Chitosan-Crosslinked Low Molecular Weight PEI-Conjugated Iron Oxide Nanoparticle for Safe and Effective DNA Delivery to Breast Cancer Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plasmid DNA Preparation
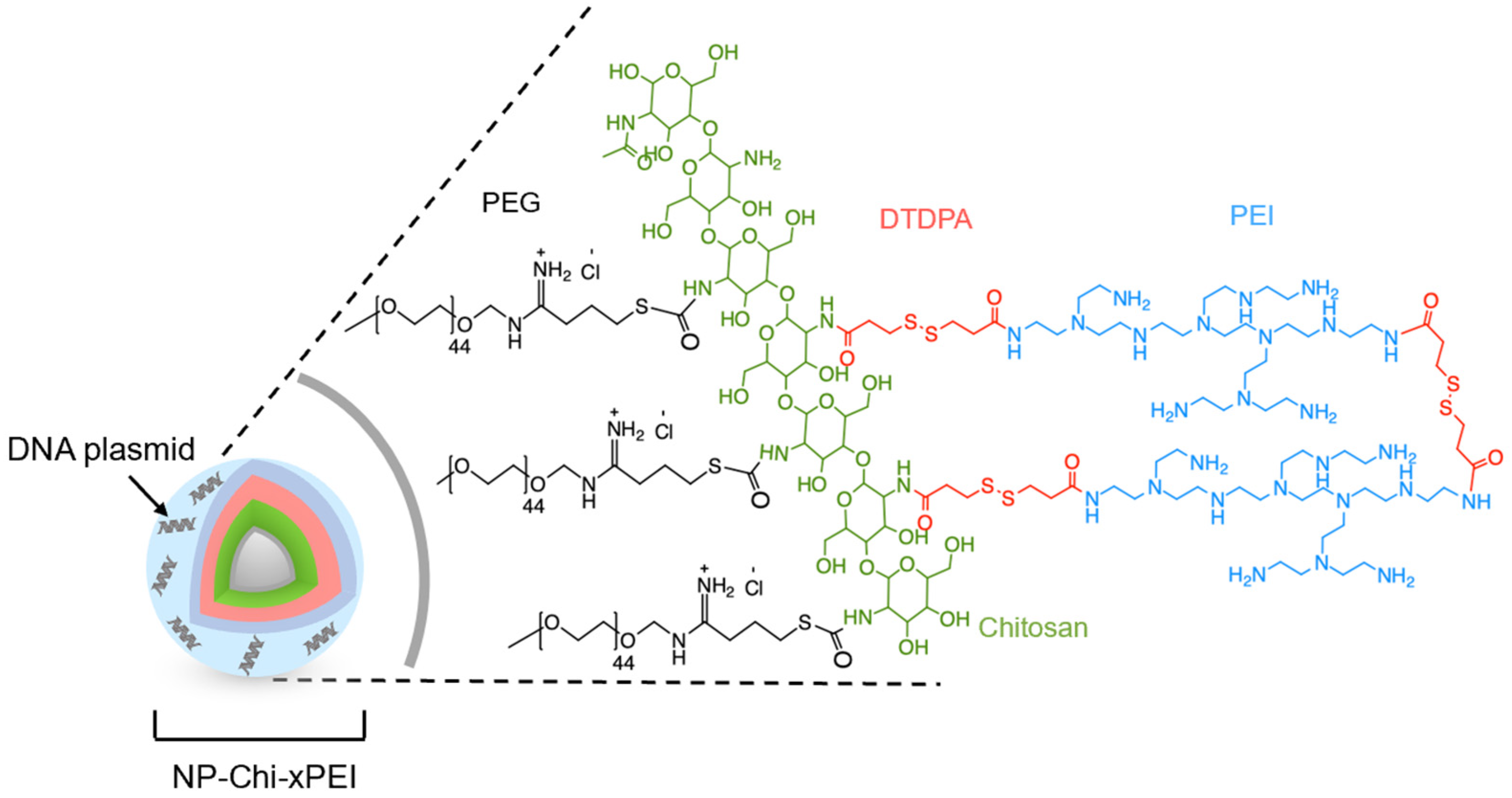
2.3. Nanoparticle and Polymer Synthesis
2.4. Quantification of Iron [Fe] Concentration in NP by Ferrozine Assay
2.5. Conjugation of Chi-xPEI onto NP
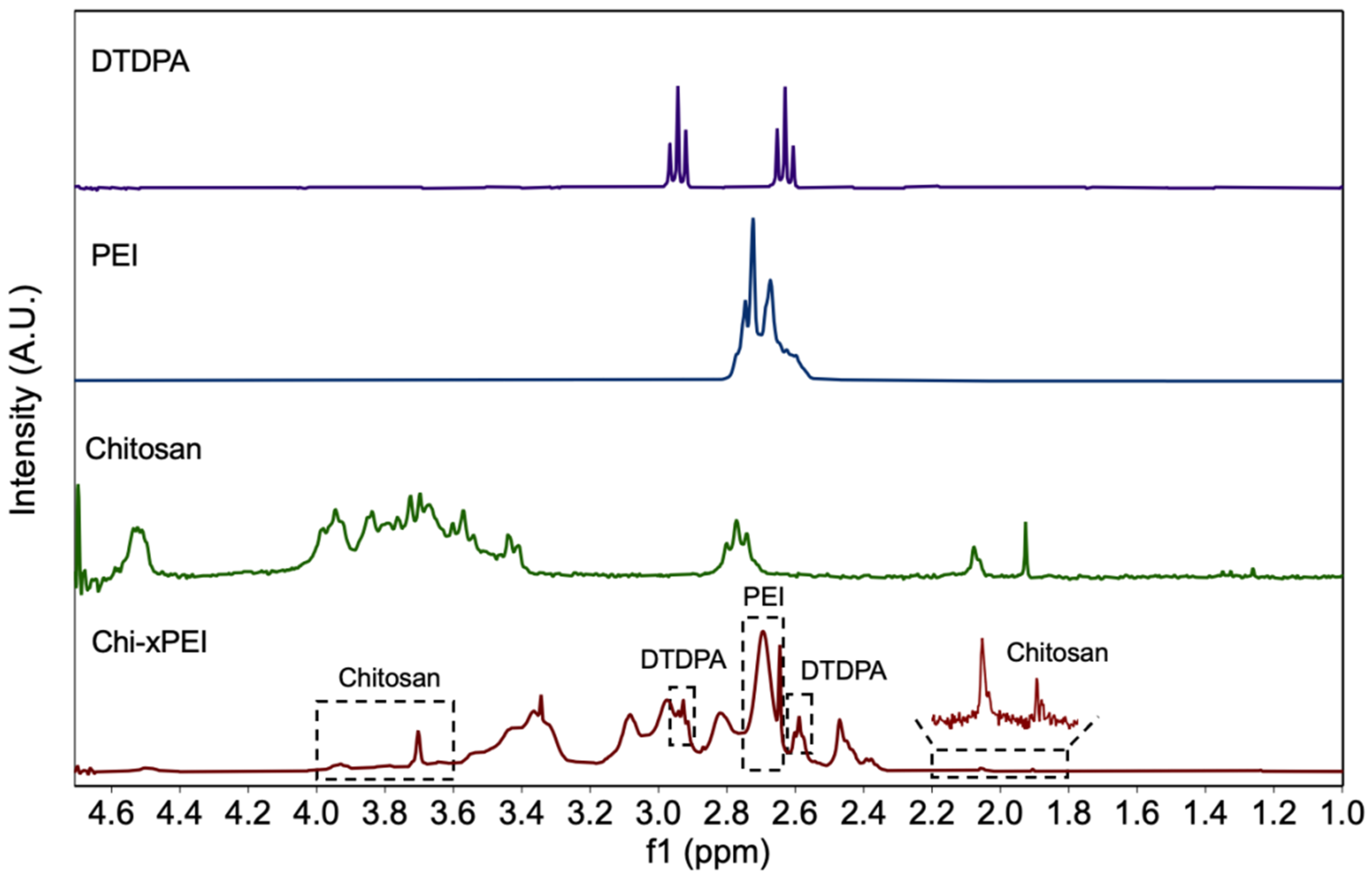
2.6. NMR Analysis
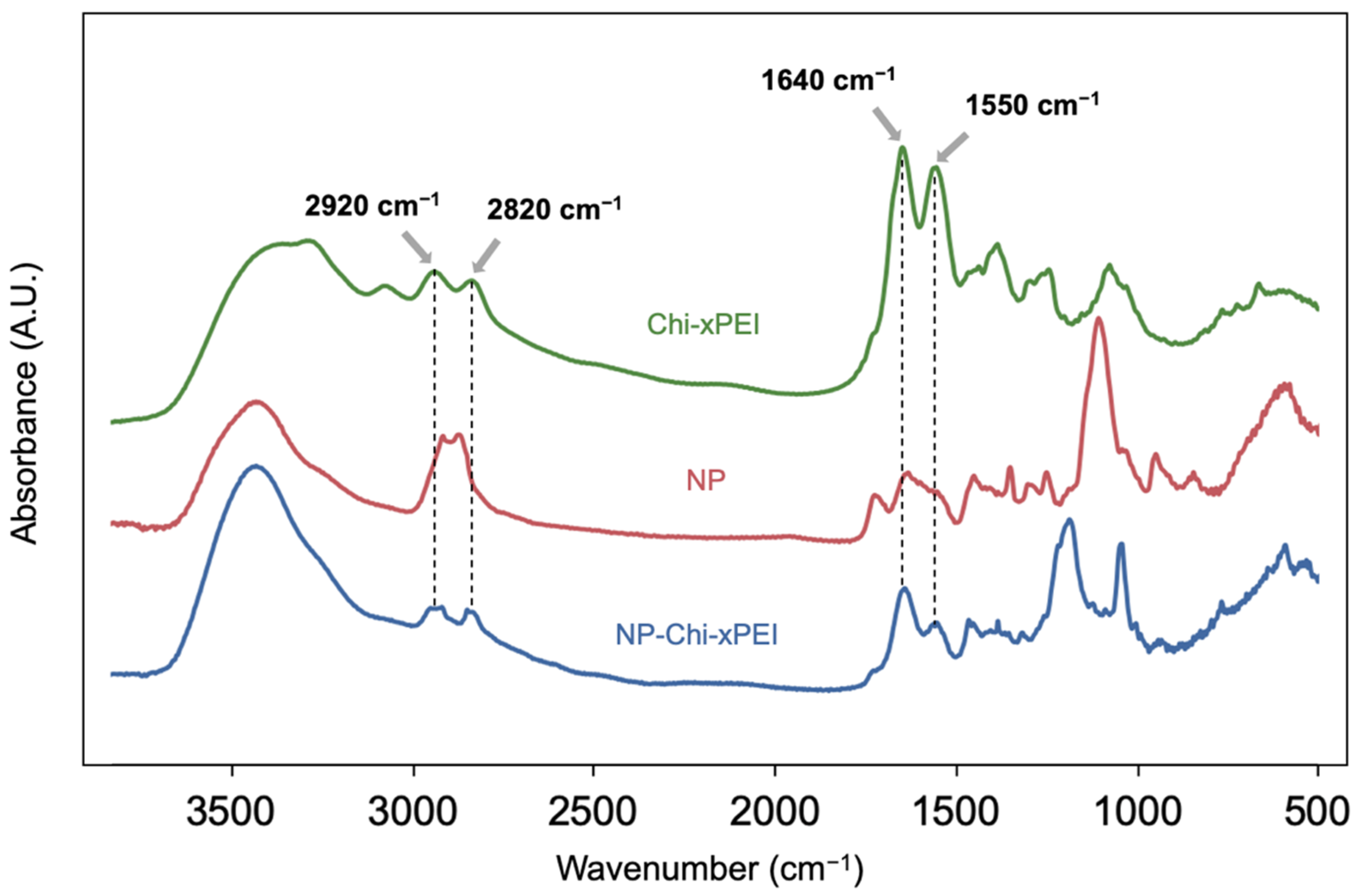
2.7. FTIR Analysis
2.8. NP-Chi-xPEI-DNA Complex Formation
2.9. TEM Imaging
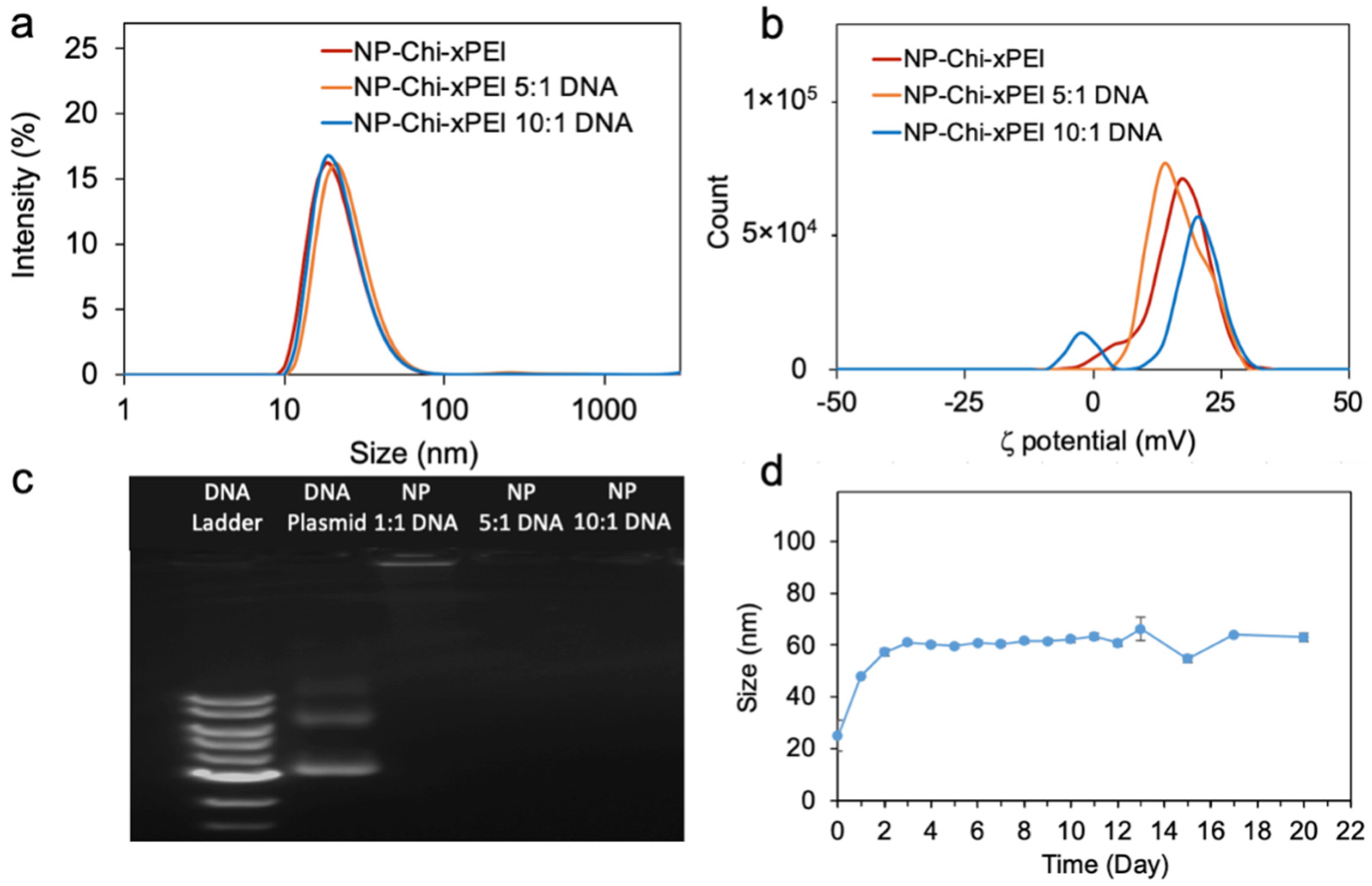
2.10. Hydrodynamic Size and Zeta Potential Measurements
2.11. Cell Culture
2.12. Fluorophore Labeling of NP-Chi-xPEI and DNA
2.13. Cellular Uptake and Intracellular Plasmid DNA Release Studies
2.14. Cell Transfections
2.15. Cell Viability Studies
2.16. Statistical Analysis
3. Results and Discussion
3.1. Synthesis and Structural Validation of NP-Chi-xPEI
3.2. Physicochemical Properties
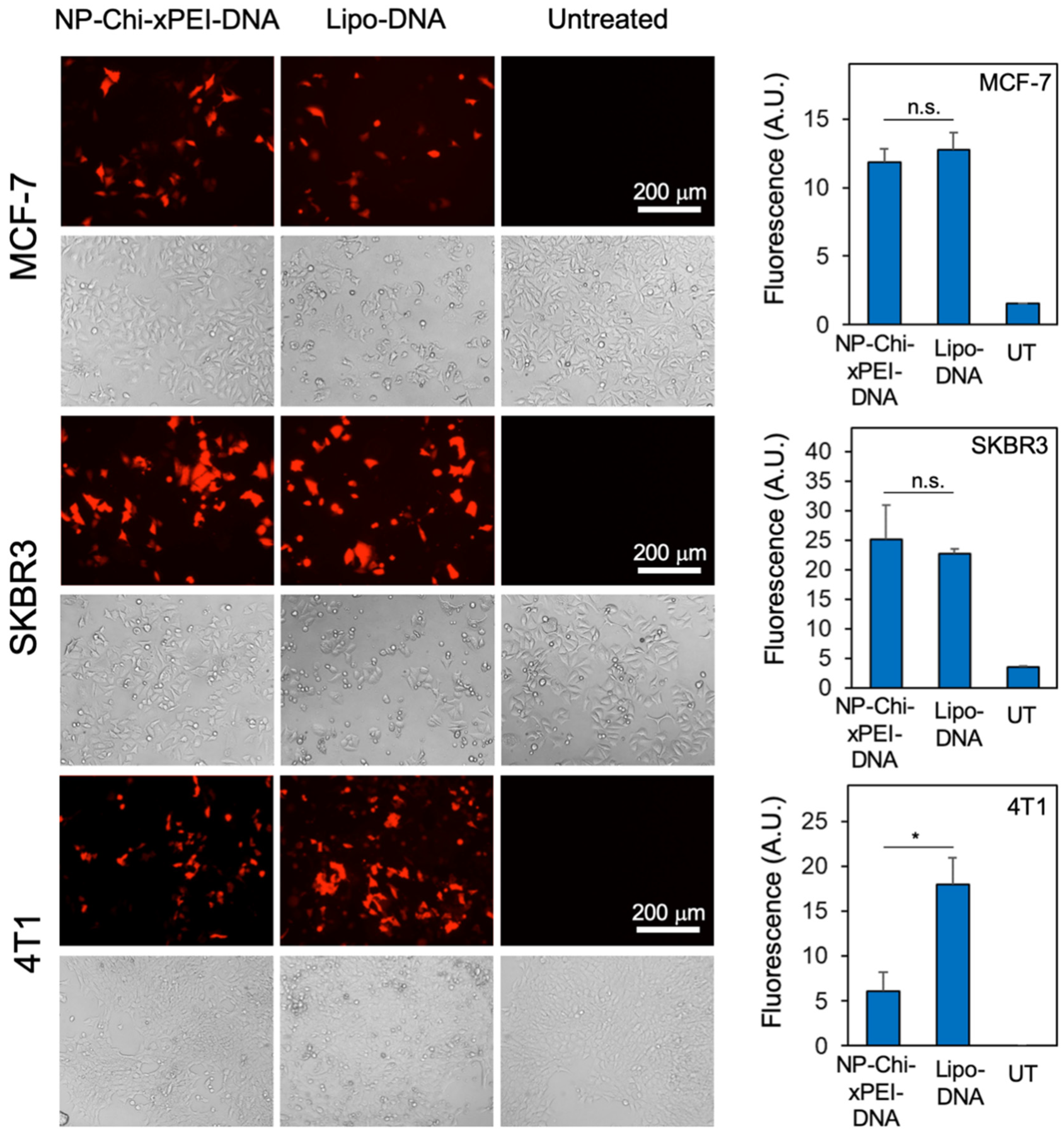
3.3. Cellular Uptake and Intracellular Plasmid DNA Release
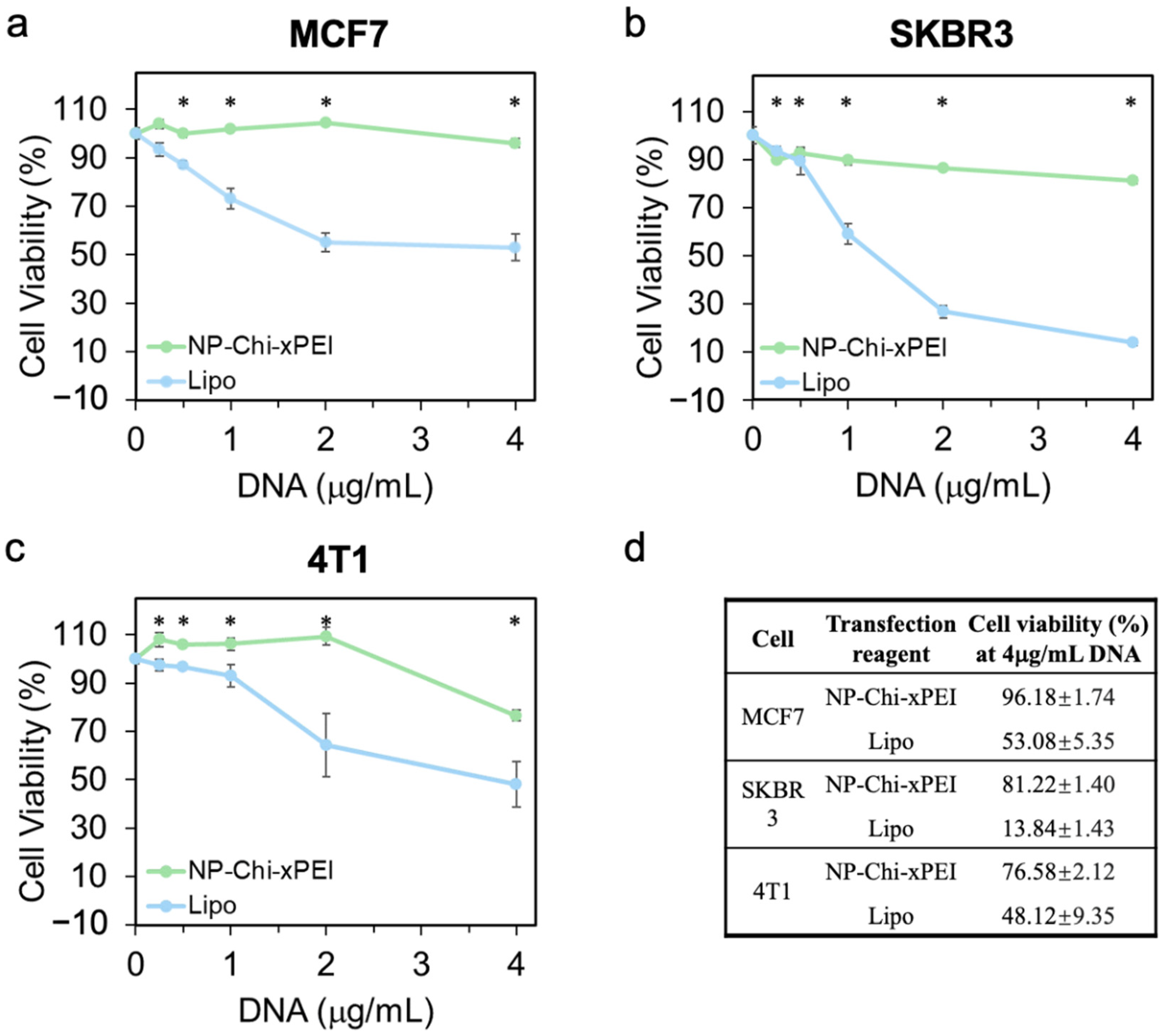
3.4. In Vitro Biocompatibility of NP-Chi-xPEI
3.5. In Vitro Transfection Efficiency of NP-Chi-xPEI-DNA
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Devulapally, R.; Lee, T.; Barghava-Shah, A.; Sekar, T.V.; Foygel, K.; Bachawal, S.V.; Willmann, J.K.; Paulmurugan, R. Ultrasound-guided delivery of thymidine kinase-nitroreductase dual therapeutic genes by PEGylated-PLGA/PEI nanoparticles for enhanced triple negative breast cancer therapy. Nanomedicine 2018, 13, 1051–1066. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Song, L.; Liu, S.; Jiang, Q.; Liu, Q.; Li, N.; Wang, Z.-G.; Ding, B. A DNA-Based Nanocarrier for Efficient Gene Delivery and Combined Cancer Therapy. Nano Lett. 2018, 18, 3328–3334. [Google Scholar] [CrossRef] [PubMed]
- Rybakova, Y.; Kowalski, P.S.; Huang, Y.; Gonzalez, J.T.; Heartlein, M.W.; DeRosa, F.; Delcassian, D.; Anderson, D.G. mRNA Delivery for Therapeutic Anti-HER2 Antibody Expression In Vivo. Mol. Ther. 2019, 27, 1415–1423. [Google Scholar] [CrossRef]
- Jahanafrooz, Z.; Baradaran, B.; Mosafer, J.; Hashemzaei, M.; Rezaei, T.; Mokhtarzadeh, A.; Hamblin, M.R. Comparison of DNA and mRNA vaccines against cancer. Drug Discov. Today 2020, 25, 552. [Google Scholar] [CrossRef]
- Liu, M.A. A Comparison of Plasmid DNA and mRNA as Vaccine Technologies. Vaccines 2019, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- Pollard, C.; Rejman, J.; De Haes, W.; Verrier, B.; Van Gulck, E.; Naessens, T.; De Smedt, S.; Bogaert, P.; Grooten, J.; Vanham, G.; et al. Type I IFN Counteracts the Induction of Antigen-Specific Immune Responses by Lipid-Based Delivery of mRNA Vaccines. Mol. Ther. 2013, 21, 251–259. [Google Scholar] [CrossRef] [Green Version]
- Lin, G.; Li, L.; Panwar, N.; Wang, J.; Tjin, S.C.; Wang, X.; Yong, K.T. Non-viral gene therapy using multifunctional nanoparticles: Status, challenges, and opportunities. Coord. Chem. Rev. 2018, 374, 133–152. [Google Scholar] [CrossRef]
- Unal, O.; Akkoc, Y.; Kocak, M.; Nalbat, E.; Isin Dogan-Ekici, A.; Yagci Acar, H.; Gozuacik, D. Treatment of breast cancer with autophagy inhibitory microRNAs carried by AGO2-conjugated nanoparticles. J. Nanobiotechnol. 2020, 18, 65. [Google Scholar] [CrossRef]
- Nayak, S.; Herzog, R.W. Progress and prospects: Immune responses to viral vectors. Gene Ther. 2010, 17, 295–304. [Google Scholar] [CrossRef] [Green Version]
- Godbey, W.T.; Wu, K.K.; Mikos, A.G. Poly(ethylenimine)-mediated gene delivery affects endothelial cell function and viability. Biomaterials 2001, 22, 471–480. [Google Scholar] [CrossRef]
- Monnery, B.D.; Wright, M.; Cavill, R.; Hoogenboom, R.; Shaunak, S.; Steinke, J.H.G.; Thanou, M. Cytotoxicity of polycations: Relationship of molecular weight and the hydrolytic theory of the mechanism of toxicity. Int. J. Pharm. 2017, 521, 249–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dokka, S.; Toledo, D.; Shi, X.; Castranova, V.; Rojanasakul, Y. Oxygen Radical-Mediated Pulmonary Toxicity Induced by Some Cationic Liposomes. Pharm. Res. 2000, 17, 521–525. [Google Scholar] [CrossRef] [PubMed]
- Mohammadinejad, R.; Dehshahri, A.; Sagar Madamsetty, V.; Zahmatkeshan, M.; Tavakol, S.; Makvandi, P.; Khorsandi, D.; Pardakhty, A.; Ashrafizadeh, M.; Ghasemipour Afshar, E.; et al. In vivo gene delivery mediated by non-viral vectors for cancer therapy. J. Control. Release 2020, 325, 249–275. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.Q.; Wei, J.; Fu, Y.R.; Shao, J.; Liang, Y.W.; Lin, Y.H.; Liu, J.; Zhu, Z.H. Toxicity of cationic liposome Lipofectamine 2000 in human pancreatic cancer Capan-2 cells. J. South. Med. Univ. 2008, 28, 1981–1984. [Google Scholar]
- Zhi, D.; Zhang, S.; Cui, S.; Zhao, Y.; Wang, Y.; Zhao, D. The Headgroup Evolution of Cationic Lipids for Gene Delivery. Bioconjug. Chem. 2013, 24, 487–519. [Google Scholar] [CrossRef]
- Xiong, M.P.; Laird Forrest, M.; Ton, G.; Zhao, A.; Davies, N.M.; Kwon, G.S. Poly(aspartate-g-PEI800), a polyethylenimine analogue of low toxicity and high transfection efficiency for gene delivery. Biomaterials 2007, 28, 4889–4900. [Google Scholar] [CrossRef]
- Zuris, J.A.; Thompson, D.B.; Shu, Y.; Guilinger, J.P.; Bessen, J.L.; Hu, J.H.; Maeder, M.L.; Joung, J.K.; Chen, Z.-Y.; Liu, D.R. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat. Biotechnol. 2014, 33, 73–80. [Google Scholar] [CrossRef] [Green Version]
- Venault, A.; Huang, Y.-C.; Lo, J.W.; Chou, C.-J.; Chinnathambi, A.; Higuchi, A.; Chen, W.-S.; Chen, W.-Y.; Chang, Y. Tunable PEGylation of branch-type PEI/DNA polyplexes with a compromise of low cytotoxicity and high transgene expression: In vitro and in vivo gene delivery. J. Mater. Chem. B 2017, 5, 4732–4744. [Google Scholar] [CrossRef]
- Liu, C.; Xie, Y.; Li, X.; Yao, X.; Wang, X.; Wang, M.; Li, Z.; Cao, F. Folic Acid/Peptides Modified PLGA–PEI–PEG Polymeric Vectors as Efficient Gene Delivery Vehicles: Synthesis, Characterization and Their Biological Performance. Mol. Biotechnol. 2020, 63, 63–79. [Google Scholar] [CrossRef]
- Sadeghpour, H.; Khalvati, B.; Entezar-Almahdi, E.; Savadi, N.; Hossaini Alhashemi, S.; Raoufi, M.; Dehshahri, A. Double domain polyethylenimine-based nanoparticles for integrin receptor mediated delivery of plasmid DNA. Sci. Rep. 2018, 8, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasiliu, T.; Florin Craciun, B.; Neamtu, A.; Clima, L.; Lucian Isac, D.; Maier, S.S.; Pinteala, M.; Mocci, F.; Laaksonen, A. In silico study of PEI-PEG-squalene-dsDNA polyplex formation: The delicate role of the PEG length in the binding of PEI to DNA. Biomater. Sci. 2021, 9, 6623–6640. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, F.; Feng, L.; Li, M.; Zhang, J.; Zhang, N. The targeted co-delivery of DNA and doxorubicin to tumor cells via multifunctional PEI-PEG based nanoparticles. Biomaterials 2013, 34, 2547–2564. [Google Scholar] [CrossRef] [PubMed]
- Lè Ne Pollard, H.; Remy, J.-S.; Loussouarn, G.; Demolombe, S.; Behr, J.-P.; Escande, D. Polyethylenimine but Not Cationic Lipids Promotes Transgene Delivery to the Nucleus in Mammalian Cells. J. Biol. Chem. 1998, 273, 7507–7511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zabner, J.; Fasbender, A.J.; Moninger, T.; Poellinger, K.A.; Welsh, M.J. Cellular and Molecular Barriers to Gene Transfer by a Cationic Lipid. J. Biol. Chem. 1995, 270, 18997–19007. [Google Scholar] [CrossRef] [Green Version]
- Jana, P.; Ghosh, S.; Sarkar, K. Low molecular weight polyethyleneimine conjugated guar gum for targeted gene delivery to triple negative breast cancer. Int. J. Biol. Macromol. 2020, 161, 1149–1160. [Google Scholar] [CrossRef]
- Wu, P.; Yin, S.; Liu, T.; Ding, D.; Wang, K. “Building-block crosslinking” micelles for enhancing cellular transfection of biocompatible polycations. Sci. China Mater. 2021, 64, 241–251. [Google Scholar] [CrossRef]
- Choi, S.; Lee, K.D. Enhanced gene delivery using disulfide-crosslinked low molecular weight polyethylenimine with listeriolysin o-polyethylenimine disulfide conjugate. J. Control. Release 2008, 131, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Fang, Z.; Yuan, M.; Li, W.; Yang, Y.; Jiang, M.; Ouyang, Y.; Yuan, W. Biodegradable Carriers for Delivery of VEGF Plasmid DNA for the Treatment of Critical Limb Ischemia. Front. Pharmacol. 2017, 8, 528. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Jiang, X.; Liu, J.; Yang, Q.; Zhuo, R. Reduction-degradable brushed polyethylenimine derivative synthesized by click chemistry for gene delivery. J. Control. Release 2013, 172, e118–e119. [Google Scholar] [CrossRef]
- Karimov, M.; Appelhans, D.; Ewe, A.; Aigner, A. The combined disulfide cross-linking and tyrosine-modification of very low molecular weight linear PEI synergistically enhances transfection efficacies and improves biocompatibility. Eur. J. Pharm. Biopharm. 2021, 161, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Bonner, D.K.; Zhao, X.; Buss, H.; Langer, R.; Hammond, P.T. Crosslinked linear polyethylenimine enhances delivery of DNA to the cytoplasm. J. Control. Release 2013, 167, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Ding, X.; Budry, R.; Mao, G. Layer-by-layer DNA films incorporating highly transfecting bioreducible poly(amido amine) and polyethylenimine for sequential gene delivery. Int. J. Nanomed. 2018, 13, 4943–4960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, H.; Jin, G.W.; Kang, H.; Lee, Y.; Nam, K.; Zhe Bai, C.; Park, J.S. Biodegradable branched poly(ethylenimine sulfide) for gene delivery. Biomaterials 2010, 31, 988–997. [Google Scholar] [CrossRef] [PubMed]
- Mintzer, M.A.; Simanek, E.E. Nonviral Vectors for Gene Delivery. Chem. Rev. 2008, 109, 259–302. [Google Scholar] [CrossRef] [PubMed]
- Stephen, Z.R.; Dayringer, C.J.; Lim, J.J.; Revia, R.A.; Halbert, M.V.; Jeon, M.; Bakthavatsalam, A.; Ellenbogen, R.G.; Zhang, M. Approach to Rapid Synthesis and Functionalization of Iron Oxide Nanoparticles for High Gene Transfection. ACS Appl. Mater. Interfaces 2016, 8, 6320–6328. [Google Scholar] [CrossRef]
- Lo, Y.L.; Chou, H.L.; Liao, Z.X.; Huang, S.J.; Ke, J.H.; Liu, Y.S.; Chiu, C.C.; Wang, L.F. Chondroitin sulfate-polyethylenimine copolymer-coated superparamagnetic iron oxide nanoparticles as an efficient magneto-gene carrier for microRNA-encoding plasmid DNA delivery. Nanoscale 2015, 7, 8554–8565. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Y.; Yu, J.C.; Chen, Y.Y.; Chan, K.M. Assembly of polyethylenimine-functionalized iron oxide nanoparticles as agents for DNA transfection with magnetofection technique. J. Mater. Chem. B 2014, 2, 7936–7944. [Google Scholar] [CrossRef]
- Lin, G.; Revia, R.A.; Zhang, M. Inorganic Nanomaterial-Mediated Gene Therapy in Combination with Other Antitumor Treatment Modalities. Adv. Funct. Mater. 2021, 31, 2007096. [Google Scholar] [CrossRef]
- Liu, W.M.; Xue, Y.N.; Peng, N.; He, W.T.; Zhuo, R.X.; Huang, S.W. Dendrimer modified magnetic iron oxide nanoparticle/DNA/PEI ternary magnetoplexes: A novel strategy for magnetofection. J. Mater. Chem. 2011, 21, 13306–13315. [Google Scholar] [CrossRef]
- Namgung, R.; Singha, K.; Yu, M.K.; Jon, S.; Kim, Y.S.; Ahn, Y.; Park, I.K.; Kim, W.J. Hybrid superparamagnetic iron oxide nanoparticle-branched polyethylenimine magnetoplexes for gene transfection of vascular endothelial cells. Biomaterials 2010, 31, 4204–4213. [Google Scholar] [CrossRef] [PubMed]
- Zakeri, A.; Kouhbanani, M.A.J.; Beheshtkhoo, N.; Beigi, V.; Mousavi, S.M.; Hashemi, S.A.R.; Zade, A.K.; Amani, A.M.; Savardashtaki, A.; Mirzaei, E.; et al. Polyethylenimine-based nanocarriers in co-delivery of drug and gene: A developing horizon. Nano Rev. Exp. 2018, 9, 1488497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ullah, I.; Muhammad, K.; Akpanyung, M.; Nejjari, A.; Luis Neve, A.; Guo, J.; Feng, Y.; Shi, C. Bioreducible, hydrolytically degradable and targeting polymers for gene delivery. J. Mater. Chem. B 2017, 5, 3253–3276. [Google Scholar] [CrossRef]
- Chuan, D.; Jin, T.; Fan, R.; Zhou, L.; Guo, G. Chitosan for gene delivery: Methods for improvement and applications. Adv. Colloid Interface Sci. 2019, 268, 25–38. [Google Scholar] [CrossRef]
- Lin, G.; Mu, Q.; Revia, R.; Stephen, Z.; Jeon, M.; Zhang, M. A highly selective iron oxide-based imaging nanoparticle for long-term monitoring of drug-induced tumor cell apoptosis. Biomater. Sci. 2021, 9, 471–481. [Google Scholar] [CrossRef] [PubMed]
- Coates, J. Interpretation of infrared spectra, a practical approach. In Encyclopedia of Analytical Chemistry; Meyers, R.A., Ed.; John Wiley & Sons Ltd.: Chichester, UK, 2000; pp. 10815–10837. [Google Scholar]
- Prabha, S.; Arya, G.; Chandra, R.; Ahmed, B.; Nimesh, S. Effect of size on biological properties of nanoparticles employed in gene delivery. Artif. Cells Nanomed. Biotechnol. 2014, 44, 83–91. [Google Scholar] [CrossRef]
- Kievit, F.M.; Veiseh, O.; Fang, C.; Bhattarai, N.; Lee, D.; Ellenbogen, R.G.; Zhang, M. Chlorotoxin Labeled Magnetic Nanovectors for Targeted Gene Delivery to Glioma. ACS Nano 2010, 4, 4587–4594. [Google Scholar] [CrossRef] [Green Version]
- Fröhlich, E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012, 7, 5577. [Google Scholar] [CrossRef] [Green Version]
- Kievit, F.M.; Wang, K.; Ozawa, T.; Tarudji, A.W.; Silber, J.R.; Holland, E.C.; Ellenbogen, R.G.; Zhang, M. Nanoparticle-mediated knockdown of DNA repair sensitizes cells to radiotherapy and extends survival in a genetic mouse model of glioblastoma. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 2131–2139. [Google Scholar] [CrossRef]
- Wang, K.; Kievit, F.M.; Jeon, M.; Silber, J.R.; Ellenbogen, R.G.; Zhang, M. Nanoparticle-Mediated Target Delivery of TRAIL as Gene Therapy for Glioblastoma. Adv. Healthc. Mater. 2015, 4, 2719–2726. [Google Scholar] [CrossRef] [Green Version]
- Zhan, Y.-R.; Yu, Q.-Y.; Zhang, J.; Liu, Y.-H.; Xiao, Y.-P.; Zhang, J.-H.; He, X.; Yu, X.-Q. Glutathione modified low molecular weight PEI for highly improved gene transfection ability and biocompatibility. New J. Chem. 2019, 43, 12109–12117. [Google Scholar] [CrossRef]
- Xu, F.; Zhong, H.; Chang, Y.; Li, D.; Jin, H.; Zhang, M.; Wang, H.; Jiang, C.; Shen, Y.; Huang, Y. Targeting death receptors for drug-resistant cancer therapy: Codelivery of pTRAIL and monensin using dual-targeting and stimuli-responsive self-assembling nanocomposites. Biomaterials 2018, 158, 56–73. [Google Scholar] [CrossRef] [PubMed]
- Bansal, R.; Kiran, P.; Kumar, P. Synthesis, characterization and evaluation of diglycidyl-1,2-cyclohexanedicarboxylate crosslinked polyethylenimine nanoparticles as efficient carriers of DNA. New J. Chem. 2016, 40, 5044–5052. [Google Scholar] [CrossRef]
- Magro, M.; Martinello, T.; Bonaiuto, E.; Gomiero, C.; Baratella, D.; Zoppellaro, G.; Cozza, G.; Patruno, M.; Zboril, R.; Vianello, F. Covalently bound DNA on naked iron oxide nanoparticles: Intelligent colloidal nano-vector for cell transfection. Biochim. et Biophys. Acta Gen. Subj. 2017, 1861, 2802–2810. [Google Scholar] [CrossRef]
- Jiang, S.; Eltoukhy, A.A.; Love, K.T.; Langer, R.; Anderson, D.G. Lipidoid-coated iron oxide nanoparticles for efficient DNA and siRNA delivery. Nano Lett. 2013, 13, 1059–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Jin, Y.; Zhao, C.; Pan, E.; Jia, M. Fe3O4 nanoparticle/graphene aerogel composite with enhanced lithium storage performance. Appl. Surf. Sci. 2018, 458, 1035–1042. [Google Scholar] [CrossRef]
- Khurshid, H.; Li, W.; Chandra, S.; Phan, M.H.; Hadjipanayis, G.C.; Mukherjee, P.; Srikanth, H. Mechanism and controlled growth of shape and size variant core/shell FeO/Fe3O4 nanoparticles. Nanoscale 2013, 5, 7942–7952. [Google Scholar] [CrossRef]
- Liang, C.; Zhai, T.; Wang, W.; Chen, J.; Zhao, W.; Lu, X.; Tong, Y. Fe3O4/reduced graphene oxide with enhanced electrochemical performance towards lithium storage. J. Mater. Chem. A 2014, 2, 7214–7220. [Google Scholar] [CrossRef]
- Ren, W.; Liu, Y.; Wan, S.; Fei, C.; Wang, W.; Chen, Y.; Zhang, Z.; Wang, T.; Wang, J.; Zhou, L.; et al. BMP9 Inhibits Proliferation and Metastasis of HER2-Positive SK-BR-3 Breast Cancer Cells through ERK1/2 and PI3K/AKT Pathways. PLoS ONE 2014, 9, e96816. [Google Scholar] [CrossRef]
- Saceda, M.; Lippman, M.E.; Chambon, P.; Lindsey, R.L.; Ponglikitmongkol, M.; Puente, M.; Martin, M.B. Regulation of the estrogen receptor in MCF-7 cells by estradiol. Mol. Endocrinol. 1988, 2, 1157–1162. [Google Scholar] [CrossRef] [Green Version]
- Arroyo-Crespo, J.J.; Armiñán, A.; Charbonnier, D.; Deladriere, C.; Palomino-Schätzlein, M.; Lamas-Domingo, R.; Forteza, J.; Pineda-Lucena, A.; Vicent, M.J. Characterization of triple-negative breast cancer preclinical models provides functional evidence of metastatic progression. Int. J. Cancer 2019, 145, 2267. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.K.; Hwang, G.B.; Seu, Y.B.; Choi, J.S.; Jin, K.S.; Doh, K.O. DOTAP/DOPE ratio and cell type determine transfection efficiency with DOTAP-liposomes. Biochim. Biophys. Acta-Biomembr. 2015, 1848, 1996–2001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, K.L.; Piccirillo, C.A.; Tabrizian, M. Cell line-dependent internalization pathways and intracellular trafficking determine transfection efficiency of nanoparticle vectors. Eur. J. Pharm. Biopharm. 2008, 68, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Narain, R. Cell line dependent uptake and transfection efficiencies of PEI–anionic glycopolymer systems. Biomaterials 2013, 34, 4368–4376. [Google Scholar] [CrossRef] [PubMed]
- Izumisawa, T.; Hattori, Y.; Date, M.; Toma, K.; Maitani, Y. Cell line-dependent internalization pathways determine DNA transfection efficiency of decaarginine-PEG-lipid. Int. J. Pharm. 2011, 404, 264–270. [Google Scholar] [CrossRef]
- Moghimi, S.M.; Symonds, P.; Murray, J.C.; Hunter, A.C.; Debska, G.; Szewczyk, A. A two-stage poly(ethylenimine)-mediated cytotoxicity: Implications for gene transfer/therapy. Mol. Ther. 2005, 11, 990–995. [Google Scholar] [CrossRef]


| Non-Viral DNA Carrier Material | Hydrodynamic Size | Zeta Potential | Reference |
|---|---|---|---|
| NP-Chi-xPEI-DNA | 45.8 nm | 18.3 mV | N/A |
| 2,6-pyridinedicarboxaldehyde-Crosslinked 1.8k PEI | 160–250 nm | 17–27 mV | [29] |
| Disulfide-crosslinked 2k PEI modified with tyrosine | 135.6 nm | 62.1 mV | [31] |
| Disulfide-crosslinked 2.5k PEI | 350–500 nm | 20–40 mV | [32] |
| Disulfide and bisepoxide crosslinked 6k PEI | 200 nm | 20 mV | [34] |
| Oxidized glutathione crosslinked 0.6k PEI | 200–400 nm | 30 mV | [52] |
| Disulfide-crosslinked 1.8k PEI modified with cyclodextrin and poly-glutamic acid | 250 nm | 20 mV | [53] |
| Diglycidyl-1,2-cyclohexanedicarboxylate-crosslinked 10k PEI | 125–201 nm | 11–20 mV | [54] |
| Non-Viral DNA Carrier Material | Hydrodynamic Size | Zeta Potential | Reference |
|---|---|---|---|
| NP-Chi-xPEI-DNA | 45.8 nm | 18.3 mV | N/A |
| IONP-Catechol-Chitosan-25k PEI-DNA | 54.3 nm | 16.2 mV | [36] |
| IONP-Chondroitin-10k PEI-DNA | 136 nm | 15 mV | [37] |
| IONP-25k PEI-DNA | 250 nm | 19.2 mV | [38] |
| IONP-PAMAM Denrimer-DNA-25k PEI | 190–285 nm | 45 mV | [40] |
| IONP-1.8k PEI-DNA | 50 nm (dry size) | 10 mV | [41] |
| IONP covalently bound to DNA | 241 nm | −26.4 mV | [55] |
| IONP-Lipids-DNA | 50–100 nm | 20 mV | [56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, G.; Huang, J.; Zhang, M.; Chen, S.; Zhang, M. Chitosan-Crosslinked Low Molecular Weight PEI-Conjugated Iron Oxide Nanoparticle for Safe and Effective DNA Delivery to Breast Cancer Cells. Nanomaterials 2022, 12, 584. https://doi.org/10.3390/nano12040584
Lin G, Huang J, Zhang M, Chen S, Zhang M. Chitosan-Crosslinked Low Molecular Weight PEI-Conjugated Iron Oxide Nanoparticle for Safe and Effective DNA Delivery to Breast Cancer Cells. Nanomaterials. 2022; 12(4):584. https://doi.org/10.3390/nano12040584
Chicago/Turabian StyleLin, Guanyou, Jianxi Huang, Mengyuan Zhang, Shanshan Chen, and Miqin Zhang. 2022. "Chitosan-Crosslinked Low Molecular Weight PEI-Conjugated Iron Oxide Nanoparticle for Safe and Effective DNA Delivery to Breast Cancer Cells" Nanomaterials 12, no. 4: 584. https://doi.org/10.3390/nano12040584
APA StyleLin, G., Huang, J., Zhang, M., Chen, S., & Zhang, M. (2022). Chitosan-Crosslinked Low Molecular Weight PEI-Conjugated Iron Oxide Nanoparticle for Safe and Effective DNA Delivery to Breast Cancer Cells. Nanomaterials, 12(4), 584. https://doi.org/10.3390/nano12040584






