol. 2014, 11, 59. [Google Scholar] [CrossRef]
- Hadrup, N.; Zhernovkov, V.; Jacobsen, N.R.; Voss, C.; Strunz, M.; Ansari, M.; Schiller, H.B.; Halappanavar, S.; Poulsen, S.S.; Kholodenko, B.; et al. Acute Phase Response as a Biological Mechanism-of-Action of (Nano)particle-Induced Cardiovascular Disease. Small 2020, 16, e1907476. [Google Scholar] [CrossRef]
- Li, Z.; Hulderman, T.; Salmen, R.; Chapman, R.; Leonard, S.S.; Young, S.-H.; Shvedova, A.; Luster, M.I.; Simeonova, P.P. Cardiovascular Effects of Pulmonary Exposure to Single-Wall Carbon Nanotubes. Environ. Health Perspect. 2007, 115, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.C.; Wilson, P.G.; Shridas, P.; Ji, A.; de Beer, M.; de Beer, F.C.; Webb, N.R.; Tannock, L.R. Serum amyloid A3 is pro-atherogenic. Atherosclerosis 2018, 268, 32–35. [Google Scholar] [CrossRef]
- Vest Christophersen, D.; Møller, P.; Thomsen, M.B.; Lykkesfeldt, J.; Loft, S.; Wallin, H.; Vogel, U.; Jacobsen, N.R. Accelerated atherosclerosis caused by serum amyloid A response in lungs of ApoE−/− mice. FASEB J. 2021, 35, e21307. [Google Scholar] [CrossRef]
- Donaldson, K.; Poland, C.A.; Murphy, F.A.; MacFarlane, M.; Chernova, T.; Schinwald, A. Pulmonary toxicity of carbon nanotubes and asbestos—Similarities and differences. Adv. Drug Deliv. Rev. 2013, 65, 2078–2086. [Google Scholar] [CrossRef]
- RAC; Committee for Risk Assessment. Opinion Proposing Harmonised Classification and Labelling at EU Level of Multi-Walled Carbon Tubes (Synthetic Graphite in Tubular Shape) with a Geometric Tube Diameter Range ≥ 30 nm to < 3 μm and a Length ≥ 5 μm and Aspect Ratio ≥ 3:1, Including Multi-Walled Carbon Nanotubes, MWC(N)T. 2022. Available online: https://www.echa.europa.eu (accessed on 29 August 2025).
- Wolff, H.; Vogel, U. Recent progress in the EU classification of the health hazards associated with certain multiwall carbon nanotubes (MWCNTs): What about the other MWCNTs? Nanotoxicology 2024, 18, 119–121. [Google Scholar] [CrossRef]
- Barthel, H.; Sébillaud, S.; Lorcin, M.; Wolff, H.; Viton, S.; Cosnier, F.; Gaté, L.; Seidel, C. Needlelike, short and thin multi-walled carbon nanotubes: Comparison of effects on wild type and p53+/− rat lungs. Nanotoxicology 2023, 17, 270–288. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, K.B.; Berthing, T.; Jackson, P.; Poulsen, S.S.; Mortensen, A.; Jacobsen, N.R.; Skaug, V.; Szarek, J.; Hougaard, K.S.; Wolff, H.; et al. Physicochemical predictors of Multi-Walled Carbon Nanotube–induced pulmonary histopathology and toxicity one year after pulmonary deposition of 11 different Multi-Walled Carbon Nanotubes in mice. Basic Clin. Pharmacol. Toxicol. 2018, 124, 211–227. [Google Scholar] [CrossRef] [PubMed]
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Some Nanomaterials and Some Fibres; International Agency for Research on Cancer: Lyon, France, 2017; IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, No. 111. 6, Evaluation. Available online: https://www.ncbi.nlm.nih.gov/books/NBK436596/ (accessed on 29 August 2025).
- Kyjovska, Z.O.; Jacobsen, N.R.; Saber, A.T.; Bengtson, S.; Jackson, P.; Wallin, H.; Vogel, U. DNA damage following pulmonary exposure by instillation to low doses of carbon black (Printex 90) nanoparticles in mice. Environ. Mol. Mutagen. 2014, 56, 41–49. [Google Scholar] [CrossRef]
- NIOSH. Current Intelligence Bulletin 65: Occupational Exposure to Carbon Nanotubes and Nanofibers; DHHS Publication No. 2013-145; U.S. Department of Health & Human Services: Washington, DC, USA, 2013; pp. 1–184. Available online: https://cdc.gov/niosh/docs/2013-145/default.html (accessed on 29 August 2025).
- Jackson, P.; Lund, S.P.; Kristiansen, G.; Andersen, O.; Vogel, U.; Wallin, H.; Hougaard, K.S. An Experimental Protocol for Maternal Pulmonary Exposure in Developmental Toxicology. Basic Clin. Pharmacol. Toxicol. 2010, 108, 202–207. [Google Scholar] [CrossRef]
- Jacobsen, N.R.; Stoeger, T.; van den Brule, S.; Saber, A.T.; Beyerle, A.; Vietti, G.; Mortensen, A.; Szarek, J.; Budtz, H.C.; Kermanizadeh, A.; et al. Acute and subacute pulmonary toxicity and mortality in mice after intratracheal instillation of ZnO nanoparticles in three laboratories. Food Chem. Toxicol. 2015, 85, 84–95. [Google Scholar] [CrossRef]
- Hadrup, N.; Guldbrandsen, M.; Terrida, E.; Bendtsen, K.M.S.; Hougaard, K.S.; Jacobsen, N.R.; Vogel, U. Intratracheal instillation for the testing of pulmonary toxicity in mice—Effects of instillation devices and feed type on inflammation. Anim. Model. Exp. Med. 2025, 8, 378–386. [Google Scholar] [CrossRef] [PubMed]
- Hadrup, N.; Bengtson, S.; Jacobsen, N.R.; Jackson, P.; Nocun, M.; Saber, A.T.; Jensen, K.A.; Wallin, H.; Vogel, U. Influence of dispersion medium on nanomaterial-induced pulmonary inflammation and DNA strand breaks: Investigation of carbon black, carbon nanotubes and three titanium dioxide nanoparticles. Mutagenesis 2017, 32, 581–597. [Google Scholar] [CrossRef] [PubMed]
- Marwah, V.S.; Scala, G.; Kinaret, P.A.S.; Serra, A.; Alenius, H.; Fortino, V.; Greco, D. eUTOPIA: solUTion for Omics data PreprocessIng and Analysis. Source Code Biol. Med. 2019, 14, 1. [Google Scholar] [CrossRef]
- Kauffmann, A.; Gentleman, R.; Huber, W. arrayQualityMetrics—A bioconductor package for quality assessment of microarray data. Bioinformatics 2009, 25, 415–416. [Google Scholar] [CrossRef]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Scala, G.; Serra, A.; Marwah, V.S.; Saarimäki, L.A.; Greco, D. FunMappOne: A tool to hierarchically organize and visually navigate functional gene annotations in multiple experiments. BMC Bioinform. 2019, 20, 79. [Google Scholar] [CrossRef] [PubMed]
- Snyder-Talkington, B.N.; Dymacek, J.; Porter, D.W.; Wolfarth, M.G.; Mercer, R.R.; Pacurari, M.; Denvir, J.; Castranova, V.; Qian, Y.; Guo, N.L. System-based identification of toxicity pathways associated with multi-walled carbon nanotube-induced pathological responses. Toxicol. Appl. Pharmacol. 2013, 272, 476–489. [Google Scholar] [CrossRef]
- Rahman, L.; Williams, A.; Gelda, K.; Nikota, J.; Wu, D.; Vogel, U.; Halappanavar, S. 21st Century Tools for Nanotoxicology: Transcriptomic Biomarker Panel and Precision-Cut Lung Slice Organ Mimic System for the Assessment of Nanomaterial-Induced Lung Fibrosis. Small 2020, 16, e2000272. [Google Scholar] [CrossRef] [PubMed]
- Gromelski, M.; Stoliński, F.; Jagiello, K.; Rybińska-Fryca, A.; Williams, A.; Halappanavar, S.; Vogel, U.; Puzyn, T. AOP173 key event associated pathway predictor—Online application for the prediction of benchmark dose lower bound (BMDLs) of a transcriptomic pathway involved in MWCNTs-induced lung fibrosis. Nanotoxicology 2022, 16, 183–194. [Google Scholar] [CrossRef]
- Jagiello, K.; Halappanavar, S.; Rybińska-Fryca, A.; Willliams, A.; Vogel, U.; Puzyn, T. Transcriptomics-Based and AOP-Informed Structure–Activity Relationships to Predict Pulmonary Pathology Induced by Multiwalled Carbon Nanotubes. Small 2021, 17, e2003465. [Google Scholar] [CrossRef]
- Merugu, S.; Jagiello, K.; Gajewicz-Skretna, A.; Halappanavar, S.; Willliams, A.; Vogel, U.; Puzyn, T. The Impact of Carbon Nanotube Properties on Lung Pathologies and Atherosclerosis Through Acute Inflammation: A New AOP-Anchored in Silico NAM. Small 2025, 21, e2501185. [Google Scholar] [CrossRef]
- Nikota, J.; Williams, A.; Yauk, C.L.; Wallin, H.; Vogel, U.; Halappanavar, S. Meta-analysis of transcriptomic responses as a means to identify pulmonary disease outcomes for engineered nanomaterials. Part. Fibre Toxicol. 2015, 13, 25. [Google Scholar] [CrossRef]
- Halappanavar, S.; Rahman, L.; Nikota, J.; Poulsen, S.S.; Ding, Y.; Jackson, P.; Wallin, H.; Schmid, O.; Vogel, U.; Williams, A. Ranking of nanomaterial potency to induce pathway perturbations associated with lung responses. NanoImpact 2019, 14, 100158. [Google Scholar] [CrossRef]
- Seidel, C.; Zhernovkov, V.; Cassidy, H.; Kholodenko, B.; Matallanas, D.; Cosnier, F.; Gaté, L. Inhaled multi-walled carbon nanotubes differently modulate global gene and protein expression in rat lungs. Nanotoxicology 2020, 15, 238–256. [Google Scholar] [CrossRef] [PubMed]
- Kinaret, P.; Ilves, M.; Fortino, V.; Rydman, E.; Karisola, P.; Lähde, A.; Koivisto, J.; Jokiniemi, J.; Wolff, H.; Savolainen, K.; et al. Inhalation and Oropharyngeal Aspiration Exposure to Rod-Like Carbon Nanotubes Induce Similar Airway Inflammation and Biological Responses in Mouse Lungs. ACS Nano 2017, 11, 291–303. [Google Scholar] [CrossRef]
- Bendtsen, K.M.; Gren, L.; Malmborg, V.B.; Shukla, P.C.; Tunér, M.; Essig, Y.J.; Krais, A.M.; Clausen, P.A.; Berthing, T.; Loeschner, K.; et al. Particle characterization and toxicity in C57BL/6 mice following instillation of five different diesel exhaust particles designed to differ in physicochemical properties. Part. Fibre Toxicol. 2020, 17, 38. [Google Scholar] [CrossRef]
- Mikkelsen, L.; Sheykhzade, M.; Jensen, K.A.; Saber, A.T.; Jacobsen, N.R.; Vogel, U.; Wallin, H.; Loft, S.; Møller, P. Modest effect on plaque progression and vasodilatory function in atherosclerosis-prone mice exposed to nanosized TiO2. Part. Fibre Toxicol. 2011, 8, 32. [Google Scholar] [CrossRef]
- Di Battista, V.; Danielsen, P.H.; Gajewicz-Skretna, A.; Kedziorski, A.; Seiffert, S.B.; Ma-Hock, L.; Berthing, T.; Mortensen, A.; Sundermann, A.; Skjolding, L.M.; et al. Oxide-Perovskites for Automotive Catalysts Biotransform and Induce Multicomponent Clearance and Hazard. ACS Nano 2024, 18, 32672–32693. [Google Scholar] [CrossRef]
- Barfod, K.K.; Bendtsen, K.M.; Berthing, T.; Koivisto, A.J.; Poulsen, S.S.; Segal, E.; Verleysen, E.; Mast, J.; Holländer, A.; Jensen, K.A.; et al. Increased surface area of halloysite nanotubes due to surface modification predicts lung inflammation and acute phase response after pulmonary exposure in mice. Environ. Toxicol. Pharmacol. 2020, 73, 103266. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, X.; Li, L.; Yang, H.; Liu, H.; Wu, D.; Liu, Z.; Liu, B.; Shen, T. C5a-C5aR1 axis mediates lung inflammation and fibrosis induced by single-walled carbon nanotubes via promoting neutrophils recruitment. Ecotoxicol. Environ. Saf. 2025, 289, 117627. [Google Scholar] [CrossRef]
- Zhang, X.; Luo, M.; Zhang, J.; Yao, Z.; Zhu, J.; Yang, S.; Zhu, Q.; Shen, T. Carbon nanotubes promote alveolar macrophages toward M2 polarization mediated epithelial-mesenchymal transition and fibroblast-to-myofibroblast transdifferentiation. Nanotoxicology 2021, 15, 588–604. [Google Scholar] [CrossRef] [PubMed]
- Halappanavar, S.; Jackson, P.; Williams, A.; Jensen, K.A.; Hougaard, K.S.; Vogel, U.; Yauk, C.L.; Wallin, H. Pulmonary response to surface-coated nanotitanium dioxide particles includes induction of acute phase response genes, inflammatory cascades, and changes in microRNAs: A toxicogenomic study. Environ. Mol. Mutagen. 2011, 52, 425–439. [Google Scholar] [CrossRef] [PubMed]
- Husain, M.; Kyjovska, Z.O.; Bourdon-Lacombe, J.; Saber, A.T.; Jensen, K.A.; Jacobsen, N.R.; Williams, A.; Wallin, H.; Halappanavar, S.; Vogel, U.; et al. Carbon black nanoparticles induce biphasic gene expression changes associated with inflammatory responses in the lungs of C57BL/6 mice following a single intratracheal instillation. Toxicol. Appl. Pharmacol. 2015, 289, 573–588. [Google Scholar] [CrossRef] [PubMed]
- Emerging Risk Factors Collaboration; Kaptoge, S.; Di Angelantonio, E.; Lowe, G.; Pepys, M.B.; Thompson, S.G.; Collins, R.; Danesh, J. C-reactive protein concentration and risk of coronary heart disease, stroke, and mortality: An individual participant meta-analysis. Lancet 2010, 375, 132–140. [Google Scholar] [CrossRef]
- Ridker, P.M.; Hennekens, C.H.; Buring, J.E.; Rifai, N. C-Reactive Protein and Other Markers of Inflammation in the Prediction of Cardiovascular Disease in Women. N. Engl. J. Med. 2000, 342, 836–843. [Google Scholar] [CrossRef]
- Ridker, P.M.; Moorthy, M.V.; Cook, N.R.; Rifai, N.; Lee, I.-M.; Buring, J.E. Inflammation, Cholesterol, Lipoprotein(a), and 30-Year Cardiovascular Outcomes in Women. N. Engl. J. Med. 2024, 391, 2087–2097. [Google Scholar] [CrossRef] [PubMed]
- Saber, A.T.; Jacobsen, N.R.; Jackson, P.; Poulsen, S.S.; Kyjovska, Z.O.; Halappanavar, S.; Yauk, C.L.; Wallin, H.; Vogel, U. Particle-induced pulmonary acute phase response may be the causal link between particle inhalation and cardiovascular disease. WIREs Nanomed. Nanobiotechnol. 2014, 6, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]


| Diameter (nm) | Length (nm) | Specific Surface Area (m2/g) | Purity (%) | Metal Content (wt%) | |
|---|---|---|---|---|---|
| NM-401 | 67 (±24) | 4048 (±2371) | 18 | 98 | Fe (0.05) Mg (0.015) |
| NM-403 | 12 (±7) | 443 (±222) | 135 | 96.6 | Fe (0.002) Mg (0.188) Co (1.2) Ni (0.0018) Mn (0.16) |
| Dose (Mass) | Body Weight (bw) | Dose (Mass) mg/kg bw |
|---|---|---|
| 18 µg (mouse) | 18.64 g | 0.966 |
| 54 µg (mouse) | 18.64 g | 2.897 |
| 180 µg (rat) | 187.7 g | 0.959 |
| 540 µg (rat) | 187.7 g | 2.877 |
| Dose (Mass) in Mice | Dosed Surface Area (cm2) | Deposited Surface Area (cm2/g bw) |
|---|---|---|
| 18 µg NM-401 | 0.018 mg × 180 cm2/mg = 3.24 | 3.24 cm2/18.64 g = 0.174 |
| 54 µg NM-401 | 0.054 mg × 180 cm2/mg = 9.72 | 9.72 cm2/18.64 g = 0.522 |
| 18 µg NM-403 | 0.018 mg × 1350 cm2/mg = 24.3 | 24.3 cm2/18.64 g = 1.304 |
| 54 µg NM-403 | 0.054 mg × 1350 cm2/mg = 72.9 | 72.9 cm2/18.64 g = 3.911 |
| Dose (Mass) in Rats | ||
| 180 µg NM-401 | 0.18 mg × 180 cm2/mg = 32.4 | 32.4 cm2/187.7 g = 0.173 |
| 540 µg NM-401 | 0.54 mg × 180 cm2/mg = 97.2 | 97.2 cm2/187.7 g = 0.518 |
| 180 µg NM-403 | 0.18 mg × 1350 cm2/mg = 243 | 243 cm2/187.7 g = 1.294 |
| 540 µg NM-403 | 0.54 mg × 1350 cm2/mg = 729 | 729 cm2/187.7 g = 3.884 |
| Day 1 | Day 28 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NM-401 | NM-403 | NM-401 | NM-403 | ||||||||||||
| Low Dose | High Dose | Low Dose | High Dose | Low Dose | High Dose | Low Dose | High Dose | ||||||||
| Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC |
| Adora | (1.41) | ||||||||||||||
| Adora2b | (0.62) | ||||||||||||||
| Arg1 | (2.38) | Arg1 | (1.33) | ||||||||||||
| C1qb | (1.43) | ||||||||||||||
| C3 | (0.82) | C3 | (1.21) | C3 | (1.12) | C3 | (1.80) | C3 | (1.11) | ||||||
| Ccl9 | (1.29) | Ccl9 | (0.95) | Ccl9 | (1.22) | ||||||||||
| Ccl17 | (1.74) | Ccl17 | (2.19) | Ccl17 | (1.28) | Ccl17 | (1.90) | Ccl17 | (1.41) | Ccl17 | (0.92) | Ccl17 | (0.99) | ||
| Ccl2 | (1.84) | Ccl2 | (2.30) | Ccl2 | (2.32) | ||||||||||
| Ccr5 | (0.80) | Ccr5 | (1.12) | Ccr5 | (0.96) | ||||||||||
| Ctss | (1.08) | ||||||||||||||
| Fcgr2b | (1.27) | Fcgr2b | (0.69) | Fcgr2b | (0.67) | ||||||||||
| Fxyd4 | (2.72) | Fxyd4 | (3.53) | Fxyd4 | (1.53) | Fxyd4 | (1.11) | ||||||||
| Il1b | (1.04) | Il1b | (1.38) | Il1b | (0.69) | ||||||||||
| Il1rn | (0.80) | Il1rn | (1.31) | ||||||||||||
| Il6 | (1.54) | Il6 | (1.69) | Il6 | (1.28) | ||||||||||
| Irf7 | (2.10) | ||||||||||||||
| Ly86 | (0.64) | ||||||||||||||
| Mmp12 | (0.83) | Mmp12 | (1.90) | Mmp12 | (2.35) | ||||||||||
| S100a4 | (0.66) | ||||||||||||||
| Selp | (0.95) | ||||||||||||||
| Serpina3n | (1.07) | Serpina3n | (1.62) | Serpina3n | (0.85) | Serpina3n | (1.97) | Serpina3n | (0.72) | ||||||
| Slc26a4 | (3.50) | Slc26a4 | (4.36) | Slc26a4 | (1.06) | Slc26a4 | (3.09) | Slc26a4 | (3.08) | ||||||
| Socs1 | (0.79) | Socs1 | (1.10) | Socs1 | (−0.61) | ||||||||||
| Timp1 | (1.65) | Timp1 | (3.08) | Timp1 | (1.28) | Timp1 | (3.50) | Timp1 | (1.36) | ||||||
| Tnfaip3 | (0.80) | ||||||||||||||
| 8 genes | 18 genes | 7 genes | 14 genes | 0 genes | 1 gene | 1 gene | 19 genes | ||||||||
| Day 1 | Day 28 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NM-401 | NM-403 | NM-401 | NM-403 | ||||||||||||
| Low Dose | High Dose | Low Dose | High Dose | Low Dose | High Dose | Low Dose | High Dose | ||||||||
| Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC |
| Adora2b | (1.04) | ||||||||||||||
| Arg1 | (0.76) | Arg1 | (0.82) | Arg1 | (1.09) | Arg1 | (2.00) | ||||||||
| C1qb | (0.67) | C1qb | (0.72) | ||||||||||||
| C3 | (1.55) | C3 | (1.80) | C3 | (1.54) | C3 | (5.58) | C3 | (1.21) | C3 | (1.66) | C3 | (2.21) | C3 | (2.29) |
| Ccl9 | (1.71) | Ccl9 | (2.84) | Ccl9 | (1.70) | Ccl9 | (1.78) | ||||||||
| Ccl17 | (1.02) | Ccl17 | (1.20) | Ccl17 | (1.05) | Ccl17 | (3.29) | Ccl17 | (0.71) | ||||||
| Ccl2 | (1.16) | Ccl2 | (2.77) | Ccl2 | (1.63) | Ccl2 | (5.26) | Ccl2 | (1.96) | Ccl2 | (1.71) | Ccl2 | (2.13) | ||
| Ccr5 | (0.83) | Ccr5 | (3.75) | ||||||||||||
| Ch25h | (1.01) | Ch25h | (1.05) | Ch25h | (1.31) | Ch25h | (1.23) | ||||||||
| Cle4a2 | (0.64) | Clec4a2 | (0.75) | Clec4a2 | (4.10) | ||||||||||
| Ctss | (0.70) | Ctss | (0.64) | Ctss | (0.72) | ||||||||||
| Fcgr2b | (0.74) | Fcgr2b | (1.13) | Fcgr2b | (0.86) | Fcgr2b | (1.12) | ||||||||
| Il1b | (0.76) | Il1b | (1.03) | Il1b | (0.94) | ||||||||||
| Il1r1 | (0.90) | ||||||||||||||
| Il1rn | (1.26) | Il1rn | (0.72) | Il1rn | (1.40) | Il1rn | (1.03) | Il1rn | (1.12) | ||||||
| Itgb2 | (0.77) | ||||||||||||||
| Mmp12 | (1.33) | Mmp12 | (1.60) | Mmp12 | (1.28) | Mmp12 | (3.89) | Mmp12 | (1.07) | Mmp12 | (3.02) | Mmp12 | (1.87) | Mmp12 | (2.47) |
| Retnla | (2.71) | Retnla | (2.63) | Retnla | (1.53) | Retnla | (3.58) | Retnla | (1.65) | Retnla | (2.47) | Rentla | (1.92) | Rentla | (2.75) |
| Sele | (0.80) | ||||||||||||||
| Serpina3n | (0.94) | Serpina3n | (1.93) | Serpina3n | (−0.79) | Serpina3n | (−0.75) | Serpina3n | (0.91) | ||||||
| Slc26a4 | (0.89) | Slc26a4 | (1.47) | Slc26a4 | (1.17) | Slc26a4 | (4.34) | Slc26a4 | (1.26) | Slc26a4 | (2.16) | Slc26a4 | (1.96) | Slc26a4 | (2.88) |
| Timp1 | (0.83) | Timp1 | (1.47) | Timp1 | (0.61) | Timp1 | (5.25) | Timp1 | (0.60) | ||||||
| Tnf | (1.24) | Tnf | (0.76) | Tnf | (1.11) | Tnf | (1.45) | ||||||||
| 8 genes | 18 genes | 12 genes | 11 genes | 7 genes | 17 gene | 9 gene | 14 genes | ||||||||
| Day 1 | Day 28 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NM-401 | NM-403 | NM-401 | NM-403 | ||||||||||||
| Low Dose | High Dose | Low Dose | High Dose | Low Dose | High Dose | Low Dose | High Dose | ||||||||
| Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC |
| Angptl4 | (1.56) | Angptl4 | (1.83) | ||||||||||||
| Fn1 | (0.91) | ||||||||||||||
| Hamp | (−1.06) | ||||||||||||||
| Hp | (0.77) | ||||||||||||||
| Il1b | (1.04) | Il1b | (1.38) | Il1b | (0.69) | ||||||||||
| Il1rn | (0.80) | Il1rn | (1.31) | ||||||||||||
| Il6 | (1.54) | Il6 | (1.69) | Il6 | (1.28) | ||||||||||
| Itih4 | (1.06) | Itih4 | (1.09) | Itih4 | (1.40) | Itih4 | (1.63) | Itih4 | (1.60) | ||||||
| Lcn2 | (2.42) | Lcn2 | (1.80) | Lcn2 | (3.14) | Lcn2 | (2.15) | ||||||||
| Orm1 | (1.08) | Orm1 | (1.20) | Orm1 | (2.38) | Orm1 | (1.28) | ||||||||
| Orm2 | (1.17) | Orm2 | (1.47) | Orm2 | (2.46) | Orm2 | (1.01) | ||||||||
| Reg3g | (2.82) | ||||||||||||||
| Selp | (0.95) | ||||||||||||||
| Serpina3n | (1.07) | Serpina3n | (1.62) | Serpina3n | (0.85) | Serpina3n | (1.97) | Serpina3n | (0.72) | ||||||
| Saa1 | (3.17) | Saa1 | (4.45) | Saa1 | (1.20) | ||||||||||
| Saa2 | (1.77) | Saa2 | (1.50) | Saa2 | (3.48) | ||||||||||
| Saa3 | (4.57) | Saa3 | (2.24) | Saa3 | (5.43) | Saa3 | (2.93) | ||||||||
| 2 genes | 12 genes | 8 genes | 15 genes | 0 genes | 0 genes | 0 genes | 10 genes | ||||||||
| Day 1 | Day 28 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NM-401 | NM-403 | NM-401 | NM-403 | ||||||||||||
| Low Dose | High Dose | Low Dose | High Dose | Low Dose | High Dose | Low Dose | High Dose | ||||||||
| Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC | Gene | LogFC |
| Fn1 | (1.06) | Fn1 | (0.67) | ||||||||||||
| Hp | (0.78) | Hp | (0.84) | Hp | (0.78) | Hp | (1.49) | Hp | (1.80) | ||||||
| Il1rn | (1.23) | Il1rn | (0.72) | Il1rn | (1.40) | Il1rn | (1.03) | Il1rn | (1.12) | ||||||
| Itih4 | (0.61) | Itih4 | (0.76) | ||||||||||||
| Kng1 | (0.69) | Kng1 | (1.16) | Kng1 | (1.54) | ||||||||||
| Lbp | (0.75) | Lbp | (1.03) | ||||||||||||
| Lcn2 | (1.82) | Lcn2 | (2.79) | Lcn2 | (3.53) | Lcn2 | (3.46) | Lcn2 | (1.90) | Lcn2 | (2.50) | Lcn2 | (3.35) | Lcn2 | (4.17) |
| Orm1 | (1.24) | Orm1 | (1.52) | Orm1 | (0.76) | Orm1 | (3.00) | Orm1 | (1.45) | ||||||
| Plscr1 | (−0.61) | Plscr1 | (−0.74) | Plscr1 | (−0.88) | ||||||||||
| Serpina1 | (−0.93) | ||||||||||||||
| Sting1 | (0.82) | ||||||||||||||
| Tf | (0.80) | ||||||||||||||
| Tfrc | (0.74) | Tfrc | (0.84) | Tfrc | (0.59) | ||||||||||
| Tnf | (1.74) | Tnf | (0.76) | Tnf | (1.11) | Tnf | (1.45) | ||||||||
| 3 genes | 8 genes | 5 genes | 6 genes | 3 genes | 5 genes | 5 genes | 10 genes | ||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saarimäki, L.A.; Danielsen, P.H.; Knudsen, K.B.; Poulsen, S.S.; Halappanavar, S.; Wolff, H.; Kinaret, P.A.S.; Greco, D.; Vogel, U. Comparison of Lung Inflammatory and Transcriptional Responses in Mice and Rats Following Pulmonary Exposure to a Fiber Paradigm-Compatible and Non-Compatible MWCNT. Nanomaterials 2025, 15, 1364. https://doi.org/10.3390/nano15171364
Saarimäki LA, Danielsen PH, Knudsen KB, Poulsen SS, Halappanavar S, Wolff H, Kinaret PAS, Greco D, Vogel U. Comparison of Lung Inflammatory and Transcriptional Responses in Mice and Rats Following Pulmonary Exposure to a Fiber Paradigm-Compatible and Non-Compatible MWCNT. Nanomaterials. 2025; 15(17):1364. https://doi.org/10.3390/nano15171364
Chicago/Turabian StyleSaarimäki, Laura Aliisa, Pernille Høgh Danielsen, Kristina Bram Knudsen, Sarah Søs Poulsen, Sabina Halappanavar, Henrik Wolff, Pia Anneli Sofia Kinaret, Dario Greco, and Ulla Vogel. 2025. "Comparison of Lung Inflammatory and Transcriptional Responses in Mice and Rats Following Pulmonary Exposure to a Fiber Paradigm-Compatible and Non-Compatible MWCNT" Nanomaterials 15, no. 17: 1364. https://doi.org/10.3390/nano15171364
APA StyleSaarimäki, L. A., Danielsen, P. H., Knudsen, K. B., Poulsen, S. S., Halappanavar, S., Wolff, H., Kinaret, P. A. S., Greco, D., & Vogel, U. (2025). Comparison of Lung Inflammatory and Transcriptional Responses in Mice and Rats Following Pulmonary Exposure to a Fiber Paradigm-Compatible and Non-Compatible MWCNT. Nanomaterials, 15(17), 1364. https://doi.org/10.3390/nano15171364








