Effect of Local Topography on Cell Division of Staphylococcus spp.
Abstract
:1. Introduction
2. Results and Discussion
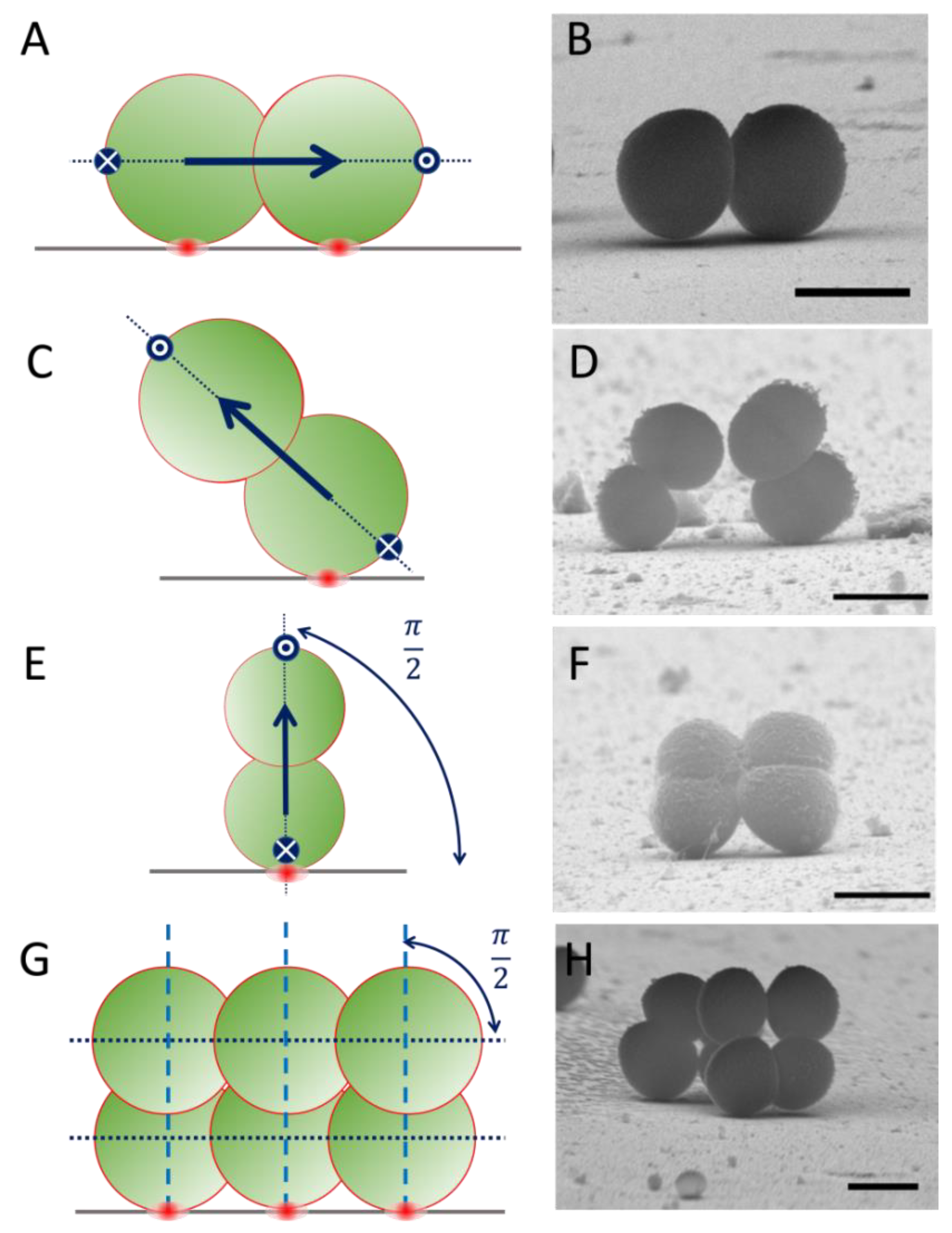
2.1. Staphylococcus spp. on Flat Si: Lateral Freedom to Grow Away from the Surface
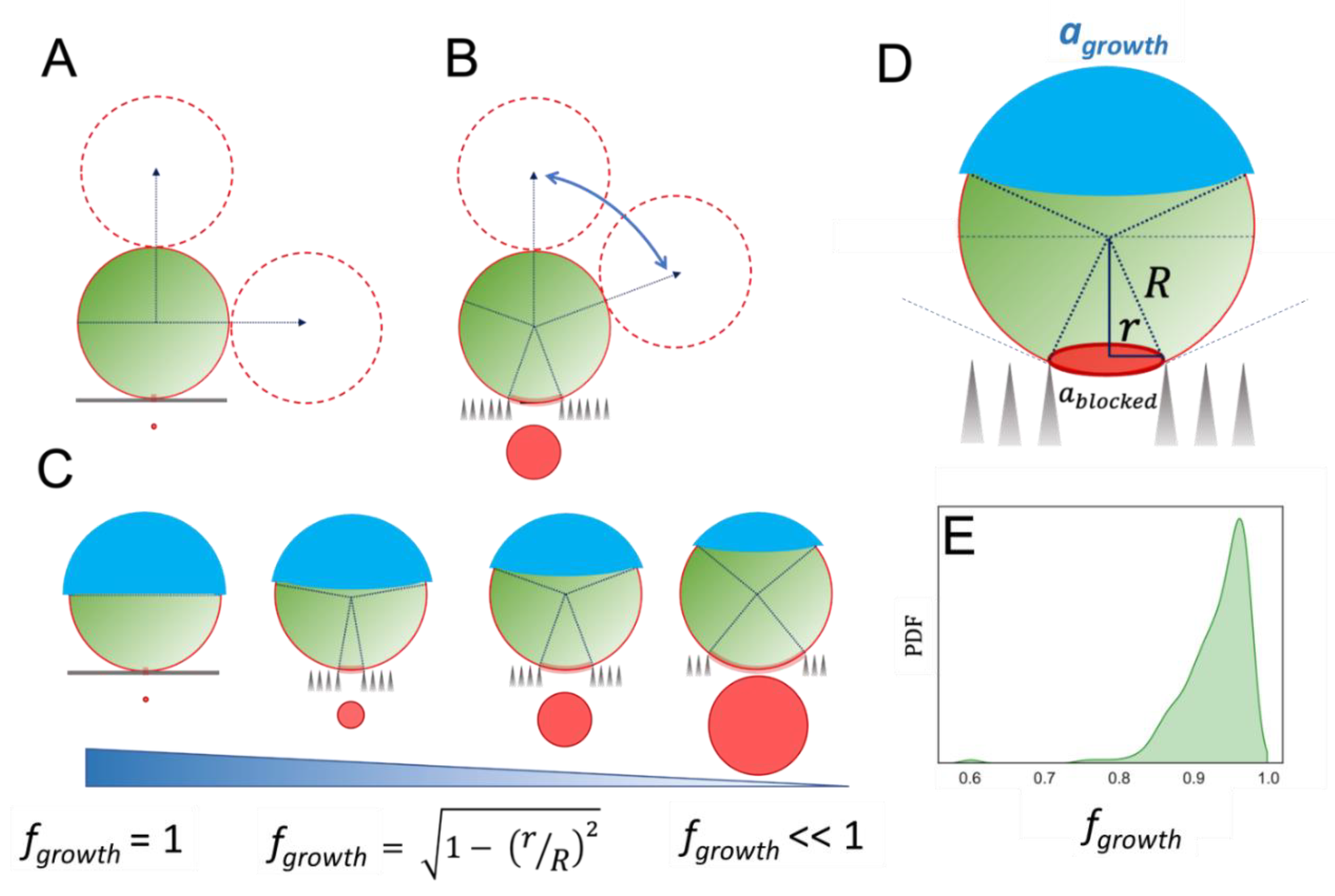
2.2. Staphylococcus spp. on SiNW Surfaces: The Effect of the Local Topography
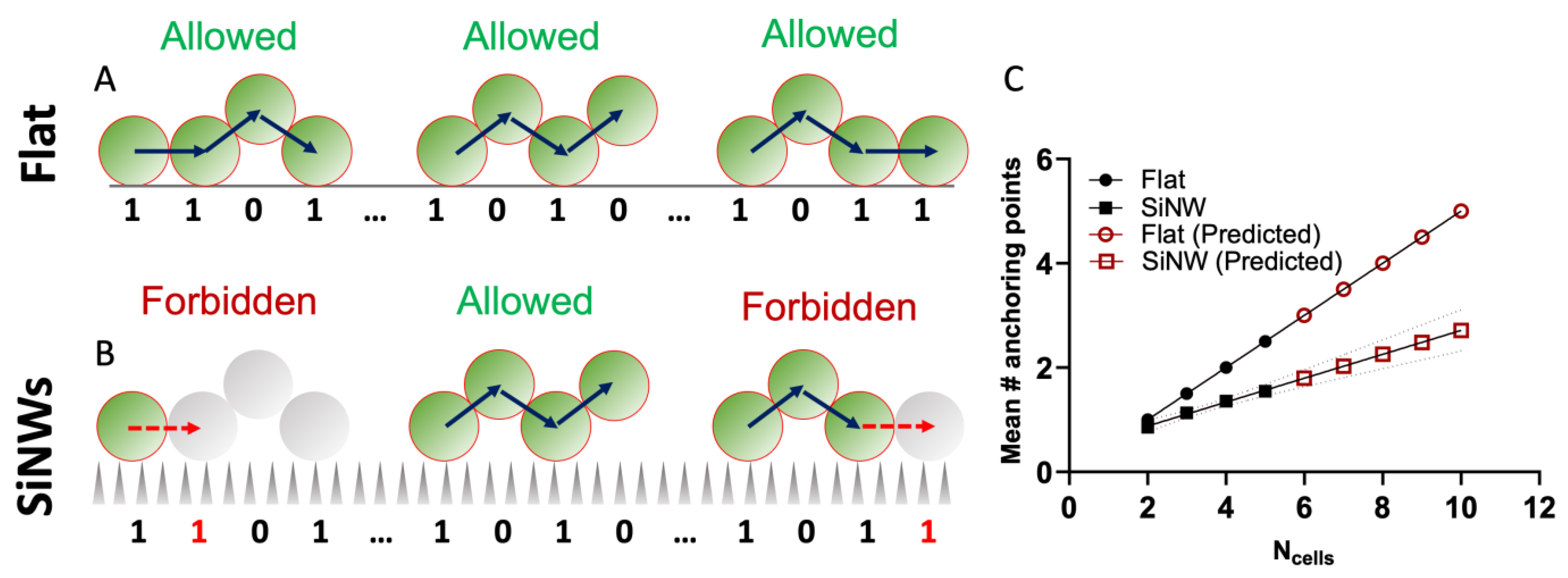
2.3. Local Propagation of Topological Information across Staphylococcal Micro-Colonies
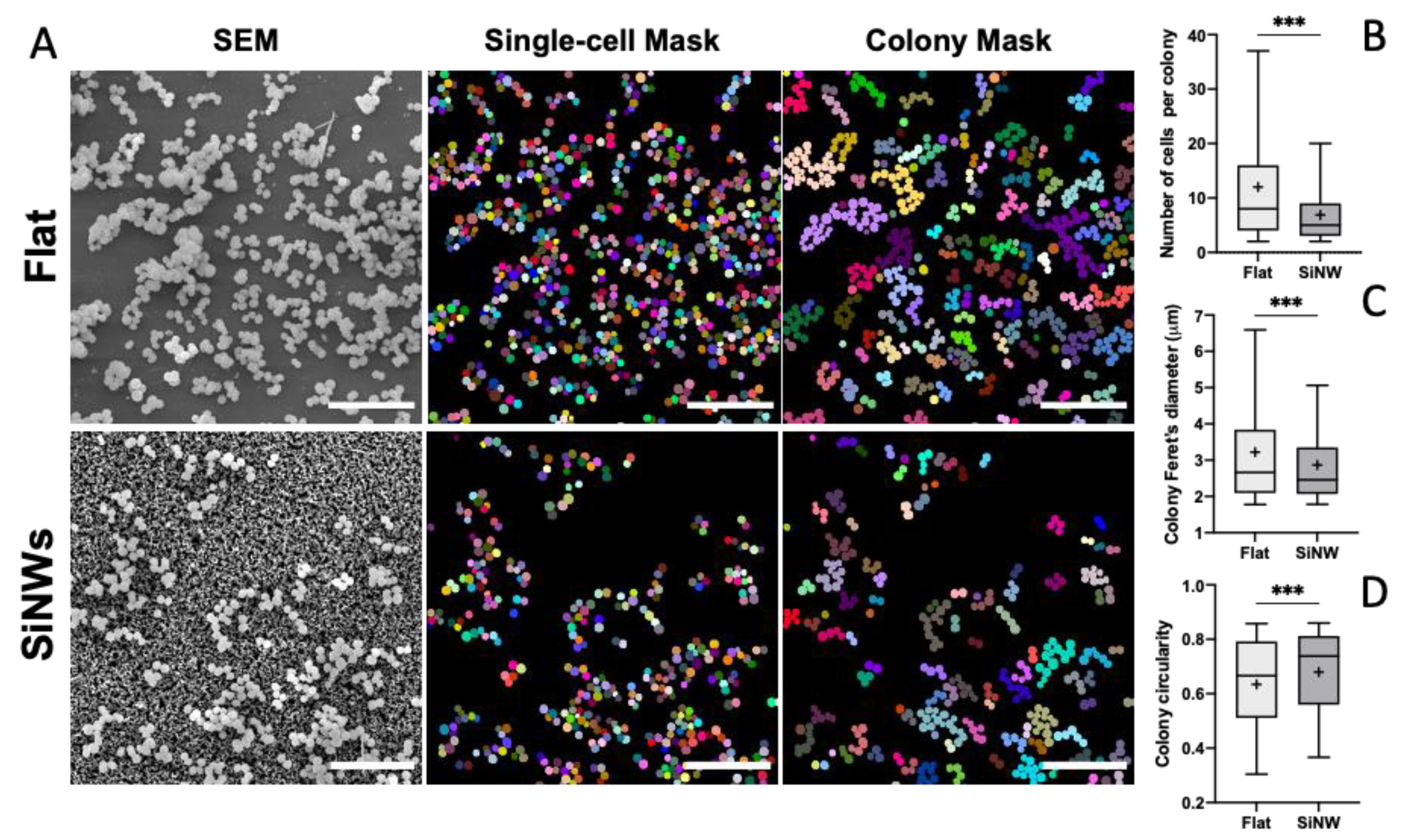
2.4. Effect of SiNW Topography on the Morphology of Staphylococcal Colonies
3. Materials and Methods
3.1. Fabrication of SiNW Surfaces
3.2. Imaging of Sessile Bacteria on Flat Si and SiNW Surfaces
3.3. Viability of Sessile Bacteria
3.4. Scanning Electron Microscopy (SEM) Imaging
3.5. Quantification of Pore Size of SiNW Surfaces
3.6. Characterisation of Colony Morphology
3.7. Data Analysis
3.8. Computations of Graph Adjacency Matrixes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cabeen, M.; Jacobs-Wagner, C. Bacterial cell shape. Nat. Rev. Microbiol. 2005, 3, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Egan, A.J.F.; Errington, J.; Vollmer, W. Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 2020, 18, 446–460. [Google Scholar] [CrossRef] [PubMed]
- Jun, S.; Wright, A. Entropy as the driver of chromosome segregation. Nat. Rev. Microbiol. 2010, 8, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Pinho, M.; Kjos, M.; Veening, J.-W. How to get (a)round: Mechanisms controlling growth and division of coccoid bacteria. Nat. Rev. Genet. 2013, 11, 601–614. [Google Scholar] [CrossRef] [Green Version]
- Veiga, H.; Jorge, A.M.; Pinho, M.G. Absence of nucleoid occlusion effector Noc impairs formation of orthogonal FtsZ rings during Staphylococcus aureus cell division. Mol. Microbiol. 2011, 80, 1366–1380. [Google Scholar] [CrossRef]
- Monteiro, J.M.; Fernandes, P.B.; Vaz, F.; Pereira, A.R.; Tavares, A.C.; Ferreira, M.T.; Pereira, P.M.; Veiga, H.; Kuru, E.; VanNieuwenhze, M.S.; et al. Cell shape dynamics during the staphylococcal cell cycle. Nat. Commun. 2015, 6, 8055. [Google Scholar] [CrossRef]
- Lund, V.A.; Wacnik, K.; Turner, R.D.; E Cotterell, B.; Walther, C.G.; Fenn, S.J.; Grein, F.; Wollman, A.J.; Leake, M.C.; Olivier, N.; et al. Molecular coordination of Staphylococcus aureus cell division. eLife 2018, 7, e32057. [Google Scholar] [CrossRef]
- Jenkins, J.; Mantell, J.; Neal, C.; Gholinia, A.; Verkade, P.; Nobbs, A.H.; Su, B. Antibacterial effects of nanopillar surfaces are mediated by cell impedance, penetration and induction of oxidative stress. Nat. Commun. 2020, 11, 1626. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Hasan, J.; Webb, H.K.; Truong, V.K.; Watson, G.S.; Watson, J.A.; Baulin, V.A.; Pogodin, S.; Wang, J.Y.; Tobin, M.J.; et al. Natural Bactericidal Surfaces: Mechanical Rupture of Pseudomonas aeruginosa Cells by Cicada Wings. Small 2012, 8, 2489–2494. [Google Scholar] [CrossRef]
- Ivanova, E.P.; Hasan, J.; Webb, H.K.; Gervinskas, G.; Juodkazis, S.; Truong, V.K.; Wu, A.H.F.; Lamb, R.N.; Baulin, V.A.; Watson, G.S.; et al. Bactericidal activity of black silicon. Nat. Commun. 2013, 4, 2838. [Google Scholar] [CrossRef]
- Fadeeva, E.; Truong, V.K.; Stiesch, M.; Chichkov, B.N.; Crawford, R.J.; Wang, J.; Ivanova, E.P. Bacterial Retention on Superhydrophobic Titanium Surfaces Fabricated by Femtosecond Laser Ablation. Langmuir 2011, 27, 3012–3019. [Google Scholar] [CrossRef] [PubMed]
- Hizal, F.; Rungraeng, N.; Lee, J.; Jun, S.; Busscher, H.J.; Van der Mei, H.C.; Choi, C.H. Nanoengineered Superhydrophobic Surfaces of Aluminum with Extremely Low Bacterial Adhesivity. ACS Appl. Mater. Interfaces 2017, 9, 12118–12129. [Google Scholar] [CrossRef] [PubMed]
- Sharifikolouei, E.; Najmi, Z.; Cochis, A.; Scalia, A.C.; Aliabadi, M.; Perero, S.; Rimondini, L. Generation of cytocompatible superhydrophobic Zr–Cu–Ag metallic glass coatings with antifouling properties for medical textiles. Mater. Today Bio 2021, 12, 100148. [Google Scholar] [CrossRef] [PubMed]
- Pogodin, S.; Hasan, J.; Baulin, V.A.; Webb, H.K.; Truong, V.K.; Nguyen, T.H.P.; Boshkovikj, V.; Fluke, C.J.; Watson, G.S.; Watson, J.; et al. Biophysical Model of Bacterial Cell Interactions with Nanopatterned Cicada Wing Surfaces. Biophys. J. 2013, 104, 835–840. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Z.; Nero, T.; Mukherjee, S.; Olson, R.; Yan, J. Searching for the Secret of Stickiness: How Biofilms Adhere to Surfaces. Front. Microbiol. 2021, 12, 686793. [Google Scholar] [CrossRef]
- Ponnuvel, S.; Sankar, S.; Ponnuraj, K. Analyzing the adhesion mechanism of FnBPA, a surface adhesin from Staphylococcus aureus on its interaction with nanoparticle. Microb. Pathog. 2020, 146, 104239. [Google Scholar] [CrossRef]
- Milles, L.F.; Schulten, K.; Gaub, H.E.; Bernardi, R.C. Molecular mechanism of extreme mechanostability in a pathogen adhesin. Science 2018, 359, 1527–1533. [Google Scholar] [CrossRef] [Green Version]
- Tomlinson, K.L.; Lung, T.W.F.; Dach, F.; Annavajhala, M.K.; Gabryszewski, S.J.; Groves, R.A.; Drikic, M.; Francoeur, N.J.; Sridhar, S.H.; Smith, M.L.; et al. Staphylococcus aureus induces an itaconate-dominated immunometabolic response that drives biofilm formation. Nat. Commun. 2021, 12, 1399. [Google Scholar] [CrossRef]
- Uneputty, A.; Dávila-Lezama, A.; Garibo, D.; Oknianska, A.; Bogdanchikova, N.; Hernández-Sánchez, J.; Susarrey-Arce, A. Strategies applied to modify structured and smooth surfaces: A step closer to reduce bacterial adhesion and biofilm formation. Colloids Interface Sci. Commun. 2021, 46, 100560. [Google Scholar] [CrossRef]
- Kim, M.K.; Zhao, A.; Wang, A.; Brown, Z.Z.; Muir, T.W.; Stone, H.A.; Bassler, B.L. Surface-attached molecules control Staphylococcus aureus quorum sensing and biofilm development. Nat. Microbiol. 2017, 2, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Cox, C.D.; Bavi, N.; Martinac, B. Bacterial Mechanosensors. Annu. Rev. Physiol. 2018, 80, 71–93. [Google Scholar] [CrossRef] [PubMed]
- Rodesney, C.A.; Roman, B.; Dhamani, N.; Cooley, B.J.; Katira, P.; Touhami, A.; Gordon, V.D. Mechanosensing of shear by Pseudomonas aeruginosa leads to increased levels of the cyclic-di-GMP signal initiating biofilm development. Proc. Natl. Acad. Sci. USA 2017, 114, 5906–5911. [Google Scholar] [CrossRef] [Green Version]
- Ellison, C.; Brun, Y.V. Mechanosensing: A Regulation Sensation. Curr. Biol. 2015, 25, R113–R115. [Google Scholar] [CrossRef] [Green Version]
- Dufrêne, Y.F.; Persat, A. Mechanomicrobiology: How bacteria sense and respond to forces. Nat. Rev. Genet. 2020, 18, 227–240. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, X.; Xi, H.; Sun, J.; Liang, X.; Wei, J.; Xiao, X.; Liu, Z.; Li, S.; Liang, Z.; et al. Interpretation of adhesion behaviors between bacteria and modified basalt fiber by surface thermodynamics and extended DLVO theory. Colloids Surf. B Biointerfaces 2019, 177, 454–461. [Google Scholar] [CrossRef] [PubMed]
- Hermansson, M. The DLVO theory in microbial adhesion. Colloids Surfaces B Biointerfaces 1999, 14, 105–119. [Google Scholar] [CrossRef]
- Zhang, B.-C.; Jie, J.; Zhang, X.; Ou, X.; Zhang, X. Large-Scale Fabrication of Silicon Nanowires for Solar Energy Applications. ACS Appl. Mater. Interfaces 2017, 9, 34527–34543. [Google Scholar] [CrossRef] [PubMed]
- Susarrey-Arce, A.; Sorzabal-Bellido, I.; Oknianska, A.; McBride, F.; Beckett, A.J.; Gardeniers, J.G.E.; Raval, R.; Tiggelaar, R.M.; Fernandez, Y.A.D. Bacterial viability on chemically modified silicon nanowire arrays. J. Mater. Chem. B 2016, 4, 3104–3112. [Google Scholar] [CrossRef] [Green Version]
- Chartier, C.; Bastide, S.; Lévy-Clément, C. Metal-assisted chemical etching of silicon in HF–H2O2. Electrochim. Acta 2008, 53, 5509–5516. [Google Scholar] [CrossRef]
- Leonardi, A.; Faro, M.; Irrera, A. Silicon Nanowires Synthesis by Metal-Assisted Chemical Etching: A Review. Nanomaterials 2021, 11, 383. [Google Scholar] [CrossRef]
- Sorzabal-Bellido, I.; Diaz-Fernandez, Y.A.; Arce, A.S.; Skelton, A.A.; McBride, F.; Beckett, A.J.; Prior, I.A.; Raval, R. Exploiting Covalent, H-Bonding, and π–π Interactions to Design Antibacterial PDMS Interfaces That Load and Release Salicylic Acid. ACS Appl. Bio Mater. 2019, 2, 4801–4811. [Google Scholar] [CrossRef] [PubMed]
- Pallavicini, P.; Bassi, B.; Chirico, G.; Collini, M.; Dacarro, G.; Fratini, E.; Grisoli, P.; Patrini, M.; Sironi, L.; Taglietti, A.; et al. Modular approach for bimodal antibacterial surfaces combining photo-switchable activity and sustained biocidal release. Sci. Rep. 2017, 7, 5259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Available online: https://github.com/BIOP/ijp-max-inscribed-circles (accessed on 15 January 2022).
- Sorzabal, B.I.; Barbieri, L.; Beckett, A.J.; Prior, I.A.; Susarrey-Arce, A.; Tiggelaar, R.M.; Forthergill, J.; Raval, R.; Diaz, F.Y.A. Effect of local topography on cell division of Staphylococcus sp. Zenodo 2021, 4765599. [Google Scholar] [CrossRef]
- Schmidt, U.; Weigert, M.; Broaddus, C.; Myers, G. Cell Detection with Star-Convex Polygons. In Medical Image Computing and Computer Assisted Intervention—MICCAI 2018; Frangi, A.F., Schnabel, J.A., Davatzikos, C., Alberola-López, C., Fichtinger, G., Eds.; Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2018; Volume 11071. [Google Scholar] [CrossRef] [Green Version]
- von Chamier, L.; Laine, R.F.; Jukkala, J.; Spahn, C.; Krentzel, D.; Nehme, E.; Lerche, M.; Hernández-Pérez, S.; Mattila, P.K.; Karinou, E.; et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat. Commun. 2021, 12, 1–18. [Google Scholar] [CrossRef]
- Haase, R.; Royer, L.A.; Steinbach, P.; Schmidt, D.; Dibrov, A.; Schmidt, U.; Weigert, M.; Maghelli, N.; Tomancak, P.; Jug, F.; et al. CLIJ: GPU-accelerated image processing for everyone. Nat. Methods 2019, 17, 5–6. [Google Scholar] [CrossRef]
- Waskom, M.L. Seaborn: Statistical data visualization. J. Open Source Softw. 2021, 6, 3021. [Google Scholar] [CrossRef]




| Staphylococcus spp. | % Live Cells on Flat Si Surface | % Live Cells on SiNW Surface |
|---|---|---|
| S. aureus | 98.1% ± 3.5% | 97.7% ± 9.3% |
| S. epidermidis | 98.9% ± 4.9% | 98.5% ± 7.9% |
| Parameter | Calculation |
|---|---|
| Determined from SEM images (see Figure S5, Table S1) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sorzabal-Bellido, I.; Barbieri, L.; Beckett, A.J.; Prior, I.A.; Susarrey-Arce, A.; Tiggelaar, R.M.; Fothergill, J.; Raval, R.; Diaz Fernandez, Y.A. Effect of Local Topography on Cell Division of Staphylococcus spp. Nanomaterials 2022, 12, 683. https://doi.org/10.3390/nano12040683
Sorzabal-Bellido I, Barbieri L, Beckett AJ, Prior IA, Susarrey-Arce A, Tiggelaar RM, Fothergill J, Raval R, Diaz Fernandez YA. Effect of Local Topography on Cell Division of Staphylococcus spp. Nanomaterials. 2022; 12(4):683. https://doi.org/10.3390/nano12040683
Chicago/Turabian StyleSorzabal-Bellido, Ioritz, Luca Barbieri, Alison J. Beckett, Ian A. Prior, Arturo Susarrey-Arce, Roald M. Tiggelaar, Joanne Fothergill, Rasmita Raval, and Yuri A. Diaz Fernandez. 2022. "Effect of Local Topography on Cell Division of Staphylococcus spp." Nanomaterials 12, no. 4: 683. https://doi.org/10.3390/nano12040683
APA StyleSorzabal-Bellido, I., Barbieri, L., Beckett, A. J., Prior, I. A., Susarrey-Arce, A., Tiggelaar, R. M., Fothergill, J., Raval, R., & Diaz Fernandez, Y. A. (2022). Effect of Local Topography on Cell Division of Staphylococcus spp. Nanomaterials, 12(4), 683. https://doi.org/10.3390/nano12040683








