Application of SERS in the Detection of Fungi, Bacteria and Viruses
Abstract
1. Introduction
2. Advances of SERS in Fungi, Bacteria and Viruses
2.1. SERS Spectroscopy for Fungal Identification and Detection
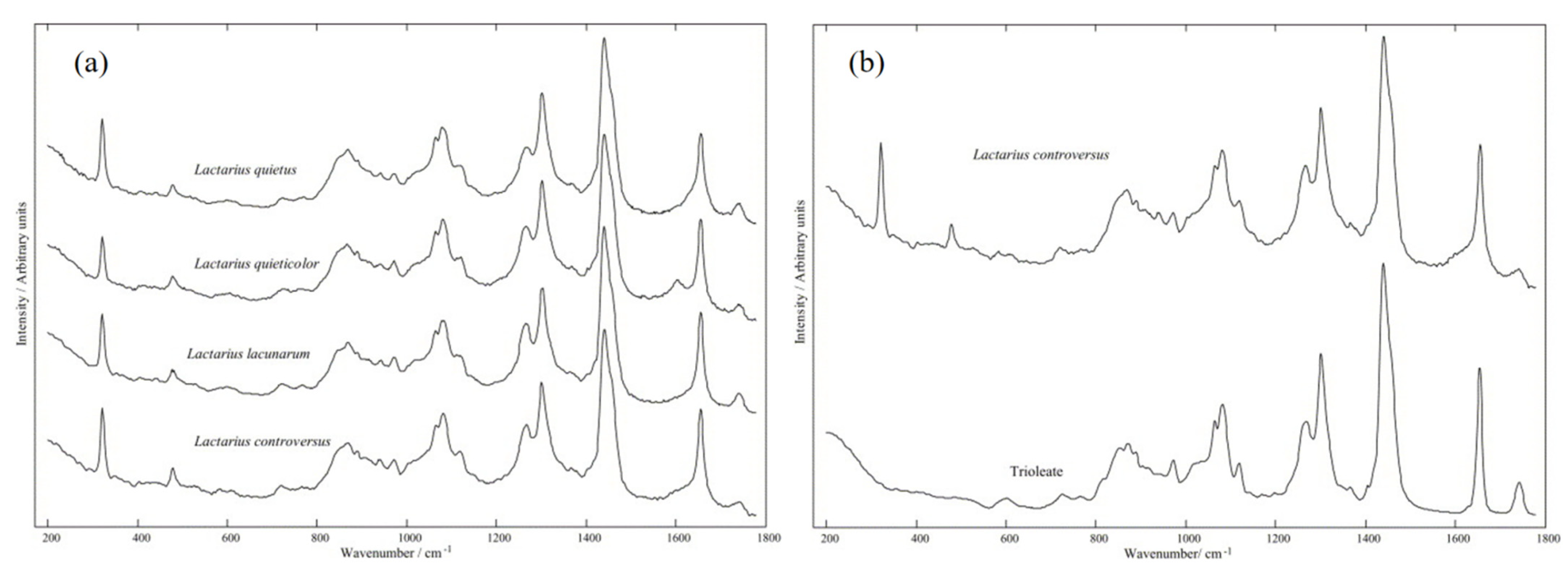
2.1.1. Feasibility Analysis of Raman/SERS Spectrum Analysis and Fungal Detection
2.1.2. Identification of Fungi Based on SERS Substrates of Various Nanostructures
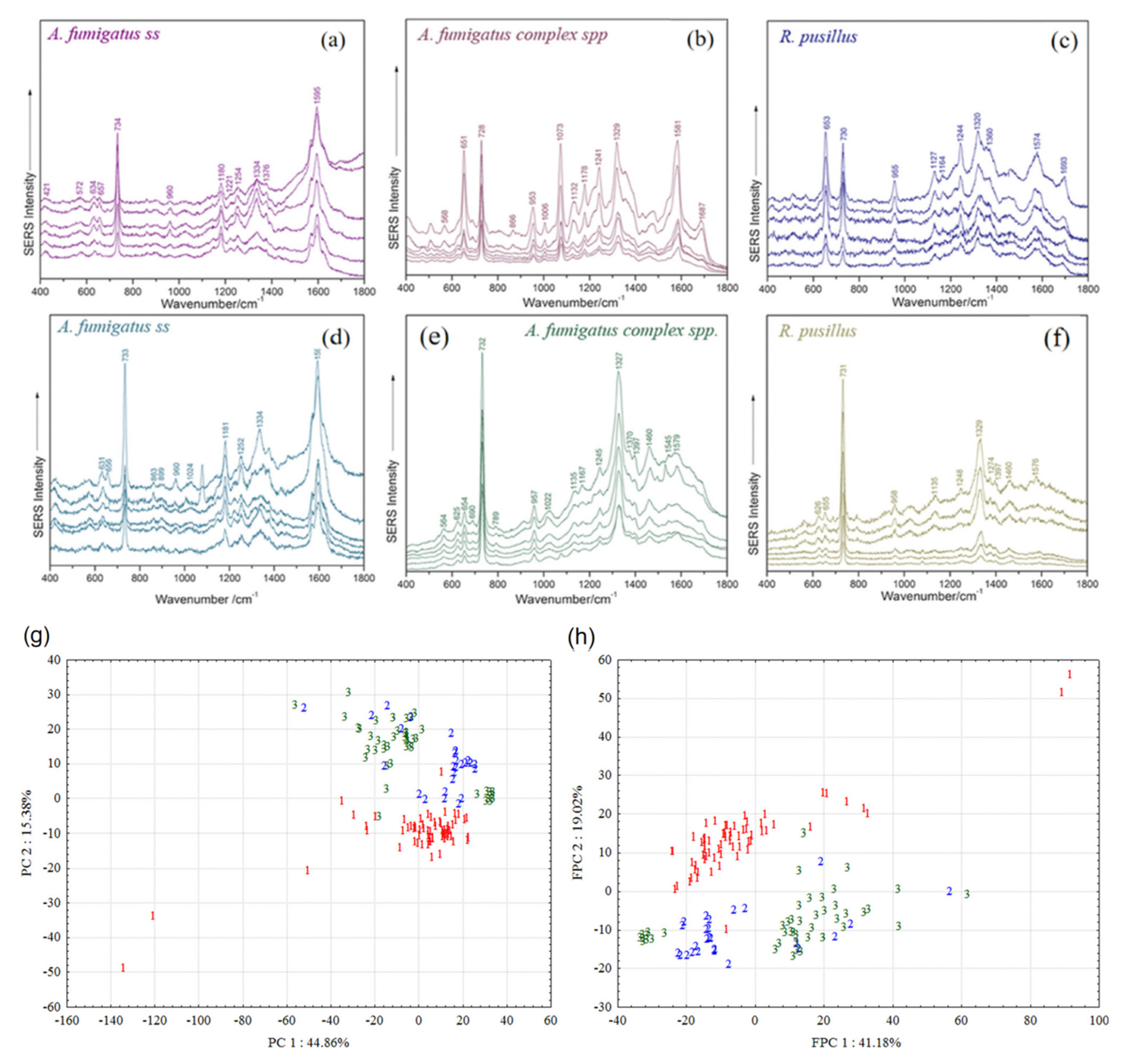
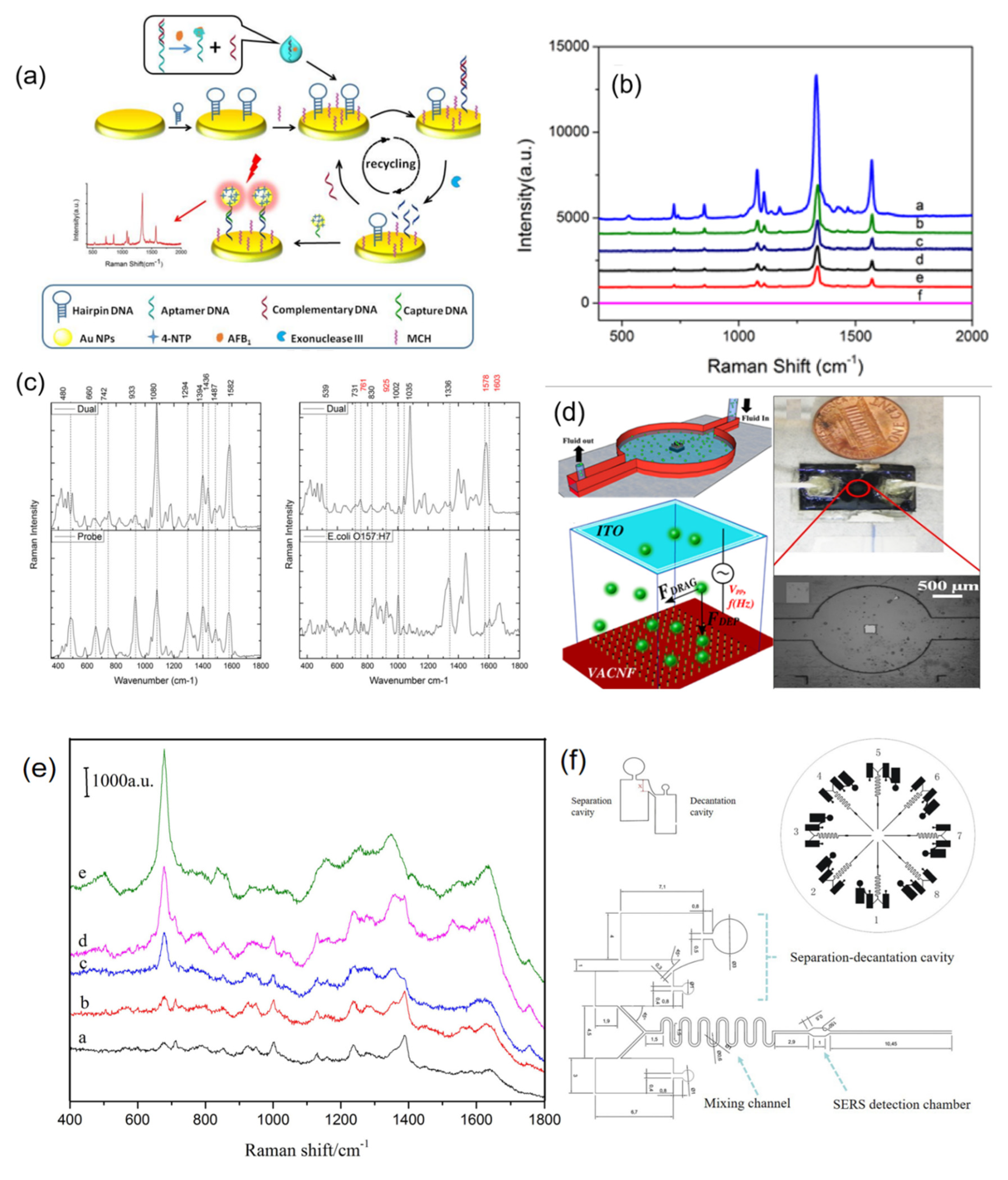
2.1.3. Efficient Quantitative Detection of Fungi Based on SERS Tag Signal Amplification Mechanism
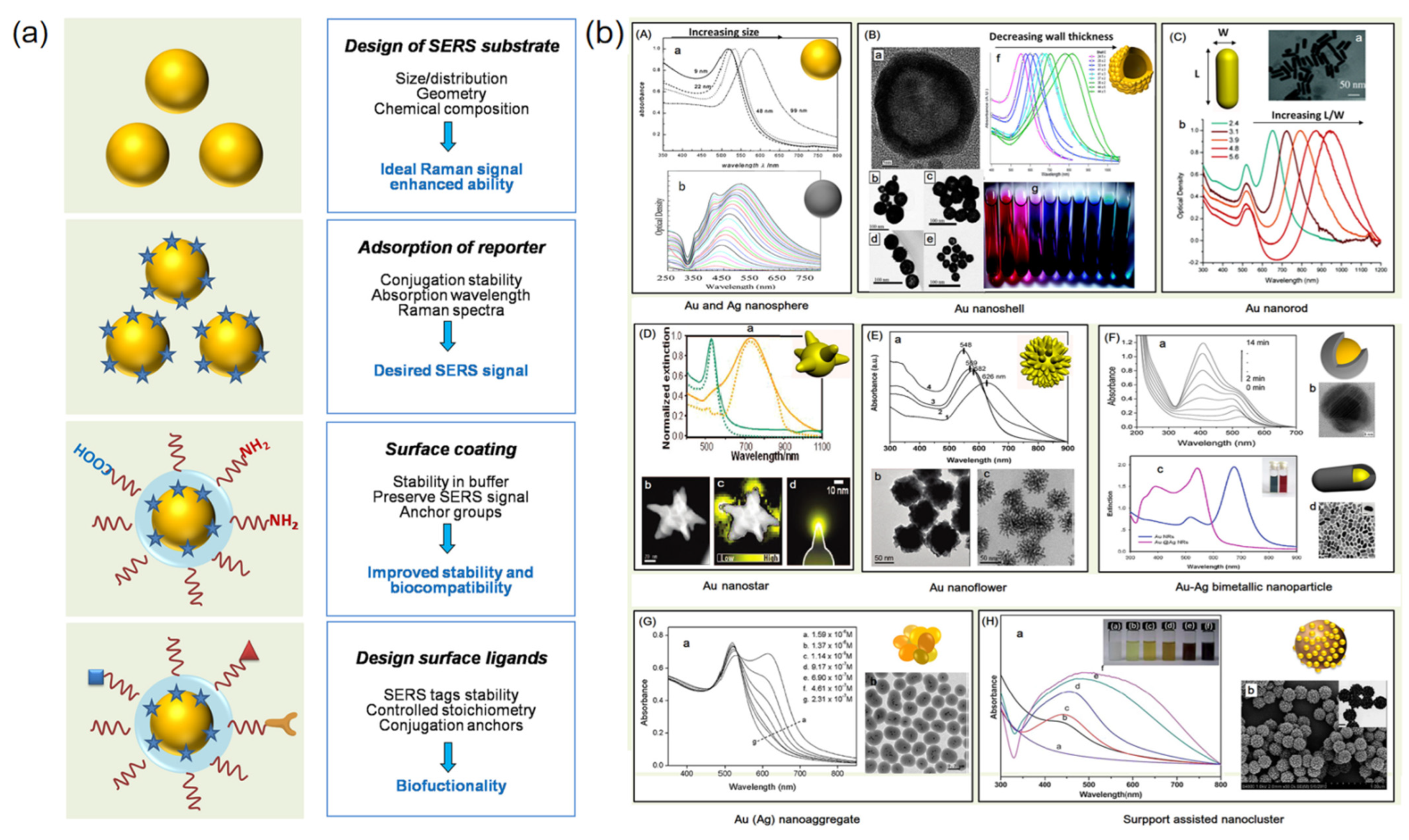
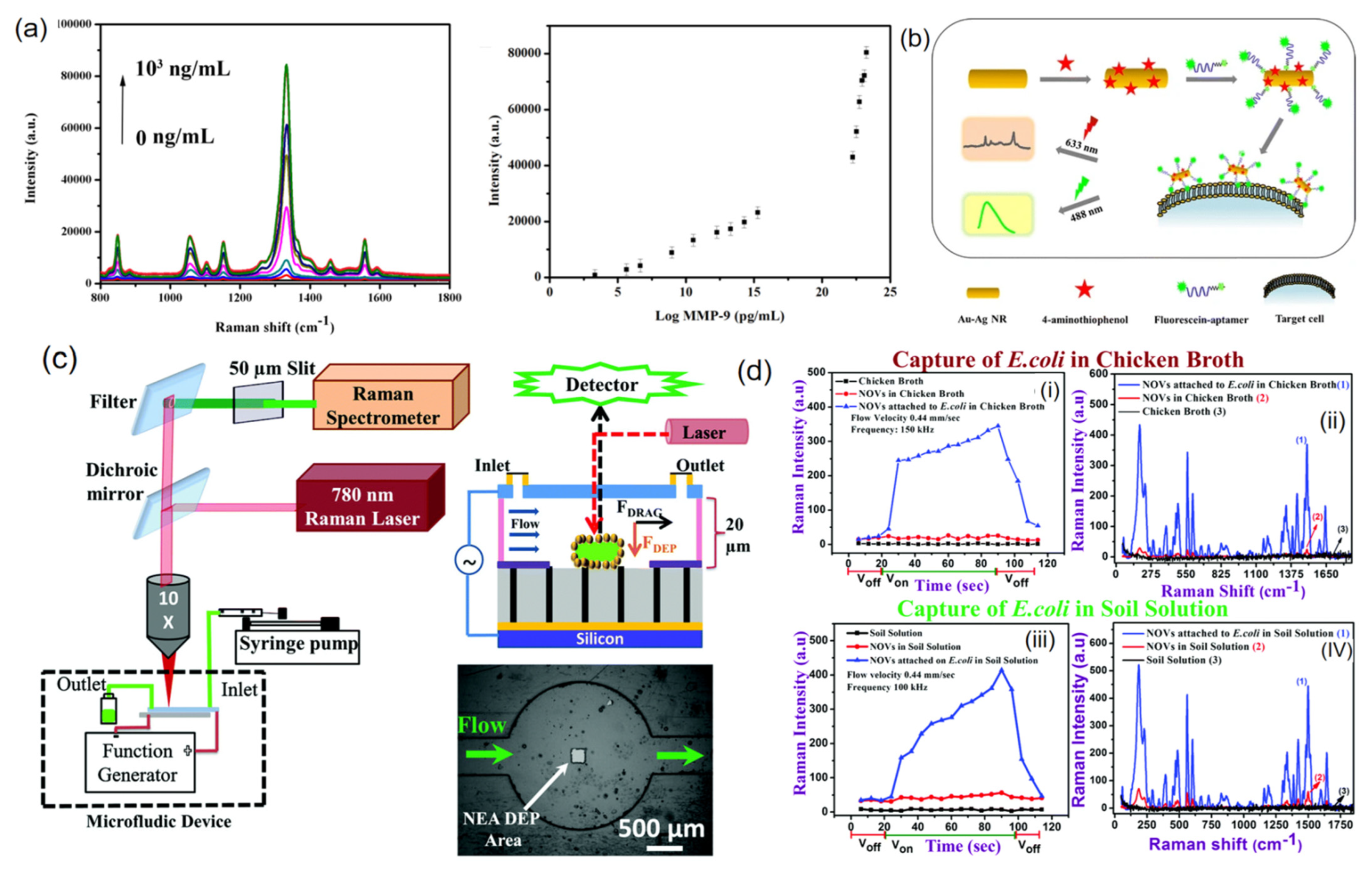
2.1.4. Efficient Detection of Fungi on Microfluidic Chip Based on Integrated SERS
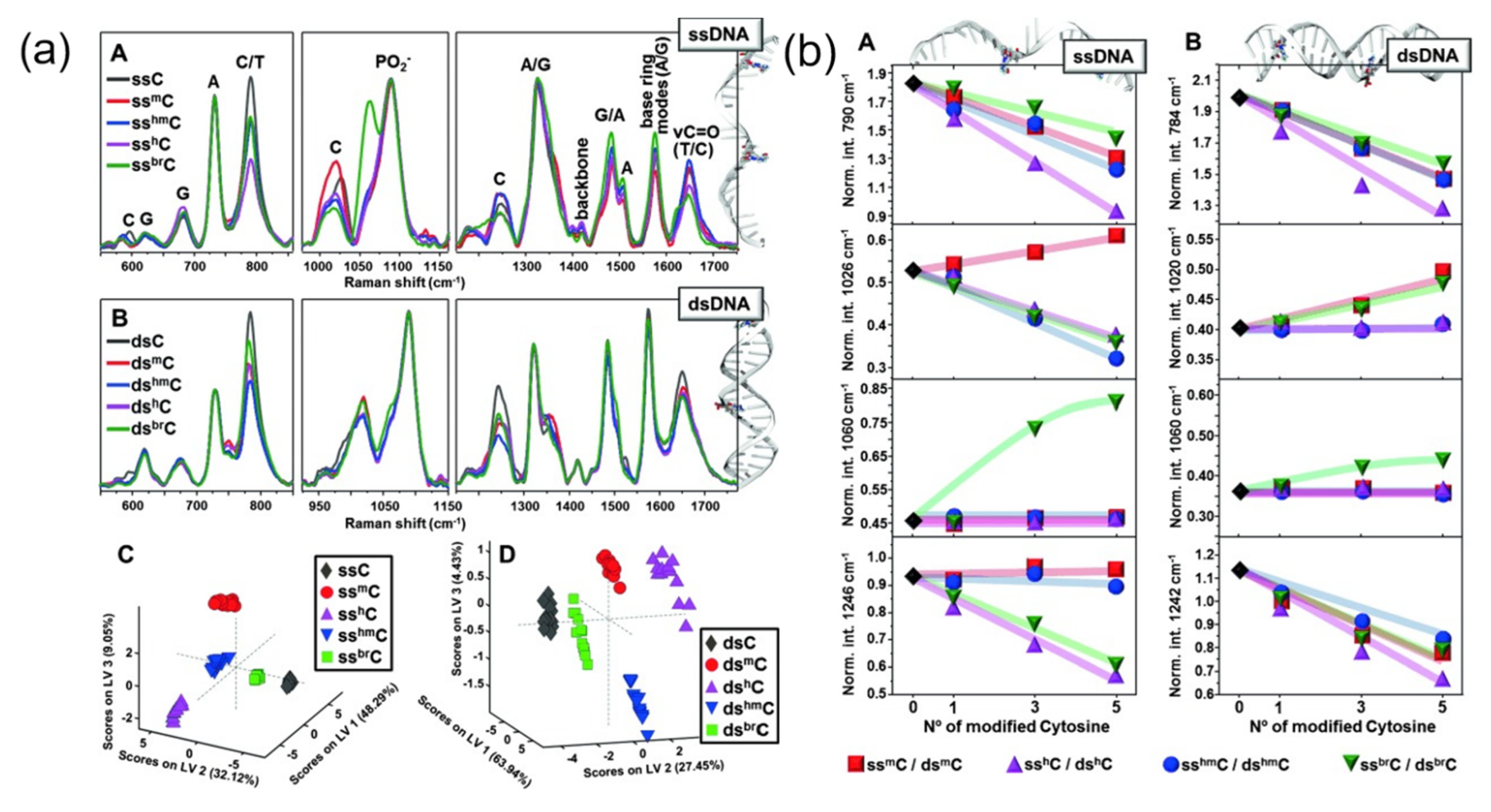
2.2. SERS Spectra Were Used for Nucleic Acid Identification and Detection
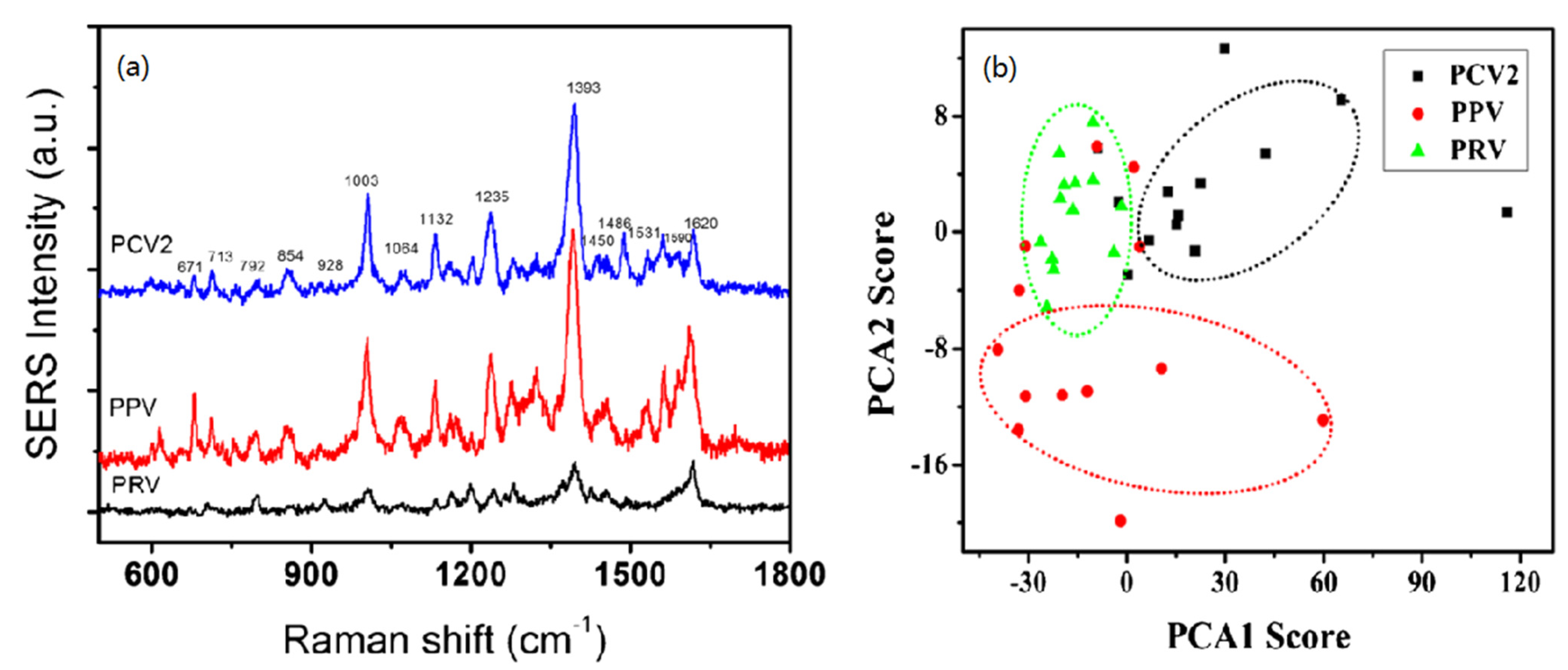
2.3. SERS Spectra Were Used for Pathogens Identification and Detection
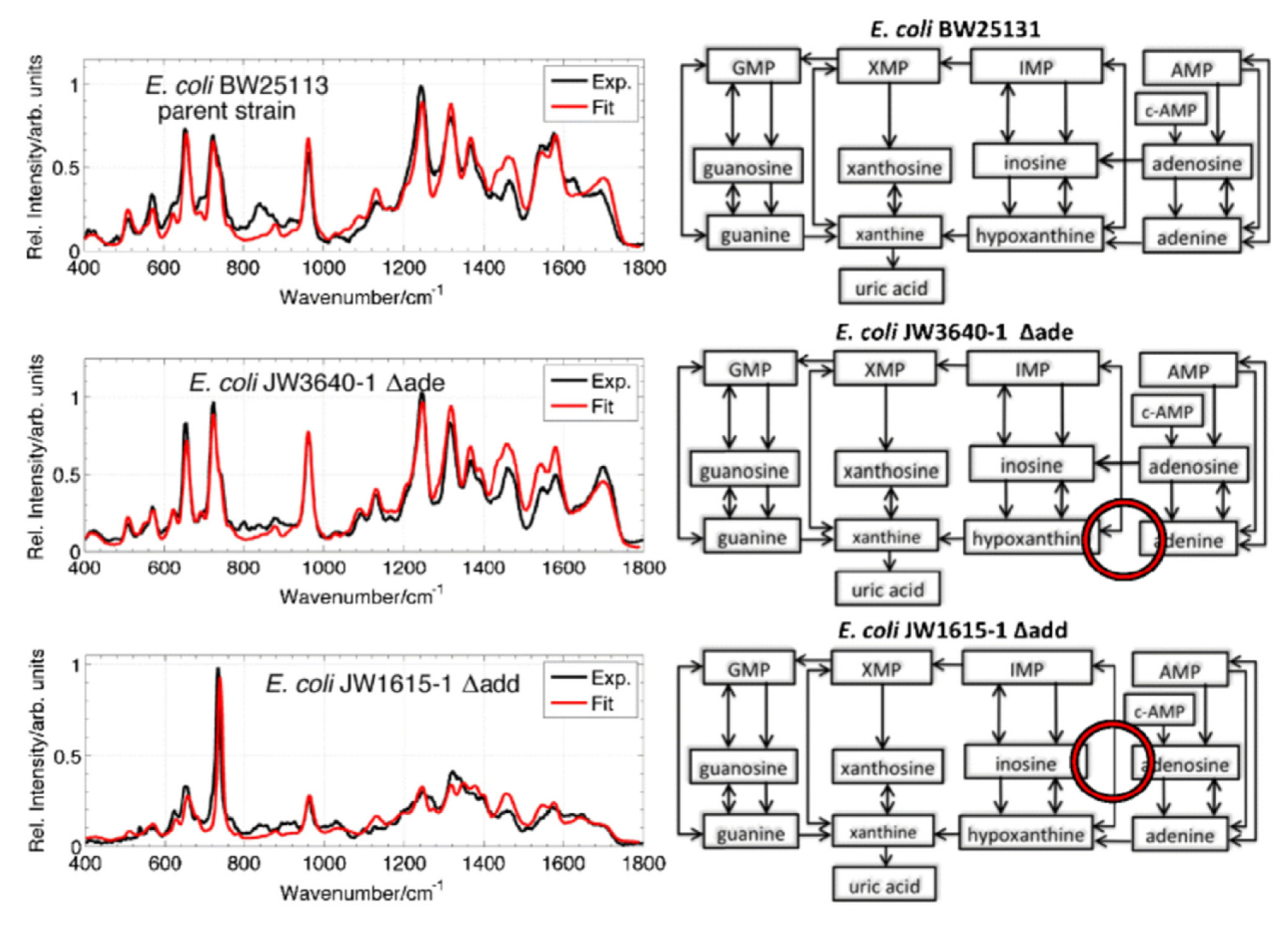
2.4. SERS-Based Microdevices for the Detection of Viruses and Bacteria
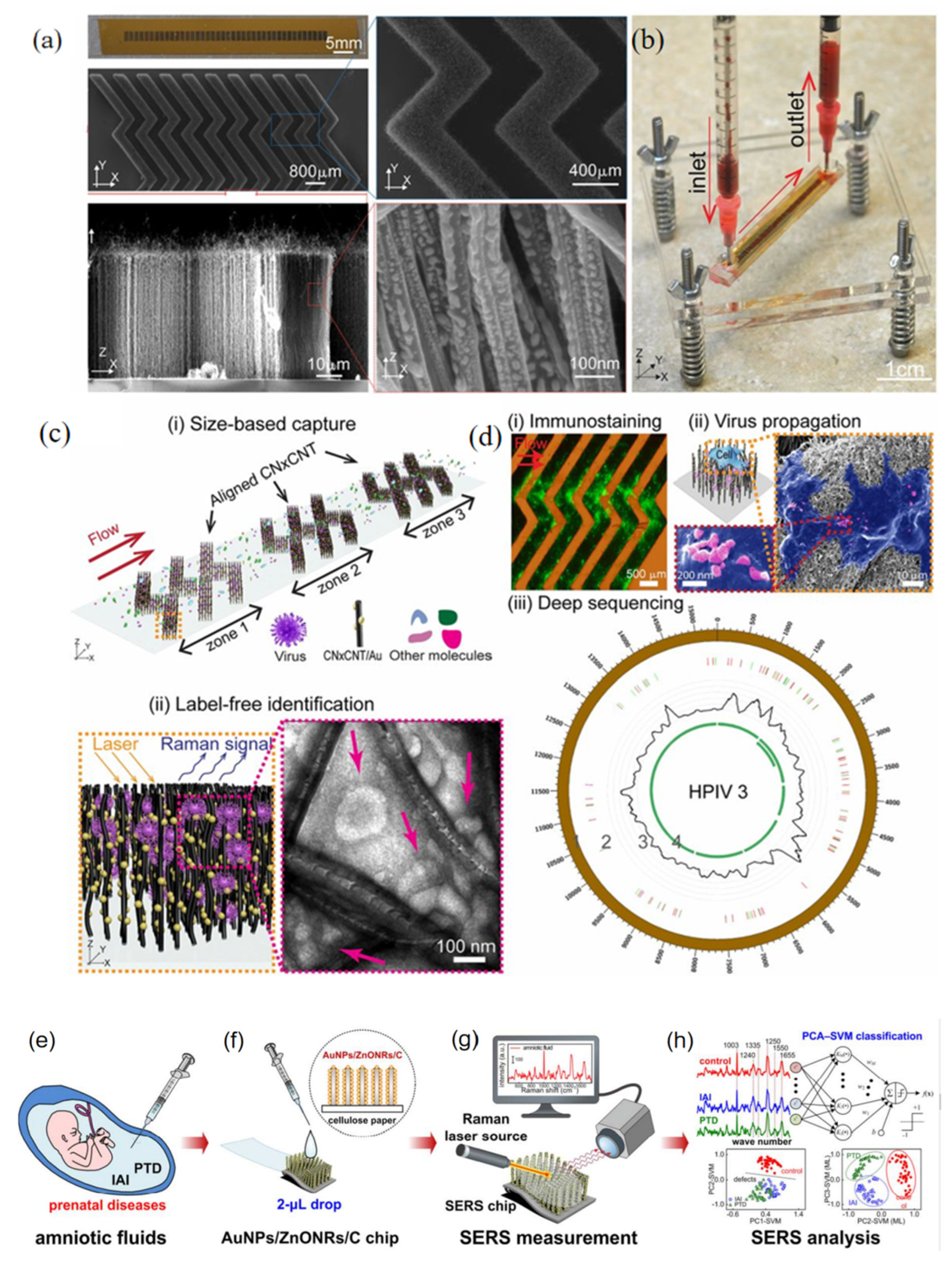
2.4.1. SERS-Based Microfluidic Devices
2.4.2. SERS-Based Three-Dimensional Nanostructured Plasmonic Substrates
3. Conclusions and Prospects
Author Contributions
Funding
Conflicts of Interest
References
- Zeng, L.; Sen, X. Application and technique advances of Raman spectroscopy in human tissues. Chin. J. Laser Med. Surg. 2011, 20, 177–182. [Google Scholar]
- Fleischmann, M.; Hendra, P.J.; McQuillan, A.J. Raman spectra of pyridine adsorbed at a silver electrode. Chem. Phys. Lett. 1974, 26, 163–166. [Google Scholar] [CrossRef]
- Albrecht, M.G.; Creighton, J.A. Anomalously intense Raman spectra of pyridine at a silver electrode. J. Am. Chem. Soc. 1977, 99, 5215–5217. [Google Scholar] [CrossRef]
- Fan, M.; Andrade, G.F.; Brolo, A.G. A review on recent advances in the applications of surface-enhanced Raman scattering in analytical chemistry. Anal. Chim. Acta 2020, 1097, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, C. Fast, portable tests come online to curb coronavirus pandemic. Nat. Biotechnol. 2020, 38, 515–518. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Zhou, H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020, 165, 112349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Wang, L.; Deng, X.; Liang, R.; Su, M.; He, C.; Hu, L.; Su, Y.; Ren, J.; Yu, F.; et al. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020, 92, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay techniques and test development for COVID-19 diagnosis. ACS Cent. Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Tong, J.; Liu, M.; Shen, Y.; Guo, D. Evaluation of coronavirus in tears and conjunctival secretions of patients with SARS-CoV-2 infection. J. Med. Virol. 2020, 92, 589–594. [Google Scholar] [CrossRef]
- Lee, T.; Mohammadniaei, M.; Zhang, H.; Yoon, J.; Choi, H.K.; Guo, S.; Guo, P.; Choi, J.W. Single Functionalized pRNA/Gold Nanoparticle for Ultrasensitive MicroRNA Detection Using Electrochemical Surface-Enhanced Raman Spectroscopy. Adv. Sci. 2020, 7, 1902477. [Google Scholar] [CrossRef]
- Wu, Y.; Choi, N.; Chen, H.; Dang, H.; Chen, L.; Choo, J. Performance evaluation of surface-enhanced Raman scattering-polymerase chain reaction sensors for future use in sensitive genetic assays. Anal. Chem. 2020, 92, 2628–2634. [Google Scholar] [CrossRef]
- Hwang, J.; Lee, S.; Choo, J. Application of a SERS-based lateral flow immunoassay strip for the rapid and sensitive detection of staphylococcal enterotoxin B. Nanoscale 2016, 8, 11418–11425. [Google Scholar] [CrossRef]
- Cardinal, M.F.; Vander Ende, E.; Hackler, R.A.; McAnally, M.O.; Stair, P.C.; Schatz, G.C.; Van Duyne, R.P. Expanding applications of SERS through versatile nanomaterials engineering. Chem. Soc. Rev. 2017, 46, 3886–3903. [Google Scholar] [CrossRef]
- Kelly, J.; Patrick, R.; Patrick, S.; Bell, S.E. Surface-Enhanced Raman Spectroscopy for the Detection of a Metabolic Product in the Headspace Above Live Bacterial Cultures. Angew. Chem. Int. Ed. 2018, 57, 15686–15690. [Google Scholar] [CrossRef]
- Bruzas, I.; Brinson, B.E.; Gorunmez, Z.; Lum, W.; Ringe, E.; Sagle, L. Surface-enhanced Raman spectroscopy of fluid-supported lipid bilayers. ACS Appl. Mater. Interfaces 2019, 11, 33442–33451. [Google Scholar] [CrossRef]
- Prinz, J.; Matković, A.; Pešić, J.; Gajić, R.; Bald, I. Hybrid structures for surface-enhanced raman scattering: DNA origami/gold nanoparticle dimer/graphene. Small 2016, 12, 5458–5467. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.; Choo, J.; Shin, S.Y.; Lee, Y.H.; Choi, H.Y.; Ha, S.; Kang, K.; Oh, C.H. Biological imaging of HEK293 cells expressing PLCγ1 using surface-enhanced Raman microscopy. Anal. Chem. 2007, 79, 916–922. [Google Scholar] [CrossRef]
- Wee, E.J.; Wang, Y.; Tsao, S.C.-H.; Trau, M. Simple, sensitive and accurate multiplex detection of clinically important melanoma DNA mutations in circulating tumour DNA with SERS nanotags. Theranostics 2016, 6, 1506. [Google Scholar] [CrossRef]
- Shanmukh, S.; Jones, L.; Driskell, J.; Zhao, Y.; Dluhy, R.; Tripp, R.A. Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Lett. 2006, 6, 2630–2636. [Google Scholar] [CrossRef]
- Galvan, D.D.; Yu, Q. Surface-Enhanced Raman Scattering for Rapid Detection and Characterization of Antibiotic-Resistant Bacteria. Adv. Healthc. Mater. 2018, 7, 1701335. [Google Scholar] [CrossRef]
- Jamil, S.; Jamil, N.; Saad, U.; Hafiz, S.; Siddiqui, S. Frequency of Candida albicans in Patients with Funguria. J. Coll. Physicians Surg. Pak. 2016, 26, 113–116. [Google Scholar]
- Tsedaley, B. A review paper on Potato virus Y (PVY) biology, economic importance and its managements. J. Biol. Agric. Healthc. 2015, 5, 110–126. [Google Scholar]
- Sexton, D.J.; Kordalewska, M.; Bentz, M.L.; Welsh, R.M.; Perlin, D.S.; Litvintseva, A.P. Direct detection of emergent fungal pathogen Candida auris in clinical skin swabs by SYBR green-based quantitative PCR assay. J. Clin. Microbiol. 2018, 56, e01337-18. [Google Scholar] [CrossRef]
- Edwards, H.G.M.; Russell, N.C.; Weinstein, R.; Wynnwilliams, D.D. Fourier transform Raman spectroscopic study of fungi. J. Raman Spectrosc. 1995, 26, 911–916. [Google Scholar] [CrossRef]
- De Gussem, K.; Vandenabeele, P.; Verbeken, A.; Moens, L. Raman spectroscopic study of Lactarius spores (Russulales, Fungi). Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2005, 61, 2896–2908. [Google Scholar] [CrossRef]
- Chen, D.; Zhang, Y.; Bessho, T.; Kudo, T.; Sang, J.; Hirahara, H.; Mori, K.; Kang, Z. Formation of reflective and conductive silver film on ABS surface via covalent grafting and solution spray. Appl. Surf. Sci. 2015, 349, 503–509. [Google Scholar] [CrossRef]
- Wang, R.; Xu, Y.; Wang, C.; Zhao, H.; Wang, R.; Liao, X.; Chen, L.; Chen, G. Fabrication of ITO-rGO/Ag NPs nanocomposite by two-step chronoamperometry electrodeposition and its characterization as SERS substrate. Appl. Surf. Sci. 2015, 349, 805–810. [Google Scholar] [CrossRef]
- Xiang, S.; Xu, Y.; Liao, X.; Zheng, X.; Chen, L.; Li, S. Dynamic Monitoring of the Oxidation Process of Phosphatidylcholine Using SERS Analysis. Anal. Chem. 2018, 90, 13751–13758. [Google Scholar] [CrossRef]
- Su, X.; Xu, Y.; Zhao, H.; Li, S.; Chen, L. Design and preparation of centrifugal microfluidic chip integrated with SERS detection for rapid diagnostics. Talanta 2019, 194, 903–909. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.; Xue, B.; Kong, X.; Liu, X.; Tu, L.; Chang, Y.; Zhang, H. A SERS nano-tag-based fiber-optic strategy for in situ immunoassay in unprocessed whole blood. Biosens. Bioelectron. 2017, 92, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Meng, G.; Chen, B.; Zhu, C.; Han, F.; Hu, X.; Wang, X.; Zhulin, H.; Guowen, M.; Bin, C.; et al. Surface-enhanced Raman scattering from Au-nanorod arrays with sub-5-nm gaps stuck out of an AAO template. J. Nanosci. Nanotechnol. 2016, 16, 934–938. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Chua, J.; Tan, E.K.M.; Kah, J.C.Y. Optimizing the SERS enhancement of a facile gold nanostar immobilized paper-based SERS substrate. RSC Adv. 2017, 7, 16264–16272. [Google Scholar] [CrossRef]
- Luo, Z.; Chen, L.; Liang, C.; Wei, Q.; Chen, Y.; Wang, J. Porous carbon films decorated with silver nanoparticles as a sensitive SERS substrate, and their application to virus identification. Microchim. Acta 2017, 184, 3505–3511. [Google Scholar] [CrossRef]
- Mabbott, S.; Thompson, D.; Sirimuthu, N.; Mcnay, G.; Faulds, K.; Graham, D. From synthetic DNA to PCR product: Detection of fungal infections using SERS. Faraday Discuss. 2016, 187, 461–472. [Google Scholar] [CrossRef]
- Sivanesan, A.; Witkowska, E.; Adamkiewicz, W.; Dziewit, Ł.; Kamińska, A.; Waluk, J. Nanostructured silver-gold bimetallic SERS substrates for selective identification of bacteria in human blood. Analyst 2014, 139, 1037–1043. [Google Scholar] [CrossRef]
- Dina, N.E.; Gherman, A.; Chis, V.; Sarbu, C.; Wieser, A.; Bauer, D.; Haisch, C. Characterization of clinically relevant fungi via SERS fingerprinting assisted by novel chemometric models. Anal. Chem. 2018, 90, 2484–2492. [Google Scholar] [CrossRef]
- Prusinkiewicz, M.A.; Farazkhorasani, F.; Dynes, J.J.; Jian, W.; Kaminskyj, S. Proof-of-principle for SERS imaging of Aspergillus nidulans hyphae using in vivo synthesis of gold nanoparticles. Analyst 2012, 137, 4934–4942. [Google Scholar] [CrossRef]
- Wang, G.i.; Wang, Z.; Chen, Z.i.; Chen, L. Rate equation of gelation of chromium(III)-polyacrylamide sol. Chin. J. Chem. 1995, 13, 97–104. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, B.; Chen, L. SERS tags: Novel optical nanoprobes for bioanalysis. Chem. Rev. 2013, 113, 1391–1428. [Google Scholar] [CrossRef]
- Koydemir, H.C.; Ozcan, A. Wearable and Implantable Sensors for Biomedical Applications. Annu. Rev. Anal. Chem. 2018, 11, 127–146. [Google Scholar] [CrossRef]
- Chen, H.; Park, S.G.; Choi, N. SERS imaging-based aptasensor for ultrasensitive and reproducible detection of influenza virus A. Biosens. Bioelectron. 2020, 167, 112496. [Google Scholar] [CrossRef]
- Zhao, P.; Li, H.-X.; Li, D.-W.; Hou, Y.-J.; Mao, L.; Yang, M.; Wang, Y. A SERS nano-tag-based magnetic-separation strategy for highly sensitive immunoassay in unprocessed whole blood. Talanta 2019, 198, 527–533. [Google Scholar] [CrossRef]
- Bamrungsap, S.; Treetong, A.; Apiwat, C.; Wuttikhun, T.; Dharakul, T. SERS-fluorescence dual mode nanotags for cervical cancer detection using aptamers conjugated to gold-silver nanorods. Microchim. Acta 2016, 183, 249–256. [Google Scholar] [CrossRef]
- Madiyar, F.R.; Bhana, S.; Swisher, L.Z.; Culbertson, C.T.; Huang, X.; Li, J. Integration of a nanostructured dielectrophoretic device and a surface-enhanced Raman probe for highly sensitive rapid bacteria detection. Nanoscale 2015, 7, 3726–3736. [Google Scholar] [CrossRef]
- Lian, Y.; Hao, S.; Wang, X.; Qi, P.; Yang, P. Carved Nanoframes of Cobalt-Iron Bimetal Phosphide as a Bifunctional Electrocatalyst for Efficient Overall Water splitting. Chem. Sci. 2019, 10, 464–474. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, C.; Xiao, R.; Tang, L.; Huang, J.; Wu, D.; Liu, S.; Wang, Y.; Zhang, D.; Wang, S. Sensitive and specific detection of clinical bacteria via vancomycin-modified Fe3O4@Au nanoparticles and aptamer-functionalized SERS tags. J. Mater. Chem. B 2018, 6, 3751–3761. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Wan, N.; Shi, L.; Wang, C.; Sun, Z.; Xiao, R.; Wang, S. Dual-recognition surface-enhanced Raman scattering (SERS) biosensor for pathogenic bacteria detection by using vancomycin-SERS tags and aptamer-Fe3O4@Au. Anal. Chim. Acta 2019, 1077, 288–296. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Qiao, Y.; Zhang, H.; Zhao, J.; Li, W.; Xie, T.; Zhong, D.; Wei, Q.; Hua, S.; Yu, Y.; et al. Gold-silver nanoshells promote wound healing from drug-resistant bacteria infection and enable monitoring via surface-enhanced Raman scattering imaging. Biomaterials 2020, 234, 119763. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.-T.; Gulino, K.; Zhang, Y.; Sabestien, A.; Chou, T.-W.; Zhou, B.; Lin, Z.; Albert, I.; Lu, H.; Swaminathan, V. A rapid and label-free platform for virus capture and identification from clinical samples. Proc. Natl. Acad. Sci. USA 2020, 117, 895–901. [Google Scholar] [CrossRef]
- Li, Q.; Lu, Z.; Tan, X.; Xiao, X.; Wang, P.; Wu, L.; Shao, K.; Yin, W.; Han, H. Ultrasensitive detection of aflatoxin B1 by SERS aptasensor based on exonuclease-assisted recycling amplification. Biosens. Bioelectron. 2017, 97, 59–64. [Google Scholar] [CrossRef]
- Wang, L.; Li, P.C.; Yu, H.-Z.; Parameswaran, A.M. Fungal pathogenic nucleic acid detection achieved with a microfluidic microarray device. Anal. Chim. Acta 2008, 610, 97–104. [Google Scholar] [CrossRef]
- Wang, C.; Madiyar, F.; Yu, C.; Li, J. Detection of extremely low concentration waterborne pathogen using a multiplexing self-referencing SERS microfluidic biosensor. J. Biol. Eng. 2017, 11, 9. [Google Scholar] [CrossRef]
- Wang, C.; Xu, Y.; Deng, C.; Liu, Z.; Wang, R.; Zhao, H. Design and preparation of a recyclable microfluidic SERS chip with integrated Au@Ag/TiO2 NTs. RSC Adv. 2016, 6, 113115–113122. [Google Scholar] [CrossRef]
- Torres-Nuñez, A.; Faulds, K.; Graham, D.; Alvarez-Puebla, R.A.; Guerrini, L. Silver colloids as plasmonic substrates for direct label-free surface-enhanced Raman scattering analysis of DNA. Analyst 2016, 141, 5170. [Google Scholar] [CrossRef]
- Luo, S.C.; Sivashanmugan, K.; Liao, J.D. Nanofabricated SERS-active substrates for single-molecule to virus detection in vitro: A review. Biosens. Bioelectron. 2014, 61, 232–240. [Google Scholar] [CrossRef]
- Morla-Folch, J.; Xie, H.n.; Gisbert-Quilis, P.; Pedro, S.G.d.; Pazos-Perez, N.; Alvarez-Puebla, R.A.; Guerrini, L. Ultrasensitive direct quantification of nucleobase modifications in DNA by surface-enhanced Raman scattering: The case of cytosine. Angew. Chem. 2015, 127, 13854–13858. [Google Scholar] [CrossRef]
- Morla-Folch, J.; Alvarez-Puebla, R.A.; Guerrini, L. Direct Quantification of DNA Base Composition by Surface-Enhanced Raman Scattering Spectroscopy. J. Phys. Chem. Lett. 2016, 7, 3037–3041. [Google Scholar] [CrossRef]
- Miljani, S.A.; Ratkaj, M.; Matkovi, M.; Piantanida, I.; Gratteri, P.; Bazzicalupi, C. Assessment of human telomeric G-quadruplex structures using surface-enhanced Raman spectroscopy. Anal. Bioanal. Chem. 2017, 409, 2285–2295. [Google Scholar] [CrossRef]
- Papadopoulou, E.; Bell, S.E.J. Label-Free Detection of Nanomolar Unmodified Single- and Double-Stranded DNA by Using Surface-Enhanced Raman Spectroscopy on Ag and Au Colloids. Chem. A Eur. J. 2012, 18, 5394–5400. [Google Scholar] [CrossRef]
- Bell, S.; Dick, S. Quantitative Surface-enhanced Raman Spectroscopy of Single Bases in Oligodeoxynucleotides. Faraday Discuss. 2017, 205, 1012–1024. [Google Scholar]
- Yüksel, S.; Schwenkbier, L.; Pollok, S.; Weber, K.; Cialla-May, D.; Popp, J. Label-free detection of Phytophthora ramorum using surface-enhanced Raman spectroscopy. Analyst 2015, 140, 7254–7262. [Google Scholar] [CrossRef]
- Meneghello, M.; Papadopoulou, E.; Ugo, P.; Bartlett, P.N. Using Electrochemical SERS to Measure the Redox Potential of Drug Molecules Bound to dsDNA—A Study of Mitoxantrone. Electrochim. Acta 2016, 187, 684–692. [Google Scholar] [CrossRef][Green Version]
- Masetti, M.; Xie, H.N.; Krpetić, Ž.; Recanatini, M.; Alvarez-Puebla, R.A.; Guerrini, L. Revealing DNA Interactions with Exogenous Agents by Surface-Enhanced Raman Scattering. J. Am. Chem. Soc. 2015, 137, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Huang, Q.; Meng, G.; Lu, Y. Highly Sensitive and Selective Surface-Enhanced Raman Spectroscopy Label-free Detection of 3,3,4,4-Tetrachlorobiphenyl Using DNA Aptamer-Modified Ag-Nanorod Arrays. ACS Appl. Mater. Interfaces 2016, 8, 5723–5728. [Google Scholar] [CrossRef]
- Morla-Folch, J.; Xie, H.N.; Alvarez-Puebla, R.A.; Guerrini, L. Fast Optical Chemical and Structural Classification of RNA. ACS Nano 2016, 10, 2834. [Google Scholar] [CrossRef]
- Morla-Folc, J.; Gisbert-Quili, P.; Masett, M.; Garcia-Rico, E.; Alvarez-Puebl, R.A.; Guerrin, L. Innentitelbild: Conformational SERS Classification of K-Ras Point Mutations for Cancer Diagnostics (Angew. Chem. 9/2017). Angew. Chem. 2017, 129, 2256. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kim, B.-C.; Byeung-Keun, O.; Choi, J.-W. Rapid and sensitive determination of HIV-1 virus based on surface enhanced raman spectroscopy. J. Biomed. Nanotechnol. 2015, 11, 2223–2230. [Google Scholar] [CrossRef] [PubMed]
- Kamińska, A.; Witkowska, E.; Kowalska, A.; Skoczyńska, A.; Ronkiewicz, P.; Szymborski, T.; Waluk, J. Rapid detection and identification of bacterial meningitis pathogens in ex vivo clinical samples by SERS method and principal component analysis. Anal. Methods 2016, 8, 4521–4529. [Google Scholar] [CrossRef]
- Cheong, Y.; Kim, Y.J.; Kang, H.; Choi, S.; Lee, H.J. Rapid label-free identification of Klebsiella pneumoniae antibiotic resistant strains by the drop-coating deposition surface-enhanced Raman scattering method. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2017, 183, 53–59. [Google Scholar] [CrossRef]
- Yang, D.; Zhou, H.; Haisch, C.; Niessner, R.; Ying, Y. Reproducible E. coli detection based on label-free SERS and mapping. Talanta 2016, 146, 457–463. [Google Scholar] [CrossRef]
- Mircescu, N.E.; Zhou, H.; Leopold, N.; Chiş, V.; Ivleva, N.P.; Niessner, R.; Wieser, A.; Haisch, C. Towards a receptor-free immobilization and SERS detection of urinary tract infections causative pathogens. Anal. Bioanal. Chem. 2014, 406, 3051–3058. [Google Scholar] [CrossRef]
- Zhou, H.; Yang, D.; Mircescu, N.E.; Ivleva, N.P.; Schwarzmeier, K.; Wieser, A.; Schubert, S.; Niessner, R.; Haisch, C. Surface-enhanced Raman scattering detection of bacteria on microarrays at single cell levels using silver nanoparticles. Microchim. Acta 2015, 182, 2259–2266. [Google Scholar] [CrossRef]
- Zhou, H.; Yang, D.; Ivleva, N.P.; Mircescu, N.E.; Niessner, R.; Haisch, C. SERS detection of bacteria in water by in situ coating with Ag nanoparticles. Anal. Chem. 2014, 86, 1525–1533. [Google Scholar] [CrossRef]
- Dina, N.; Zhou, H.; Colniţă, A.; Leopold, N.; Szoke-Nagy, T.; Coman, C.; Haisch, C. Rapid single-cell detection and identification of pathogens by using surface-enhanced Raman spectroscopy. Analyst 2017, 142, 1782–1789. [Google Scholar] [CrossRef]
- Premasiri, W.R.; Lee, J.C.; Sauer-Budge, A.; Théberge, R.; Costello, C.E.; Ziegler, L.D. The biochemical origins of the surface-enhanced Raman spectra of bacteria: A metabolomics profiling by SERS. Anal. Bioanal. Chem. 2016, 408, 4631–4647. [Google Scholar] [CrossRef]
- Premasiri, W.; Chen, Y.; Williamson, P.; Bandarage, D.; Pyles, C.; Ziegler, L. Rapid urinary tract infection diagnostics by surface-enhanced Raman spectroscopy (SERS): Identification and antibiotic susceptibilities. Anal. Bioanal. Chem. 2017, 409, 3043–3054. [Google Scholar] [CrossRef]
- Wang, C.; Wang, J.; Li, M.; Qu, X.; Zhang, K.; Rong, Z.; Xiao, R.; Wang, S. A rapid SERS method for label-free bacteria detection using polyethylenimine-modified Au-coated magnetic microspheres and Au@Ag nanoparticles. Analyst 2016, 141, 6226–6238. [Google Scholar] [CrossRef]
- Muhlig, A.; Bocklitz, T.; Labugger, I.; Dees, S.; Henk, S.; Richter, E.; Andres, S.; Merker, M.; Stockel, S.; Weber, K.; et al. LOC-SERS: A promising closed system for the identification of mycobacteria. Anal. Chem. 2016, 88, 7998–8004. [Google Scholar] [CrossRef]
- Witkowska, E.; Korsak, D.; Kowalska, A.; Księżopolska-Gocalska, M.; Niedziółka-Jönsson, J.; Roźniecka, E.; Michałowicz, W.; Albrycht, P.; Podrażka, M.; Hołyst, R.; et al. Surface-enhanced Raman spectroscopy introduced into the International Standard Organization (ISO) regulations as an alternative method for detection and identification of pathogens in the food industry. Anal. Bioanal. Chem. 2017, 409, 1555–1567. [Google Scholar] [CrossRef]
- Boardman, A.K.; Wong, W.S.; Premasiri, W.R.; Ziegler, L.D.; Lee, J.C.; Miljkovic, M.; Klapperich, C.M.; Sharon, A.; Sauer-Budge, A.F. Rapid detection of bacteria from blood with surface-enhanced Raman spectroscopy. Anal. Chem. 2016, 88, 8026–8035. [Google Scholar] [CrossRef]
- He, X.; Zhou, X.; Liu, Y.; Wang, X. Ultrasensitive, recyclable and portable microfluidic surface-enhanced raman scattering (SERS) biosensor for uranyl ions detection. Sens. Actuators B Chem. 2020, 311, 127676. [Google Scholar] [CrossRef]
- Ko, J.; Park, S.-G.; Lee, S.; Wang, X.; Mun, C.; Kim, S.; Kim, D.-H.; Choo, J. Culture-free detection of bacterial pathogens on plasmonic nanopillar arrays using rapid Raman mapping. ACS Appl. Mater. Interfaces 2018, 10, 6831–6840. [Google Scholar] [CrossRef]
- Gao, R.; Cheng, Z.; Wang, X.; Yu, L.; Guo, Z.; Zhao, G.; Choo, J. Simultaneous immunoassays of dual prostate cancer markers using a SERS-based microdroplet channel. Biosens. Bioelectron. 2018, 119, 126–133. [Google Scholar] [CrossRef]
- Choi, N.; Lee, J.; Ko, J.; Jeon, J.H.; Rhie, G.-E.; de Mello, A.J.; Choo, J. Integrated SERS-based microdroplet platform for the automated immunoassay of F1 antigens in Yersinia pestis. Anal. Chem. 2017, 89, 8413–8420. [Google Scholar] [CrossRef]
- Wang, Y.; Ruan, Q.; Lei, Z.-C.; Lin, S.-C.; Zhu, Z.; Zhou, L.; Yang, C. Highly sensitive and automated surface enhanced raman scattering-based immunoassay for H5N1 detection with digital microfluidics. Anal. Chem. 2018, 90, 5224–5231. [Google Scholar] [CrossRef]
- Kamińska, A.; Witkowska, E.; Winkler, K.; Dzięcielewski, I.; Weyher, J.L.; Waluk, J. Detection of Hepatitis B virus antigen from human blood: SERS immunoassay in a microfluidic system. Biosens. Bioelectron. 2015, 66, 461–467. [Google Scholar] [CrossRef]
- Jeon, J.; Choi, N.; Chen, H.; Moon, J.-I.; Chen, L.; Choo, J. SERS-based droplet microfluidics for high-throughput gradient analysis. Lab Chip 2019, 19, 674–681. [Google Scholar] [CrossRef]
- Kim, W.; Lee, S.H.; Kim, J.H.; Ahn, Y.J.; Kim, Y.-H.; Yu, J.S.; Choi, S. Paper-Based surface-enhanced Raman spectroscopy for diagnosing prenatal diseases in women. ACS Nano 2018, 12, 7100–7108. [Google Scholar] [CrossRef]
- Wang, X.; Park, S.G.; Ko, J.; Xiao, X.; Giannini, V.; Maier, S.A.; Kim, D.H.; Choo, J. Sensitive and reproducible immunoassay of multiple mycotoxins using surface-enhanced Raman scattering mapping on 3D plasmonic nanopillar arrays. Small 2018, 14, 1801623. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, J.; Li, W.; Sun, M.; Wang, H. Application of SERS in the Detection of Fungi, Bacteria and Viruses. Nanomaterials 2022, 12, 3572. https://doi.org/10.3390/nano12203572
Xia J, Li W, Sun M, Wang H. Application of SERS in the Detection of Fungi, Bacteria and Viruses. Nanomaterials. 2022; 12(20):3572. https://doi.org/10.3390/nano12203572
Chicago/Turabian StyleXia, Jiarui, Wenwen Li, Mengtao Sun, and Huiting Wang. 2022. "Application of SERS in the Detection of Fungi, Bacteria and Viruses" Nanomaterials 12, no. 20: 3572. https://doi.org/10.3390/nano12203572
APA StyleXia, J., Li, W., Sun, M., & Wang, H. (2022). Application of SERS in the Detection of Fungi, Bacteria and Viruses. Nanomaterials, 12(20), 3572. https://doi.org/10.3390/nano12203572





