Automated Recognition of Nanoparticles in Electron Microscopy Images of Nanoscale Palladium Catalysts
Abstract
1. Introduction
2. Results and Discussion
2.1. General Approach
2.2. Methods Comparison
- The straightforward local intensity maxima (LM) method;
- Three classical computer vision methods of blob detection: Laplacian of Gaussian (LOG), difference of Gaussians (DOG), and determinant of the Hessian (DOH) methods;
- The proposed methods in this work include exponential approximation method (Exp) and
- Deep neural network (DNN) approach.
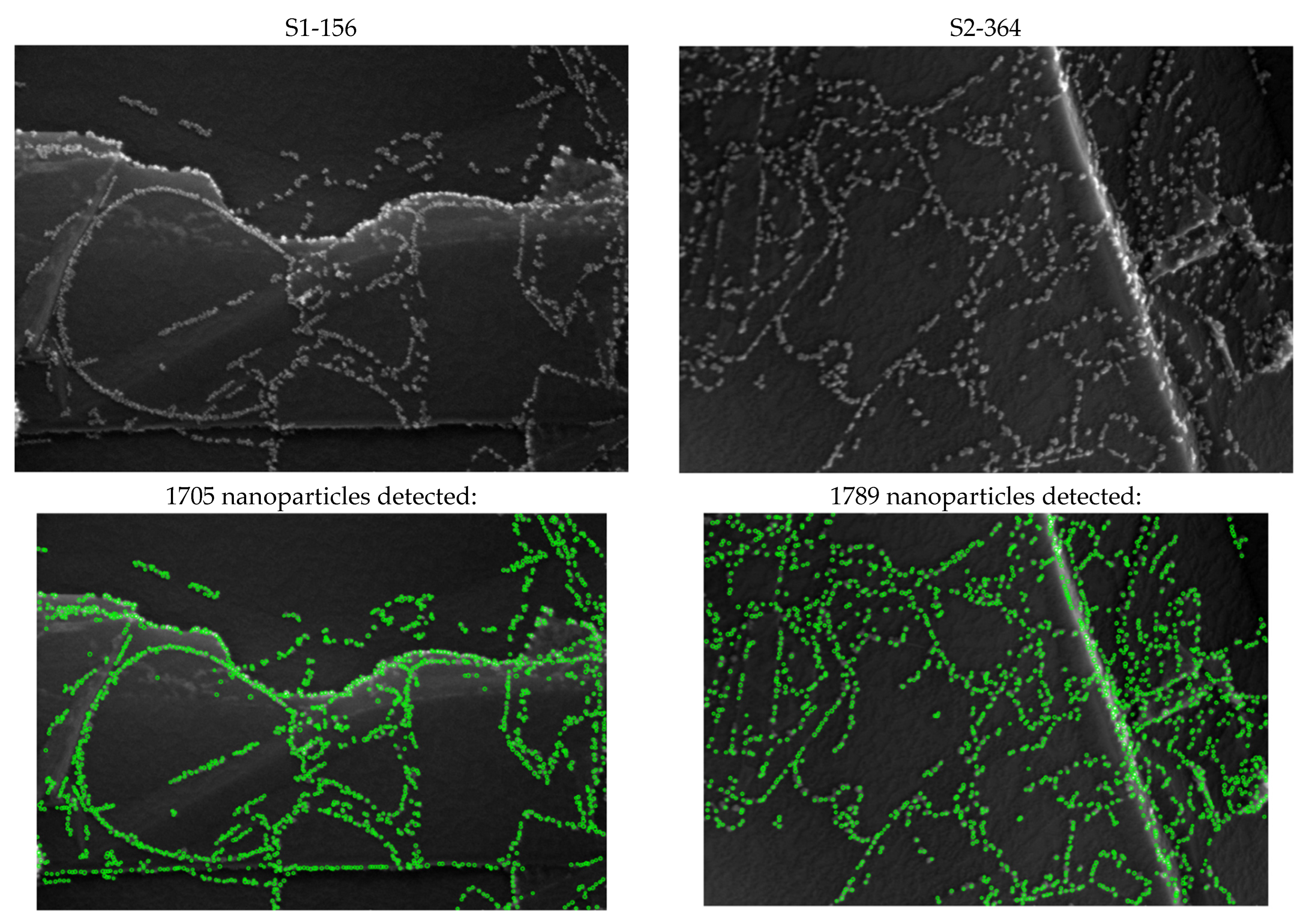
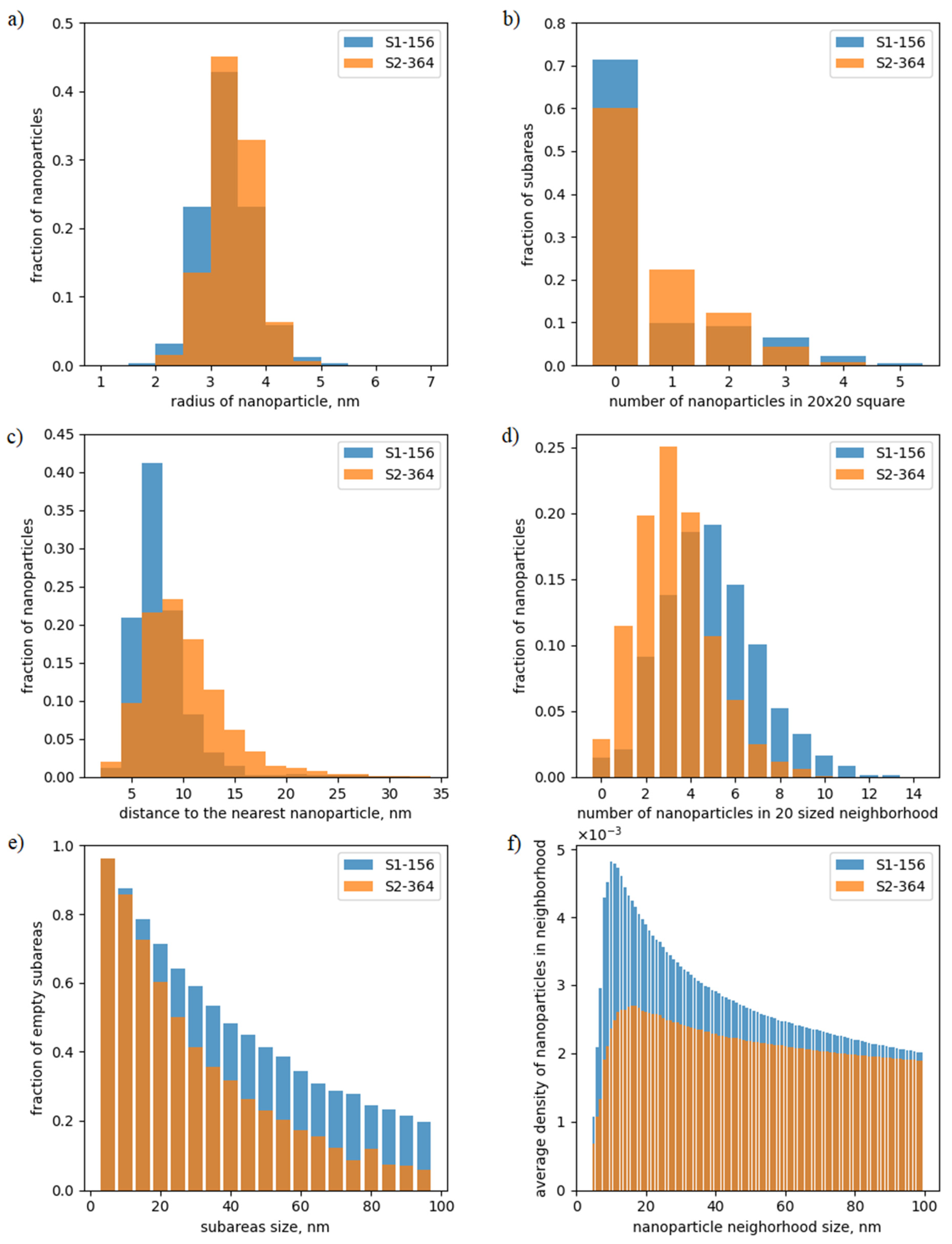
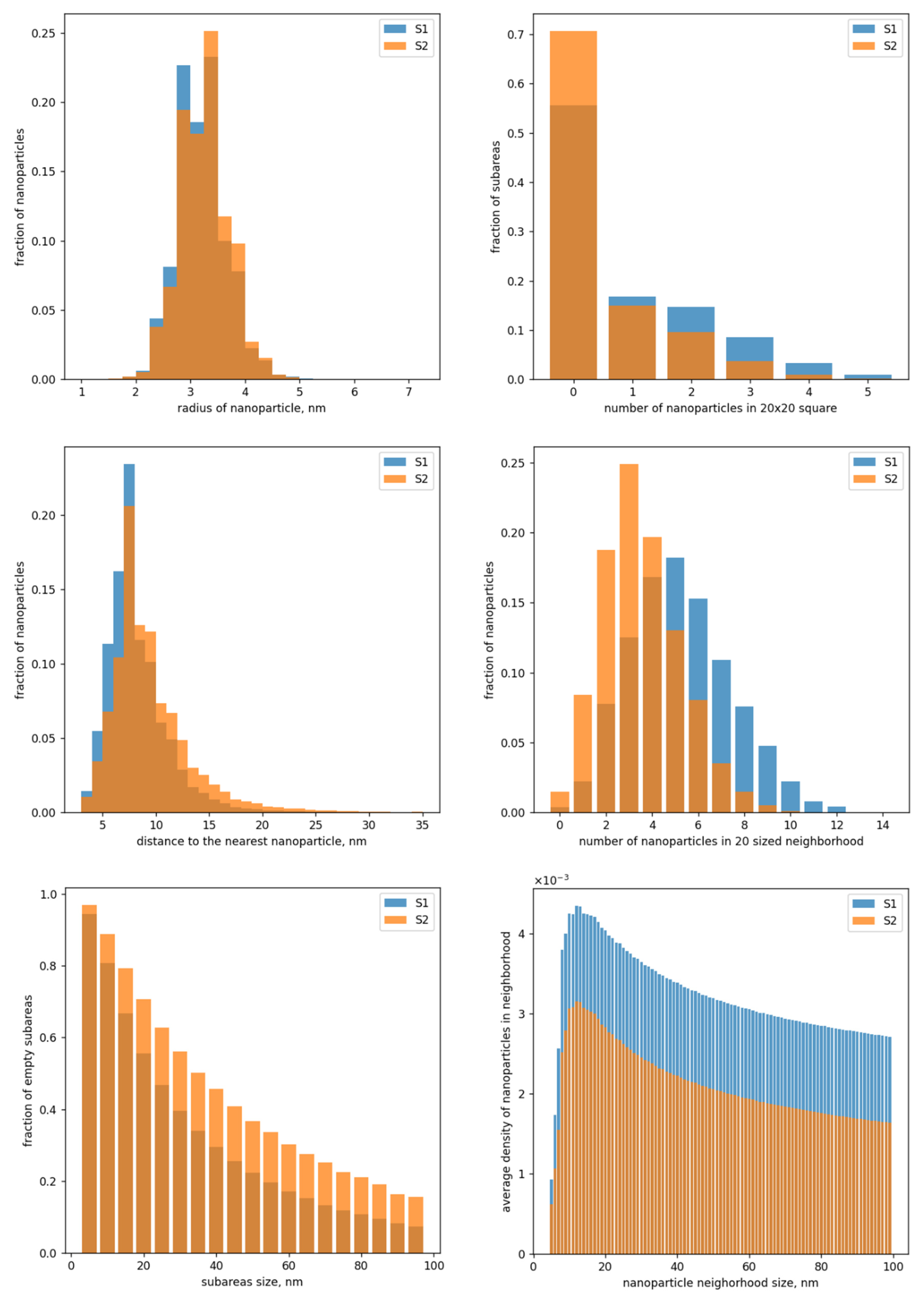
2.3. Analysis of the Data
- Size of image fragments for approximation ,
- Set of possible radius values: from 1 to 7 through 0.1,
- Values of adaptive threshold coefficients for prefiltering (stage 2) and detection (stage 4) were estimated as parameters that maximize the sums of accuracies for all four manually labeled images: , .
3. Conclusions
4. Methods
4.1. Image Labeling
4.2. Metric Calculation
4.3. Method Implementation
4.3.1. Classic Methods
4.3.2. Exponential Approximation (Exp)
- Stage 1. Preprocessing
- Stage 2. Selecting small image fragments
- Stage 3. Exponential approximation of one small image fragment
- Stage 4. Detecting fragments that contain a nanoparticle
- Stage 5. Determining the radius of a particle
4.3.3. Deep Neural Network (DNN) Predictions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mao, Z.; Gu, H.; Lin, X. Recent Advances of Pd/C-Catalyzed Reactions. Catalysts 2021, 11, 1078. [Google Scholar] [CrossRef]
- Liu, X.; Astruc, D. Development of the Applications of Palladium on Charcoal in Organic Synthesis. Adv. Synth. Catal. 2018, 360, 3426–3459. [Google Scholar] [CrossRef]
- Mori, A.; Mizusaki, T.; Miyakawa, Y.; Ohashi, E.; Haga, T.; Maegawa, T.; Monguchi, Y.; Sajiki, H. Chemoselective Hydrogenation Method Catalyzed by Pd/C Using Diphenylsulfide as a Reasonable Catalyst Poison. Tetrahedron 2006, 62, 11925–11932. [Google Scholar] [CrossRef]
- Harvey, F.M.; Bochet, C.G. Stereospecific Hydrogenolysis of Benzylic Alcohols over Pd/C. J. Org. Chem. 2020, 85, 7611–7619. [Google Scholar] [CrossRef] [PubMed]
- Felpin, F.; Ayad, T.; Mitra, S. Pd/C: An Old Catalyst for New Applications—Its Use for the Suzuki–Miyaura Reaction. Eur. J. Org. Chem. 2006, 2006, 2679–2690. [Google Scholar] [CrossRef]
- Shi, G.; Dong, Z. Palladium Supported on Porous Organic Polymer as Heterogeneous and Recyclable Catalyst for Cross Coupling Reaction. Molecules 2022, 27, 4777. [Google Scholar] [CrossRef]
- Chen, X.; Engle, K.M.; Wang, D.-H.; Yu, J.-Q. Palladium(II)-Catalyzed C-H Activation/C-C Cross-Coupling Reactions: Versatility and Practicality. Angew. Chem. Int. Ed. 2009, 48, 5094–5115. [Google Scholar] [CrossRef]
- Eremin, D.B.; Galushko, A.S.; Boiko, D.A.; Pentsak, E.O.; Chistyakov, I.V.; Ananikov, V.P. Toward Totally Defined Nanocatalysis: Deep Learning Reveals the Extraordinary Activity of Single Pd/C Particles. J. Am. Chem. Soc. 2022, 144, 6071. [Google Scholar] [CrossRef]
- Xu, F.; Zhang, Z.; Ma, J.; Ma, C.; Guan, B.-O.; Chen, K. Large-Area Ordered Palladium Nanostructures by Colloidal Lithography for Hydrogen Sensing. Molecules 2022, 27, 6100. [Google Scholar] [CrossRef]
- Yoosefian, M.; Ayoubi, E.; Atanase, L.I. Palladium-Doped Single-Walled Carbon Nanotubes as a New Adsorbent for Detecting and Trapping Volatile Organic Compounds: A First Principle Study. Nanomaterials 2022, 12, 2572. [Google Scholar] [CrossRef]
- Yurkov, G.; Koksharov, Y.; Fionov, A.; Taratanov, N.; Kolesov, V.; Kirillov, V.; Makeev, M.; Mikhalev, P.; Ryzhenko, D.; Solodilov, V. Polymer Nanocomposite Containing Palladium Nanoparticles: Synthesis, Characterization, and Properties. Polymers 2022, 14, 3795. [Google Scholar] [CrossRef]
- Bulanova, A.V.; Shafigulin, R.V.; Vinogradov, K.Y.; Tokranova, E.O.; Martynenko, E.A.; Vostrikov, S.V.; Podlipnov, V.V. Effect of Modifying Carbon Materials with Metal Phthalocynines and Palladium on Their Catalytic Activity in ORR. Catalysts 2022, 12, 1013. [Google Scholar] [CrossRef]
- Sayury Miyashiro, C.; Hamoudi, S. Palladium and Graphene Oxide Doped ZnO for Aqueous Acetamiprid Degradation under Visible Light. Catalysts 2022, 12, 709. [Google Scholar] [CrossRef]
- Solovov, R.; Ershov, B. Features of Hydrogen Reduction of Fe(CN)63−Ions in Aqueous Solutions: Effect of Hydrogen Dissolved in Palladium Nanoparticles. Nanomaterials 2021, 11, 2587. [Google Scholar] [CrossRef] [PubMed]
- Sikora, E.; Muránszky, G.; Kristály, F.; Fiser, B.; Farkas, L.; Viskolcz, B.; Vanyorek, L. Development of Palladium and Platinum Decorated Granulated Carbon Nanocomposites for Catalytic Chlorate Elimination. Int. J. Mol. Sci. 2022, 23, 10514. [Google Scholar] [CrossRef]
- Çalışkan, M.; Akay, S.; Kayan, B.; Baran, T.; Kalderis, D. Preparation and Application of a Hydrochar-Based Palladium Nanocatalyst for the Reduction of Nitroarenes. Molecules 2021, 26, 6859. [Google Scholar] [CrossRef] [PubMed]
- Prekob, Á.; Udayakumar, M.; Karacs, G.; Kristály, F.; Muránszky, G.; Leskó, A.K.; Németh, Z.; Viskolcz, B.; Vanyorek, L. Development of Highly Efficient, Glassy Carbon Foam Supported, Palladium Catalysts for Hydrogenation of Nitrobenzene. Nanomaterials 2021, 11, 1172. [Google Scholar] [CrossRef]
- Al-Fakeh, M.S.; Osman, S.O.M.; Gassoumi, M.; Rabhi, M.; Omer, M. Characterization, Antimicrobial and Anticancer Properties of Palladium Nanoparticles Biosynthesized Optimally Using Saudi Propolis. Nanomaterials 2021, 11, 2666. [Google Scholar] [CrossRef]
- Pentsak, E.O.; Kashin, A.S.; Polynski, M.V.; Kvashnina, K.O.; Glatzel, P.; Ananikov, V.P. Spatial Imaging of Carbon Reactivity Centers in Pd/C Catalytic Systems. Chem. Sci. 2015, 6, 3302–3313. [Google Scholar] [CrossRef]
- Fei, Z.; Rodin, A.S.; Gannett, W.; Dai, S.; Regan, W.; Wagner, M.; Liu, M.K.; McLeod, A.S.; Dominguez, G.; Thiemens, M.; et al. Electronic and Plasmonic Phenomena at Graphene Grain Boundaries. Nat. Nanotechnol. 2013, 8, 821–825. [Google Scholar] [CrossRef]
- Boiko, D.A.; Pentsak, E.O.; Cherepanova, V.A.; Gordeev, E.G.; Ananikov, V.P. Deep Neural Network Analysis of Nanoparticle Ordering to Identify Defects in Layered Carbon Materials. Chem. Sci. 2021, 12, 7428–7441. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Nartova, A.V.; Mashukov, M.Y.; Astakhov, R.R.; Kudinov, V.Y.; Matveev, A.V.; Okunev, A.G. Particle Recognition on Transmission Electron Microscopy Images Using Computer Vision and Deep Learning for Catalytic Applications. Catalysts 2022, 12, 135. [Google Scholar] [CrossRef]
- Okunev, A.G.; Mashukov, M.Y.; Nartova, A.V.; Matveev, A.V. Nanoparticle Recognition on Scanning Probe Microscopy Images Using Computer Vision and Deep Learning. Nanomaterials 2020, 10, 1285. [Google Scholar] [CrossRef]
- Georgantzinos, S.K. Multiscale Simulation of Composite Structures: Damage Assessment, Mechanical Analysis and Prediction. Materials 2022, 15, 6494. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, Y.; Yang, B.; Li, G.; Chen, T.; Nakajima, M.; Sun, L.; Fukuda, T. Mechatronic Development and Vision Feedback Control of a Nanorobotics Manipulation System inside SEM for Nanodevice Assembly. Sensors 2016, 16, 1479. [Google Scholar] [CrossRef] [PubMed]
- Aversa, R.; Modarres, M.H.; Cozzini, S.; Ciancio, R.; Chiusole, A. The First Annotated Set of Scanning Electron Microscopy Images for Nanoscience. Sci. Data 2018, 5, 180172. [Google Scholar] [CrossRef] [PubMed]
- DeCost, B.L.; Holm, E.A. A Large Dataset of Synthetic SEM Images of Powder Materials and Their Ground Truth 3D Structures. Data Br. 2016, 9, 727–731. [Google Scholar] [CrossRef]
- Boiko, D.A.; Pentsak, E.O.; Cherepanova, V.A.; Ananikov, V.P. Electron Microscopy Dataset for the Recognition of Nanoscale Ordering Effects and Location of Nanoparticles. Sci. Data 2020, 7, 101. [Google Scholar] [CrossRef]
- Boiko, D.A.; Pentsak, E.O.; Cherepanova, V.A.; Ananikov, V.P. Electron Microscopy Dataset for the Recognition of Nanoscale Ordering Effects and Location of Nanoparticles—Dataset 1 (Ordered). (2020). Figshare. Available online: https://figshare.com/articles/dataset/Electron_microscopy_dataset_for_the_recognition_of_nanoscale_ordering_effects_and_location_of_nanoparticles_Dataset_1_ordered_/11783661 (accessed on 12 October 2022).
- Lunts, A.L.; Brailovskii, V.L. Estimation of Features Obtained in Statistical Recognition Procedures. Izv. Akad. Nauk SSSR Ser. Tekh. Kibern. 1969, 3. [Google Scholar]
- Faloutsos, C.; Lin, K.-I. FastMap: A Fast Algorithm for Indexing, Data-Mining and Visualization of Traditional and Multimedia Datasets. In Proceedings of the 1995 ACM SIGMOD International Conference on Management of Data—SIGMOD ’95, San Jose, CA, USA, 22–25 May 1995; ACM Press: New York, NY, USA, 1995; pp. 163–174. [Google Scholar]
- Lindeberg, T. Scale-Space Theory in Computer Vision; Springer: Boston, MA, USA, 1994; ISBN 978-1-4419-5139-7. [Google Scholar]
- Marr, D.; Hildreth, E. Theory of Edge Detection. Proc. R. Soc. B 1980, 207, 187–217. [Google Scholar] [CrossRef]
- Bright, D.S.; Steel, E.B. Two-Dimensional Top Hat Filter for Extracting Spots and Spheres from Digital Images. J. Microsc. 1987, 146, 191–200. [Google Scholar] [CrossRef]
- Zeng, M.; Li, J.; Peng, Z. The Design of Top-Hat Morphological Filter and Application to Infrared Target Detection. Infrared Phys. Technol. 2006, 48, 67–76. [Google Scholar] [CrossRef]
- Wang, C.; Wang, L. Multidirectional Ring Top-Hat Transformation for Infrared Small Target Detection. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 8077–8088. [Google Scholar] [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Advances in Neural Information Processing Systems 32; Wallach, H., Larochelle, H., Beygelzimer, A., Alché-Buc, F., Fox, E., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2019; pp. 8024–8035. [Google Scholar]
- Kashin, A.S.; Boiko, D.A.; Ananikov, V.P. Neural Network Analysis of Electron Microscopy Video Data Reveals the Temperature-Driven Microphase Dynamics in the Ions/Water System. Small 2021, 17, 2007726. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar] [CrossRef]
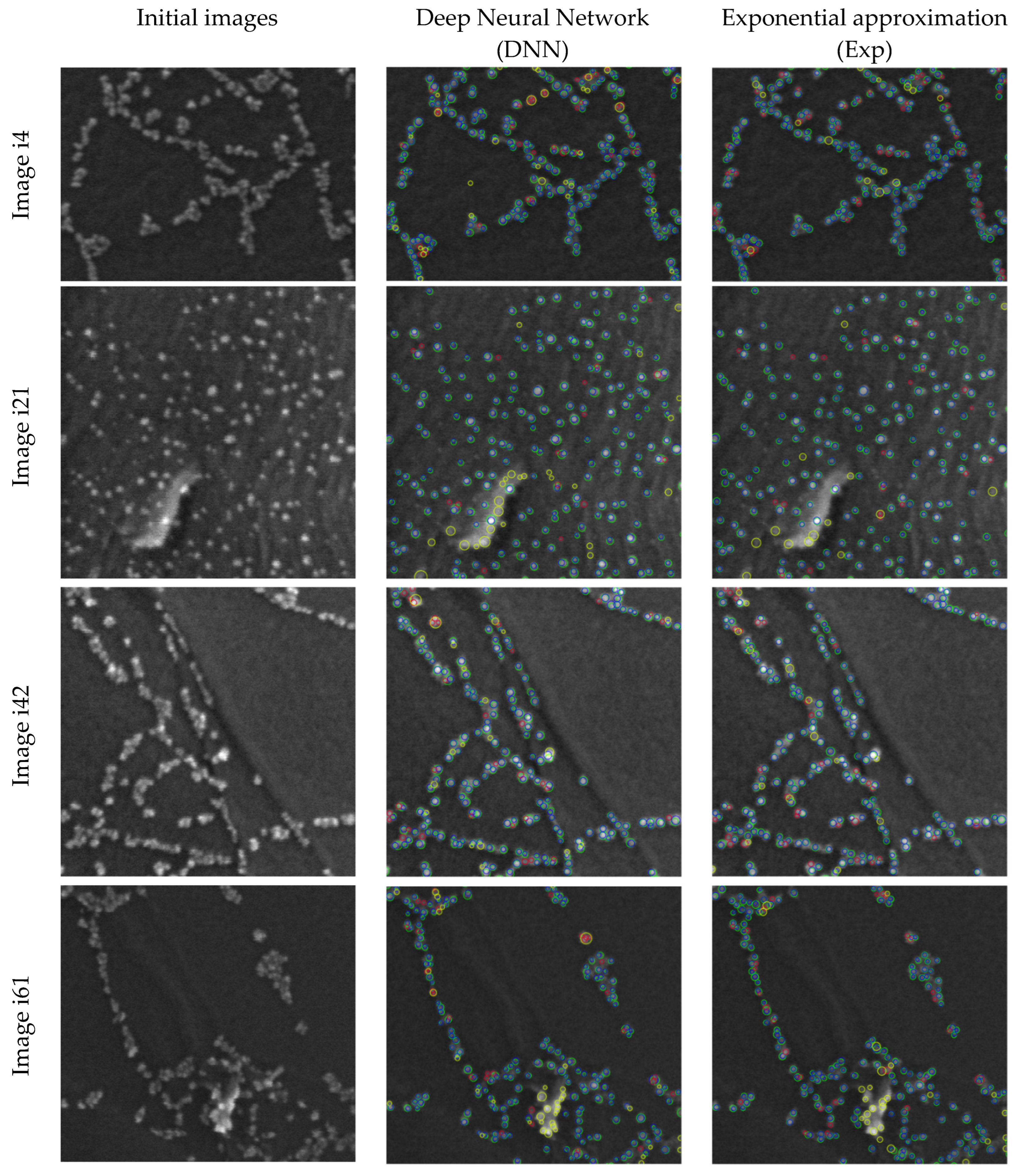
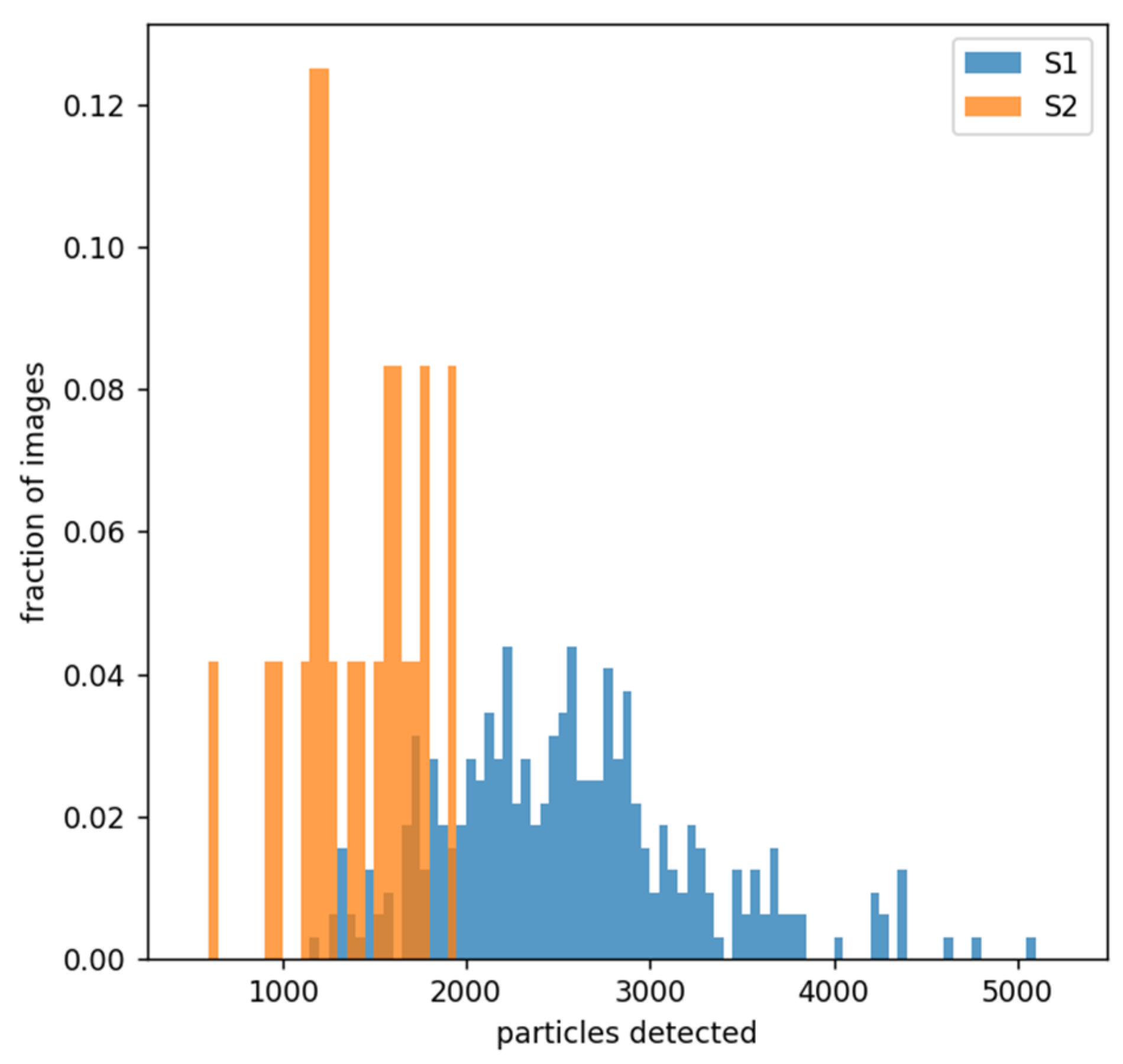
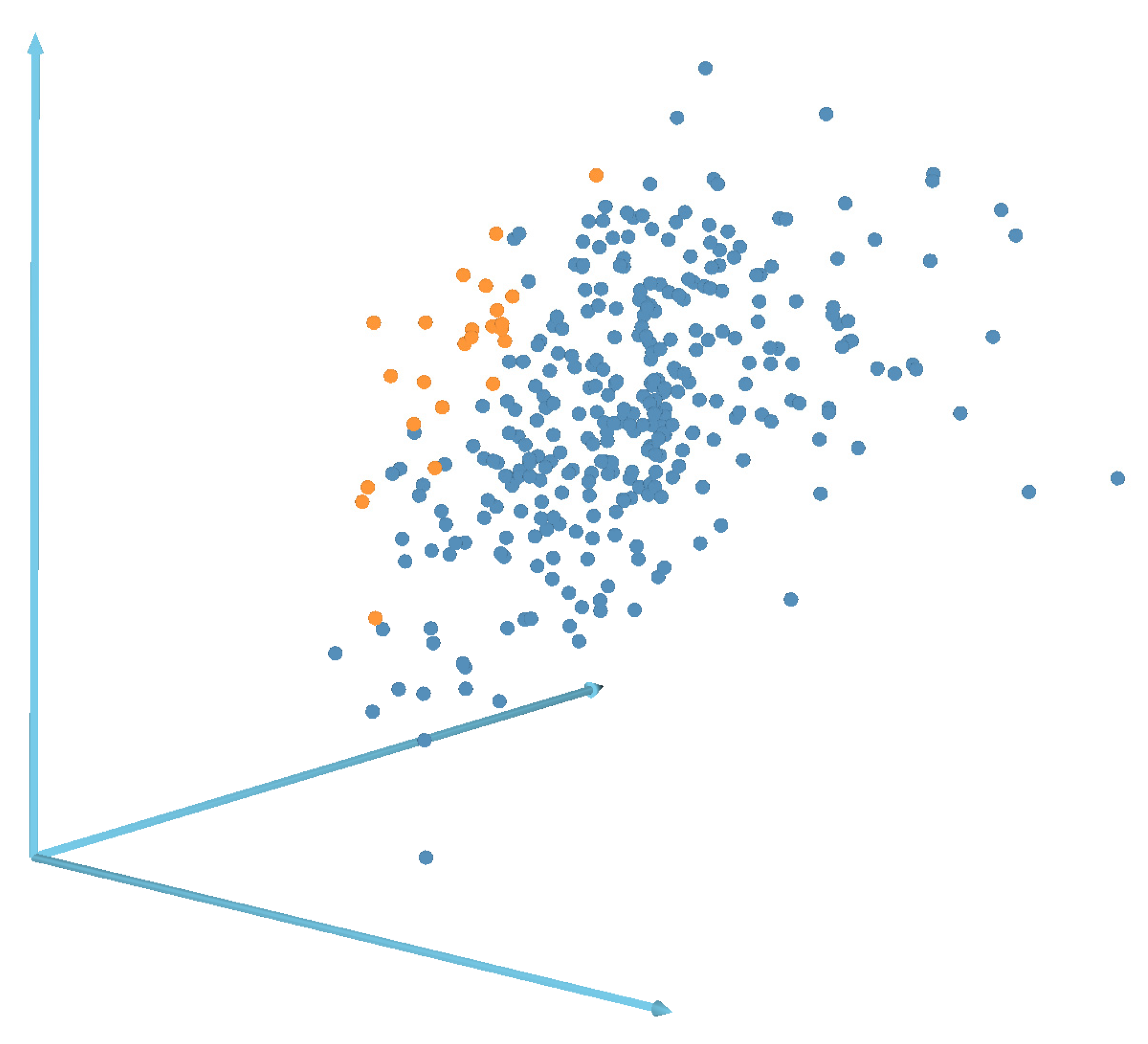
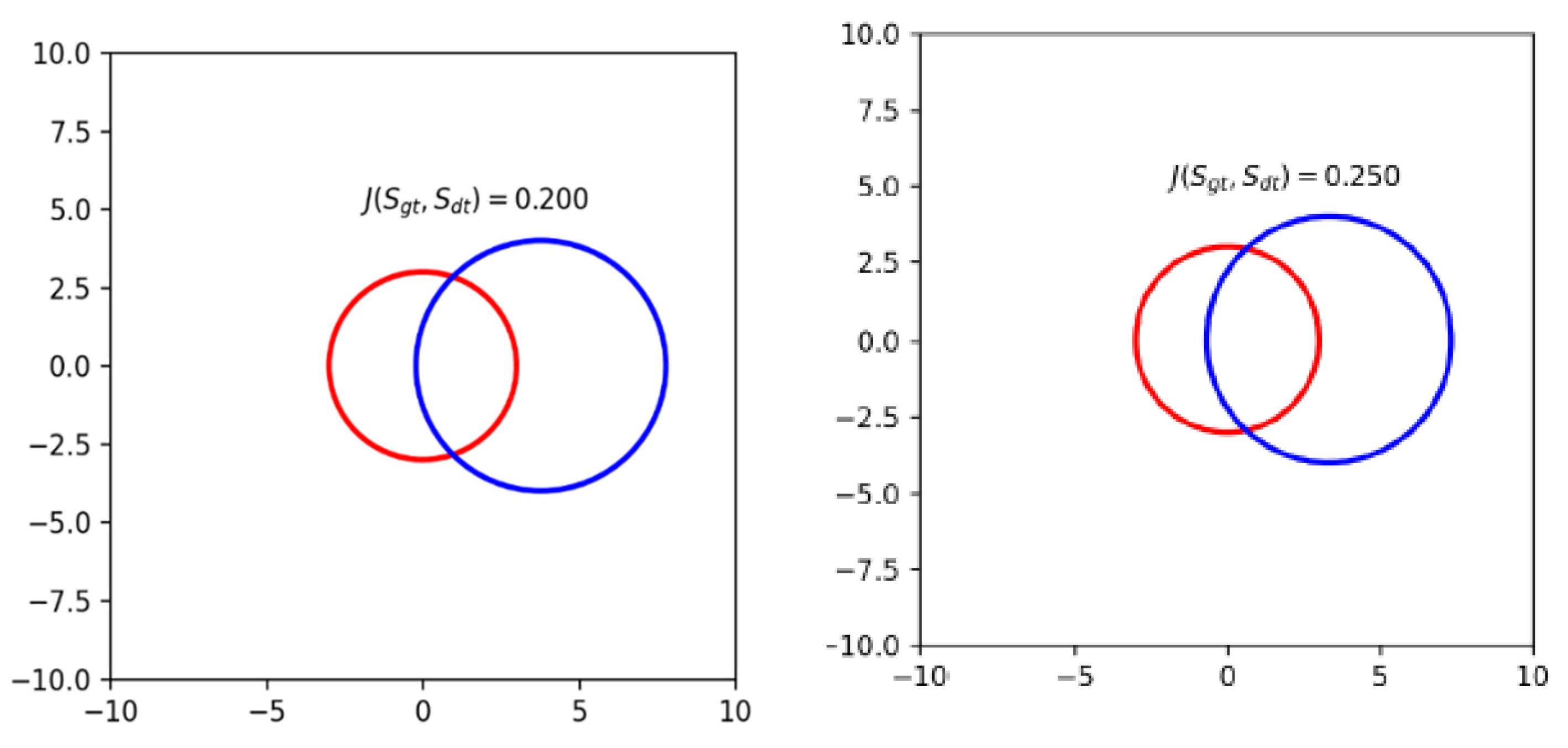
| Method | Image | Mean ± SD | |||
|---|---|---|---|---|---|
| i4 | i61 | i42 | i21 | ||
| LoG | 75.79 | 66.40 | 76.23 | 84.62 | 75.76 ± 6.45 |
| DoG | 78.42 | 72.81 | 79.15 | 82.43 | 78.20 ± 3.46 |
| DoH | 57.93 | 57.38 | 62.92 | 81.70 | 64.98 ± 9.89 |
| LM | 73.90 | 67.52 | 69.92 | 79.46 | 72.70 ± 4.52 |
| Exp | 75.52 | 74.77 | 77.42 | 84.30 | 78.00 ± 3.76 |
| DNN | 73.15 | 69.49 | 76.13 | 82.13 | 75.23 ± 4.63 |
| Method | Images for Finding Parameters/Test Image | Mean ± SD | |||
|---|---|---|---|---|---|
| i21, i42, i64/i4 | i21, i42, i4/i61 | i21, i4, i64/i42 | i4, i42, i64/i21 | ||
| LoG | 58.44 | 64.47 | 70.52 | 80.30 | 68.43 ± 8.07 |
| DoG | 64.98 | 69.06 | 75.55 | 70.18 | 69.94 ± 8.36 |
| DoH | 39.08 | 47.71 | 48.33 | 70.82 | 51.48 ± 11.75 |
| LM | 73.90 | 67.52 | 69.92 | 73.33 | 71.16 ± 2.60 |
| Exp | 73.75 | 72.93 | 75.58 | 83.48 | 76.44 ± 4.18 |
| DNN | 69.88 | 67.95 | 76.09 | 79.59 | 73.38 ± 4.68 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boiko, D.A.; Sulimova, V.V.; Kurbakov, M.Y.; Kopylov, A.V.; Seredin, O.S.; Cherepanova, V.A.; Pentsak, E.O.; Ananikov, V.P. Automated Recognition of Nanoparticles in Electron Microscopy Images of Nanoscale Palladium Catalysts. Nanomaterials 2022, 12, 3914. https://doi.org/10.3390/nano12213914
Boiko DA, Sulimova VV, Kurbakov MY, Kopylov AV, Seredin OS, Cherepanova VA, Pentsak EO, Ananikov VP. Automated Recognition of Nanoparticles in Electron Microscopy Images of Nanoscale Palladium Catalysts. Nanomaterials. 2022; 12(21):3914. https://doi.org/10.3390/nano12213914
Chicago/Turabian StyleBoiko, Daniil A., Valentina V. Sulimova, Mikhail Yu. Kurbakov, Andrei V. Kopylov, Oleg S. Seredin, Vera A. Cherepanova, Evgeniy O. Pentsak, and Valentine P. Ananikov. 2022. "Automated Recognition of Nanoparticles in Electron Microscopy Images of Nanoscale Palladium Catalysts" Nanomaterials 12, no. 21: 3914. https://doi.org/10.3390/nano12213914
APA StyleBoiko, D. A., Sulimova, V. V., Kurbakov, M. Y., Kopylov, A. V., Seredin, O. S., Cherepanova, V. A., Pentsak, E. O., & Ananikov, V. P. (2022). Automated Recognition of Nanoparticles in Electron Microscopy Images of Nanoscale Palladium Catalysts. Nanomaterials, 12(21), 3914. https://doi.org/10.3390/nano12213914










