Fluorescence-Based Multimodal DNA Logic Gates
Abstract
1. Introduction
2. Materials and Methods
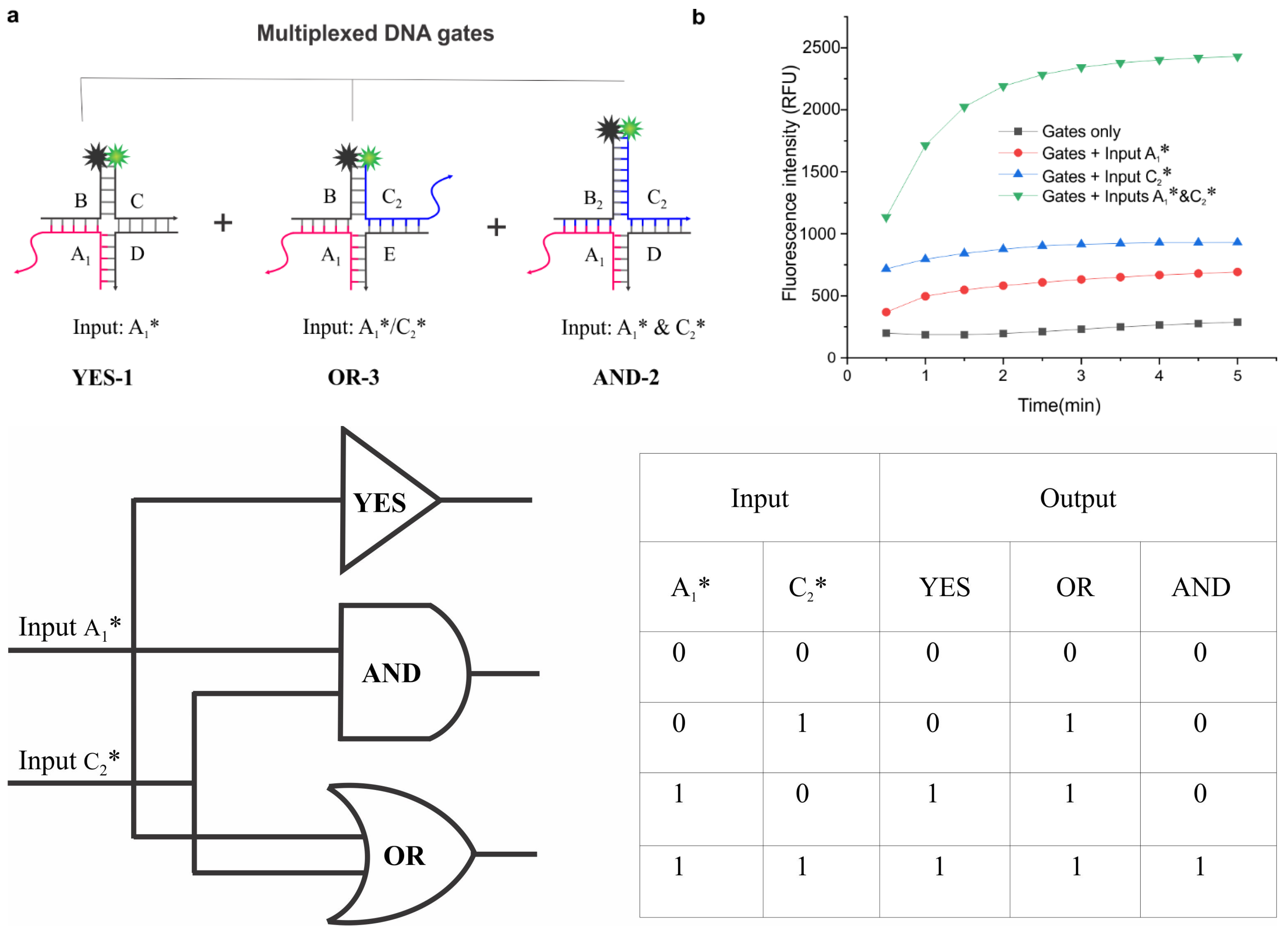
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Konry, T.; Walt, D.R. Intelligent Medical Diagnostics via Molecular Logic. J. Am. Chem. Soc. 2009, 131, 13232–13233. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhang, L.; Dong, S.; Wang, E. Four-Way Junction-Driven DNA Strand Displacement and Its Application in Building Majority Logic Circuit. ACS Nano 2013, 7, 10211–10217. [Google Scholar] [CrossRef]
- Anisul Haque, S.; Yamamoto, M.; Nakatani, R.; Endo, Y. Binary Logic Gates by Ferromagnetic Nanodots. J. Magn. Magn. Mater. 2004, 282, 380–384. [Google Scholar] [CrossRef]
- Katz, E.; Privman, V. Enzyme-Based Logic Systems for Information Processing. Chem. Soc. Rev. 2010, 39, 1835–1857. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Q.; Ke, Q.; Wang, T.; Zhang, Y.; Wei, F.; Wang, X.; Liu, G. Implementation of Novel Boolean Logic Gates for IMPLICATION and XOR Functions Using Riboregulators. Bioengineered 2022, 13, 1235–1248. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Yan, L.; Fan, Z. Multi-Responsive Fluorescent Probe Based on AIE for the Determination of Fe3+, Total Inorganic Iron, and CN- in Aqueous Medium and Its Application in Logic Gates. J. Photochem. Photobiol. Chem. 2021, 405, 112969. [Google Scholar] [CrossRef]
- Dwivedi, R.; Singh, D.P.; Singh, S.; Singh, A.K.; Chauhan, B.S.; Srikrishna, S.; Singh, V.P. Logic Gate Behavior and Intracellular Application of a Fluorescent Molecular Switch for the Detection of Fe3+ and Cascade Sensing of F− in Pure Aqueous Media. Org. Biomol. Chem. 2019, 17, 7497–7506. [Google Scholar] [CrossRef] [PubMed]
- Zou, Q.; Li, X.; Xue, T.; Zheng, J.; Su, Q. SERS Detection of Mercury (II)/Lead (II): A New Class of DNA Logic Gates. Talanta 2019, 195, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Cheng, N.; Zhu, P.; Xu, Y.; Huang, K.; Luo, Y.; Yang, Z.; Xu, W. High-Sensitivity Assay for Hg (II) and Ag (I) Ion Detection: A New Class of Droplet Digital PCR Logic Gates for an Intelligent DNA Calculator. Biosens. Bioelectron. 2016, 84, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Pan, J.; Liu, C. Versatile Sensing Platform for Cd2+ Detection in Rice Samples and Its Applications in Logic Gate Computation. Anal. Chem. 2020, 92, 6173–6180. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, Q.; Wu, W. Graphene-Based Steganographic Aptasensor for Information Computing and Monitoring Toxins of Biofilm in Food. Front. Microbiol. 2020, 10, 3139. [Google Scholar] [CrossRef] [PubMed]
- Das, D.; Alam, R.; Katarkar, A.; Ali, M. A Differentially Selective Probe for Trivalent Chemosensor upon Single Excitation with Cell Imaging Application: Potential Applications in Combinatorial Logic Circuit and Memory Devices. Photochem. Photobiol. Sci. 2019, 18, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Tavallali, H.; Deilamy-Rad, G.; Mosallanejad, N. A Reversible and Dual Responsive Sensing Approach for Determination of Ascorbate Ion in Fruit Juice, Biological, and Pharmaceutical Samples by Use of Available Triaryl Methane Dye and Its Application to Constructing a Molecular Logic Gate and a Set/Reset Memorized Device. Spectrochim Acta A Mol Biomol Spectrosc. 2019, 215, 276–289. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Jin, X.; Xing, Y.; Ni, G.; Peng, J. Construction of an NAND Logic Gate Based on Molecularly Imprinted Dual-Emission Quantum Dot Composites for the Detection of Antibiotics. Anal. Methods 2019, 11, 2033–2040. [Google Scholar] [CrossRef]
- Sanjabi, M.; Jahanian, A. Multi-Threshold and Multi-Input DNA Logic Design Style for Profiling the microRNA Biomarkers of Real Cancers. IET Nanobiotechnol. 2019, 13, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Liu, P.; Ga, L.; Ai, J. Advances in Applications of Molecular Logic Gates. ACS Omega 2021, 6, 30189–30204. [Google Scholar] [CrossRef] [PubMed]
- Bhat, M.P.; Vinayak, S.; Yu, J.; Jung, H.-Y.; Kurkuri, M. Colorimetric Receptors for the Detection of Biologically Important Anions and Their Application in Designing Molecular Logic Gate. ChemistrySelect 2020, 5, 13135–13143. [Google Scholar] [CrossRef]
- Gong, X.; Li, Z.; Hu, Q.; Zhou, R.; Shuang, S.; Dong, C. N,S,P Co-Doped Carbon Nanodot Fabricated from Waste Microorganism and Its Application for Label-Free Recognition of Manganese(VII) and l-Ascorbic Acid and AND Logic Gate Operation. ACS Appl. Mater. Interfaces 2017, 9, 38761–38772. [Google Scholar] [CrossRef]
- Zhourui, X.; Donghua, H.; Mengmeng, B.; Yali, Y.; Jinfang, N.I.E. Fluorescence Detection Based on DNA-Templated Silver Nanoclusters and the Construction of Multi-Level Logic Gate. Chem. J. Chin. Univ. 2020, 41, 102. [Google Scholar] [CrossRef]
- Yue, R.; Li, Z.; Wang, G.; Li, J.; Ma, N. Logic Sensing of MicroRNA in Living Cells Using DNA-Programmed Nanoparticle Network with High Signal Gain. ACS Sens. 2019, 4, 250–256. [Google Scholar] [CrossRef]
- Song, T.; Eshra, A.; Shah, S.; Bui, H.; Fu, D.; Yang, M.; Mokhtar, R.; Reif, J. Fast and Compact DNA Logic Circuits Based on Single-Stranded Gates Using Strand-Displacing Polymerase. Nat. Nanotechnol. 2019, 14, 1075–1081. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, A.; Tanaka, K.; Saito, I. DNA Logic Gates. J. Am. Chem. Soc. 2004, 126, 9458–9463. [Google Scholar] [CrossRef] [PubMed]
- Carbone, A.; Seeman, N.C. Circuits and Programmable Self-Assembling DNA Structures. Proc. Natl. Acad. Sci. USA 2002, 99, 12577–12582. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Winfree, E. Scaling up Digital Circuit Computation with DNA Strand Displacement Cascades. Science 2011, 332, 1196–1201. [Google Scholar] [CrossRef] [PubMed]
- McConnell, E.M.; Cozma, I.; Mou, Q.; Brennan, J.D.; Lu, Y.; Li, Y. Biosensing with DNAzymes. Chem. Soc. Rev. 2021, 50, 8954–8994. [Google Scholar] [CrossRef] [PubMed]
- Adleman, L.M. Molecular Computation of Solutions to Combinatorial Problems. Science 1994, 266, 1021–1024. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.; Li, Q.; Shi, J.; Fan, C.; Wang, F. Biocomputing Based on DNA Strand Displacement Reactions. ChemPhysChem Eur. J. Chem. Phys. Phys. Chem. 2021, 22, 1151–1166. [Google Scholar] [CrossRef]
- de Silva, A.P.; Uchiyama, S.; Vance, T.P.; Wannalerse, B. A Supramolecular Chemistry Basis for Molecular Logic and Computation. Coord. Chem. Rev. 2007, 251, 1623–1632. [Google Scholar] [CrossRef]
- Zhao, S.; Yu, L.; Yang, S.; Tang, X.; Chang, K.; Chen, M. Boolean Logic Gate Based on DNA Strand Displacement for Biosensing: Current and Emerging Strategies. Nanoscale Horiz. 2021, 6, 298–310. [Google Scholar] [CrossRef]
- Boruah, K.; Dutta, J. DNA Computing Models for Boolean Circuits and Logic Gates. In Proceedings of the 2015 IEEE International Conference on Computational Intelligence & Communication Technology, Ghaziabad, India, 13–14 February 2015; pp. 529–533. [Google Scholar] [CrossRef]
- Lin, Y.; Yang, Z.; Lake, R.J.; Zheng, C.; Lu, Y. Enzyme-Mediated Endogenous and Bioorthogonal Control of a DNAzyme Fluorescent Sensor for Imaging Metal Ions in Living Cells. Angew. Chem. Int. Ed. 2019, 58, 17061–17067. [Google Scholar] [CrossRef]
- Yin, F.; Wang, F.; Fan, C.; Zuo, X.; Li, Q. Biosensors Based on DNA Logic Gates. VIEW 2021, 2, 20200038. [Google Scholar] [CrossRef]
- Peng, R.; Zheng, X.; Lyu, Y.; Xu, L.; Zhang, X.; Ke, G.; Liu, Q.; You, C.; Huan, S.; Tan, W. Engineering a 3D DNA-Logic Gate Nanomachine for Bispecific Recognition and Computing on Target Cell Surfaces. J. Am. Chem. Soc. 2018, 140, 9793–9796. [Google Scholar] [CrossRef]
- Yang, Q.; Yang, F.; Dai, W.; Meng, X.; Wei, W.; Cheng, Y.; Dong, J.; Lu, H.; Dong, H. DNA Logic Circuits for Multiple Tumor Cells Identification Using Intracellular MicroRNA Molecular Bispecific Recognition. Adv. Healthc. Mater. 2021, 10, e2101130. [Google Scholar] [CrossRef]
- Miao, P.; Tang, Y. Cascade Strand Displacement and Bipedal Walking Based DNA Logic System for miRNA Diagnostics. ACS Cent. Sci. 2021, 7, 1036–1044. [Google Scholar] [CrossRef]
- Zhong, R.; Xiao, M.; Zhu, C.; Shen, X.; Tang, Q.; Zhang, W.; Wang, L.; Song, S.; Qu, X.; Pei, H.; et al. Logic Catalytic Interconversion of G-Molecular Hydrogel. ACS Appl. Mater. Interfaces 2018, 10, 4512–4518. [Google Scholar] [CrossRef]
- Bader, A.; Cockroft, S.L. Simultaneous G-Quadruplex DNA Logic. Chem.—Eur. J. 2018, 24, 4820–4824. [Google Scholar] [CrossRef]
- Wang, S.; Sun, J.; Zhao, J.; Lu, S.; Yang, X. Photo-Induced Electron Transfer-Based Versatile Platform with G-Quadruplex/Hemin Complex as Quencher for Construction of DNA Logic Circuits. Anal. Chem. 2018, 90, 3437–3442. [Google Scholar] [CrossRef]
- Qu, X.; Wang, S.; Ge, Z.; Wang, J.; Yao, G.; Li, J.; Zuo, X.; Shi, J.; Song, S.; Wang, L.; et al. Programming Cell Adhesion for On-Chip Sequential Boolean Logic Functions. J. Am. Chem. Soc. 2017, 139, 10176–10179. [Google Scholar] [CrossRef] [PubMed]
- Massey, M.; Medintz, I.L.; Ancona, M.G.; Algar, W.R. Time-Gated FRET and DNA-Based Photonic Molecular Logic Gates: AND, OR, NAND, and NOR. ACS Sens. 2017, 2, 1205–1214. [Google Scholar] [CrossRef] [PubMed]
- Margulies, D.; Felder, C.E.; Melman, G.; Shanzer, A. A Molecular Keypad Lock: A Photochemical Device Capable of Authorizing Password Entries. J. Am. Chem. Soc. 2007, 129, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Y.; Dong, S.; Wang, E. DNA-Based Advanced Logic Circuits for Nonarithmetic Information Processing. NPG Asia Mater. 2015, 7, e166. [Google Scholar] [CrossRef]
- Chen, T.; Fu, X.; Zhang, Q.; Mao, D.; Song, Y.; Feng, C.; Zhu, X. A DNA Logic Gate with Dual-Anchored Proximity Aptamers for the Accurate Identification of Circulating Tumor Cells. Chem. Commun. 2020, 56, 6961–6964. [Google Scholar] [CrossRef] [PubMed]
- Llopis-Lorente, A.; de Luis, B.; García-Fernández, A.; Jimenez-Falcao, S.; Orzáez, M.; Sancenón, F.; Villalonga, R.; Martínez-Máñez, R. Hybrid Mesoporous Nanocarriers Act by Processing Logic Tasks: Toward the Design of Nanobots Capable of Reading Information from the Environment. ACS Appl. Mater. Interfaces 2018, 10, 26494–26500. [Google Scholar] [CrossRef]
- Tregubov, A.A.; Nikitin, P.I.; Nikitin, M.P. Advanced Smart Nanomaterials with Integrated Logic-Gating and Biocomputing: Dawn of Theranostic Nanorobots. Chem. Rev. 2018, 118, 10294–10348. [Google Scholar] [CrossRef]
- Thubagere, A.J.; Li, W.; Johnson, R.F.; Chen, Z.; Doroudi, S.; Lee, Y.L.; Izatt, G.; Wittman, S.; Srinivas, N.; Woods, D.; et al. A Cargo-Sorting DNA Robot. Science 2017, 357, eaan6558. [Google Scholar] [CrossRef]
- Chao, J.; Wang, J.; Wang, F.; Ouyang, X.; Kopperger, E.; Liu, H.; Li, Q.; Shi, J.; Wang, L.; Hu, J.; et al. Solving Mazes with Single-Molecule DNA Navigators. Nat. Mater. 2019, 18, 273–279. [Google Scholar] [CrossRef]
- Chang, D.; Kim, K.T.; Lindberg, E.; Winssinger, N. Smartphone DNA or RNA Sensing Using Semisynthetic Luciferase-Based Logic Device. ACS Sens. 2020, 5, 807–813. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, L.; Jiang, W. Toehold-Mediated Strand Displacement Reaction-Dependent Fluorescent Strategy for Sensitive Detection of Uracil-DNA Glycosylase Activity. Biosens. Bioelectron. 2017, 89, 984–988. [Google Scholar] [CrossRef]
- Genot, A.J.; Bath, J.; Turberfield, A.J. Combinatorial Displacement of DNA Strands: Application to Matrix Multiplication and Weighted Sums. Angew. Chem.-Int. Ed. 2013, 52, 1189–1192. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhang, L.; Wang, E. Measurement of the Base Number of DNA Using a Special Calliper Made of a Split G-Quadruplex. Chem. Commun. 2012, 48, 11990–11992. [Google Scholar] [CrossRef] [PubMed]
- Seelig, G.; Soloveichik, D.; Zhang, D.Y.; Winfree, E. Enzyme-Free Nucleic Acid Logic Circuits. Science 2006, 314, 1585–1588. [Google Scholar] [CrossRef] [PubMed]
- Wijesinghe, K.M.; Sabbih, G.; Algama, C.H.; Syed, R.; Danquah, M.K.; Dhakal, S. FRET-Based Single-Molecule Detection of Pathogen Protein IsdA Using Computationally Selected Aptamers. Anal. Chem. 2023, 95, 9839–9846. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Sapkota, K.; Dhakal, S. Multiplexed Nucleic Acid Sensing with Single-Molecule FRET. ACS Sens. 2019, 4, 623–633. [Google Scholar] [CrossRef]
- Aitken, C.E.; Marshall, R.A.; Puglisi, J.D. An Oxygen Scavenging System for Improvement of Dye Stability in Single-Molecule Fluorescence Experiments. Biophys. J. 2008, 94, 1826–1835. [Google Scholar] [CrossRef]
- Swoboda, M.; Henig, J.; Cheng, H.-M.; Brugger, D.; Haltrich, D.; Plumeré, N.; Schlierf, M. Enzymatic Oxygen Scavenging for Photostability without pH Drop in Single-Molecule Experiments. ACS Nano 2012, 6, 6364–6369. [Google Scholar] [CrossRef]
- Andréasson, J.; Pischel, U. Smart Molecules at Work—Mimicking Advanced Logic Operations. Chem. Soc. Rev. 2009, 39, 174–188. [Google Scholar] [CrossRef]
- Peng, Y.; Zhou, W.; Yuan, R.; Xiang, Y. Dual-Input Molecular Logic Circuits for Sensitive and Simultaneous Sensing of Multiple microRNAs from Tumor Cells. Sens. Actuators B Chem. 2018, 264, 202–207. [Google Scholar] [CrossRef]
- Chen, X.; Liu, X.; Wang, F.; Li, S.; Chen, C.; Qiang, X.; Shi, X. Massively Parallel DNA Computing Based on Domino DNA Strand Displacement Logic Gates. ACS Synth. Biol. 2022, 11, 2504–2512. [Google Scholar] [CrossRef]
- Gao, J.; Liu, Y.; Lin, X.; Deng, J.; Yin, J.; Wang, S. Implementation of Cascade Logic Gates and Majority Logic Gate on a Simple and Universal Molecular Platform. Sci. Rep. 2017, 7, 14014. [Google Scholar] [CrossRef]
- Seeja, V.M.; Daniel Raj, A.; Sanjoy, D. Wallace Tree Multiplier Design and Simulation with DNA Logic Gates. J. VLSI Des. Signal Process. 2016, 2, 1–16. [Google Scholar]
- Faheem, H.; Mathivanan, J.; Talbot, H.; Zeghal, H.; Vangaveti, S.; Sheng, J.; Chen, A.A.; Chandrasekaran, A.R. Toehold Clipping: A Mechanism for Remote Control of DNA Strand Displacement. Nucleic Acids Res. 2023, 51, 4055–4063. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Algama, C.H.; Basir, J.; Wijesinghe, K.M.; Dhakal, S. Fluorescence-Based Multimodal DNA Logic Gates. Nanomaterials 2024, 14, 1185. https://doi.org/10.3390/nano14141185
Algama CH, Basir J, Wijesinghe KM, Dhakal S. Fluorescence-Based Multimodal DNA Logic Gates. Nanomaterials. 2024; 14(14):1185. https://doi.org/10.3390/nano14141185
Chicago/Turabian StyleAlgama, Chamika Harshani, Jamil Basir, Kalani M. Wijesinghe, and Soma Dhakal. 2024. "Fluorescence-Based Multimodal DNA Logic Gates" Nanomaterials 14, no. 14: 1185. https://doi.org/10.3390/nano14141185
APA StyleAlgama, C. H., Basir, J., Wijesinghe, K. M., & Dhakal, S. (2024). Fluorescence-Based Multimodal DNA Logic Gates. Nanomaterials, 14(14), 1185. https://doi.org/10.3390/nano14141185





