An Exploration of Nanoparticle-Based Diagnostic Approaches for Coronaviruses: SARS-CoV-2, SARS-CoV and MERS-CoV
Abstract
1. Introduction
1.1. Global Prevalence
1.2. Importance of Diagnostics in the COVID-19 Pandemic
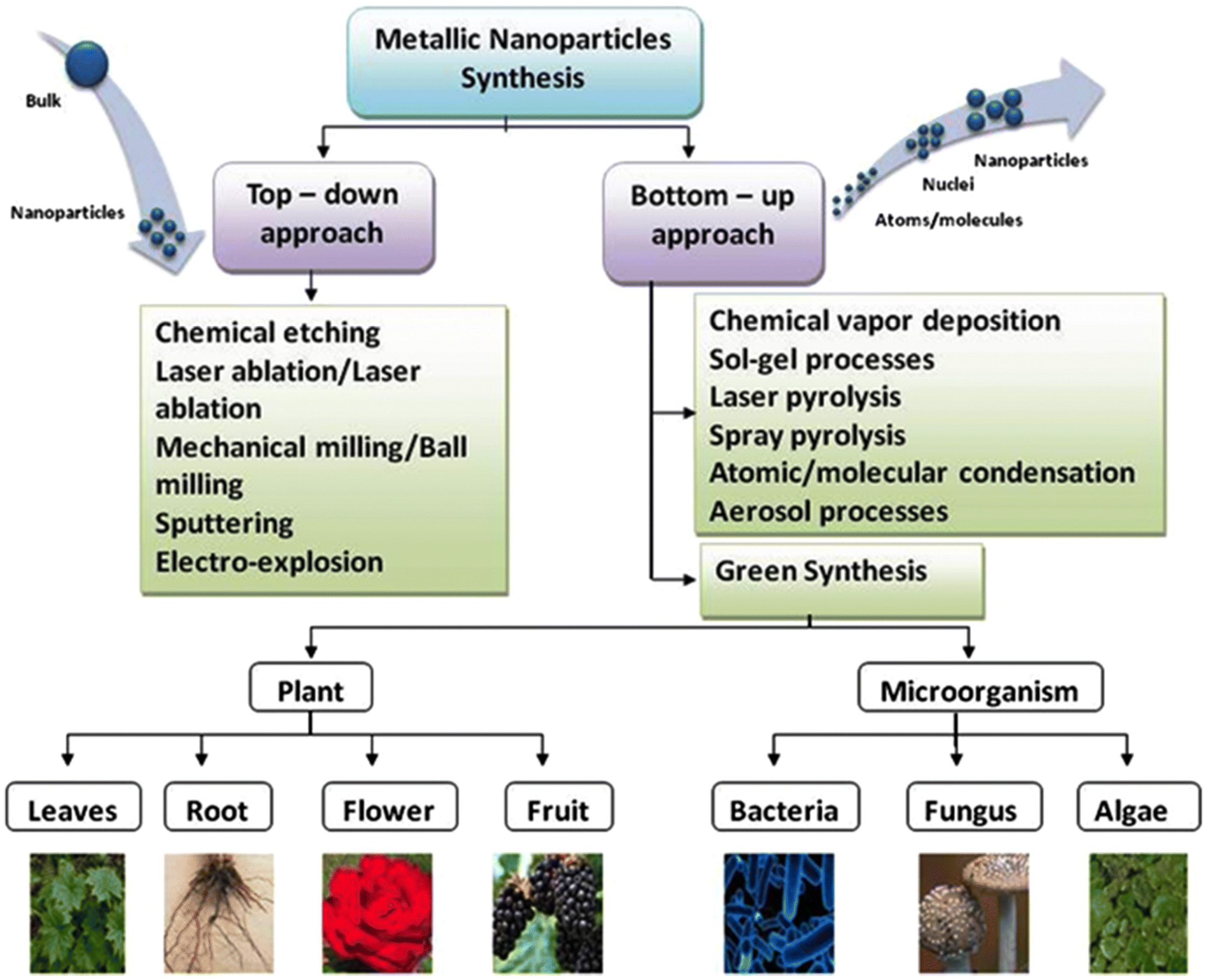
1.3. Nanoparticles
2. Types of Coronaviruses
2.1. Severe Acute Respiratory Syndrome-Associated Coronavirus (SARS-CoV)
2.2. Middle East Respiratory Syndrome Coronavavirus (MERS-CoV)
3. Structure and Pathogenesis of SARS-CoV-2
4. Current Methods of COVID-19 Diagnosis
4.1. Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
4.2. Computer Tomography (CT) Scan
4.3. Rapid Antigen Testing (RAT)
4.4. Immunoassays and Enzyme-Linked Immunosorbent Assays (ELISA)
4.5. Loop-Mediated Isothermal Amplification (LAMP)
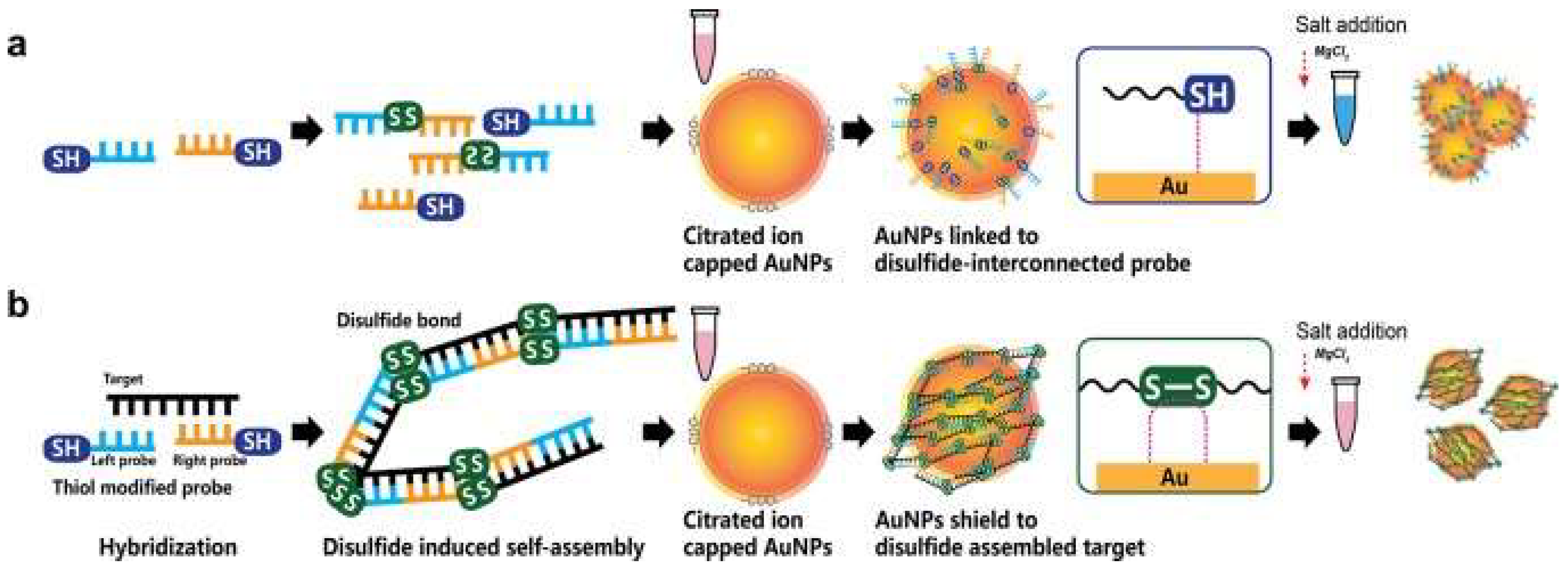
5. Usage of Nanoparticles in the Detection of Coronaviruses
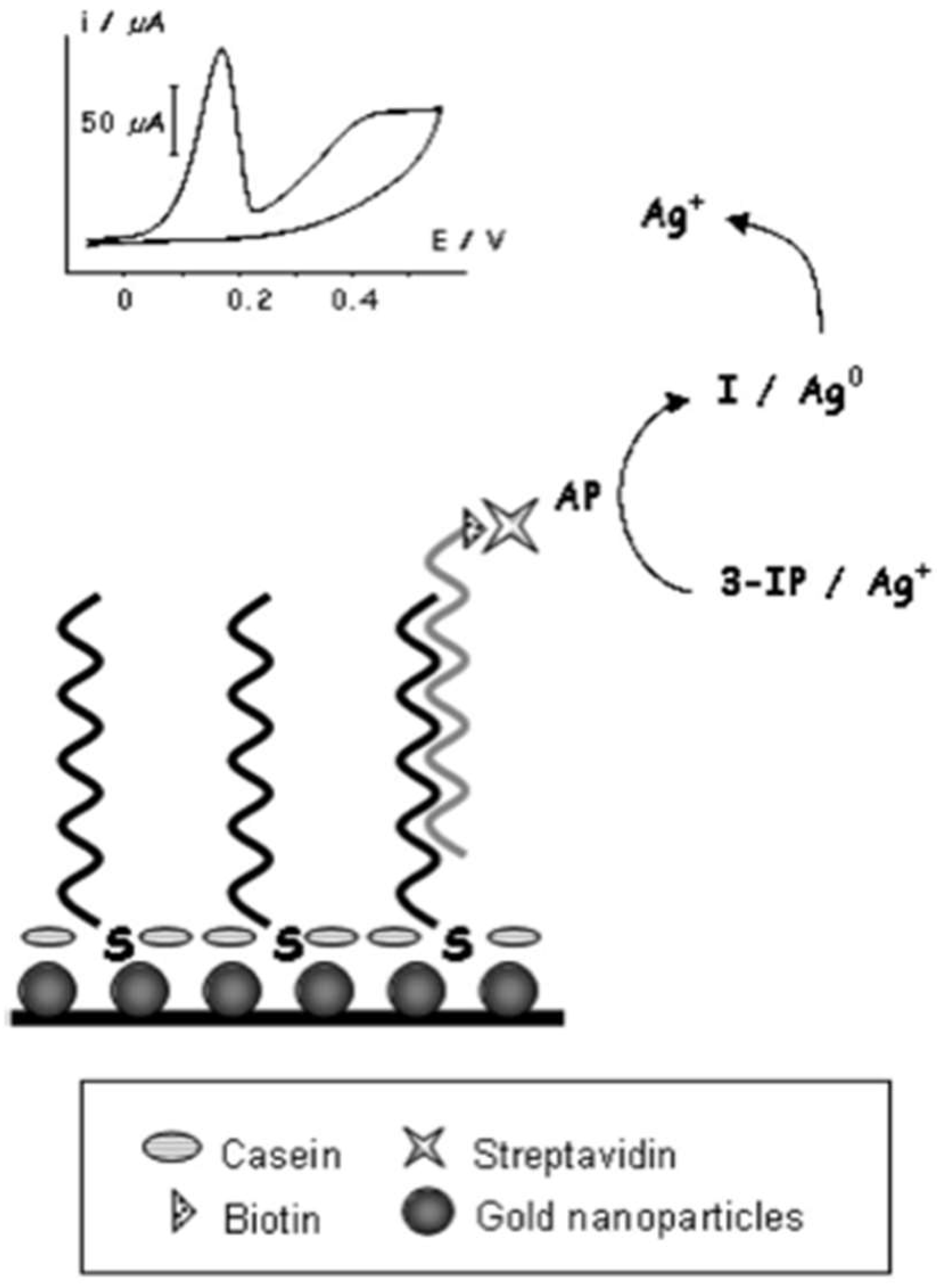
5.1. Usage of Nanoparticles in the Detection of Middle East Respiratory Syndrome Coronavirus (MERS-CoV)
5.2. Usage of Nanoparticles in the Detection of Severe Acute Respiratory Syndrome-Associated Coronavirus (SARS-CoV)
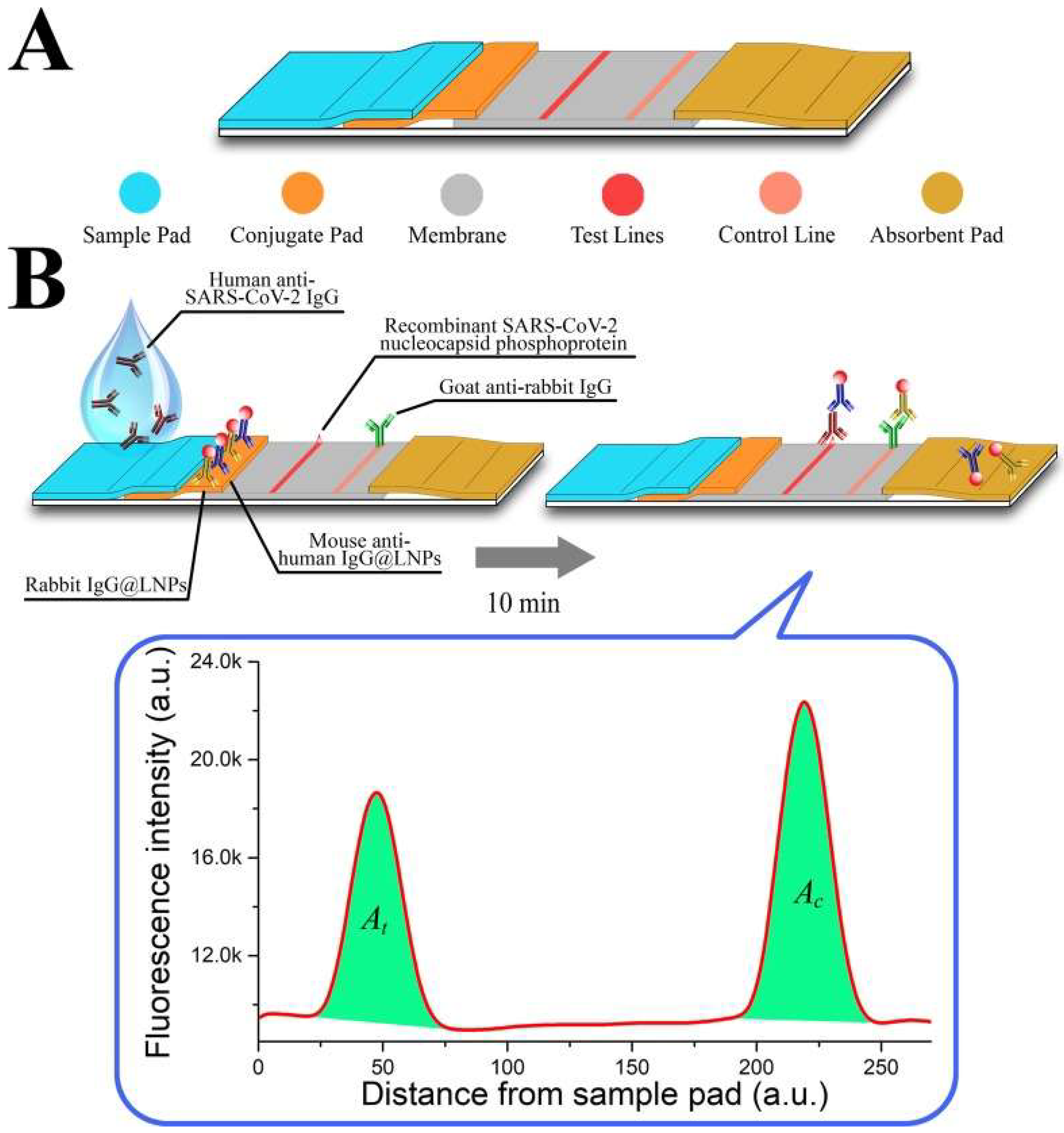
5.3. Usage of Nanoparticles in the Detection of Severe Acute Respiratory Syndrome-Associated Coronavirus-2 (SARS-CoV-2)
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- John Hopkins University. Coronavirus COVID-19 Global Cases by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University. Available online: https://coronavirus.jhu.edu/map.html (accessed on 2 August 2022).
- World Health Organization. Coronavirus Disease (COVID-19) Pandemic. Available online: https://www.who.int/europe/emergencies/situations/covid-19 (accessed on 28 September 2022).
- McKibbin, W.; Fernando, R. The Global Macroeconomic Impacts of COVID-19- Seven Scenarios. SSRN Electron. J. 2020, 20, 1–30. [Google Scholar] [CrossRef]
- Talic, S.; Shah, S.; Wild, H.; Gasevic, D.; Maharaj, A.; Ademi, Z.; Li, X.; Xu, W.; Mesa-Eguiagaray, I.; Rostron, J.; et al. Effectiveness of public health measures in reducing the incidence of COVID-19, SARS-CoV-2 transmission, and covid-19 mortality: Systematic review and meta-analysis. BMJ 2021, 375, e068302. [Google Scholar] [CrossRef]
- World Health Organization. Coronavirus Disease (COVID-19) Advice for the Public. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/advice-for-public (accessed on 20 September 2020).
- Wang, W.; Xu, Y.; Gao, R.; Lu, R.; Han, K.; Wu, G.; Tan, W. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA 2020, 323, 1843–1844. [Google Scholar] [CrossRef] [PubMed]
- Kim, H. Outbreak of novel coronavirus (COVID-19): What is the role of radiologists? Eur. Radiol. 2020, 30, 3266–3267. [Google Scholar] [CrossRef] [PubMed]
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Zhou, H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020, 165, 112349. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, X.; Han, L.; Chen, T.; Wang, L.; Li, H.; Li, S.; He, L.; Fu, X.; Chen, S.; et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020, 166, 112437. [Google Scholar] [CrossRef]
- Li, D.; Wang, D.; Dong, J.; Wang, N.; Huang, H.; Xu, H.; Xia, C. False-Negative Results of Real-Time Reverse-Transcriptase Polymerase Chain Reaction for Severe Acute Respiratory Syndrome Coronavirus 2: Role of Deep-Learning-Based CT Diagnosis and Insights from Two Cases. Korean J. Radiol. 2020, 21, 505–508. [Google Scholar] [CrossRef]
- Zu, Z.Y.; Jiang, M.D.; Xu, P.P.; Chen, W.; Ni, Q.Q.; Lu, G.M.; Zhang, L.J. Coronavirus Disease 2019 (COVID-19)- A Perspective from China. Radiology 2020, 296, E15–E25. [Google Scholar] [CrossRef]
- Fernandes, Q.; Inchakalody, V.P.; Merhi, M.; Mestiri, S.; Taib, N.; Moustafa Abo El-Ella, D.; Bedhiafi, T.; Raza, A.; Al-Zaidan, L.; Mohsen, M.O.; et al. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann. Med. 2022, 54, 524–540. [Google Scholar] [CrossRef]
- Thakur, P.; Thakur, V.; Kumar, P.; Singh Patel, S.K. Emergence of novel omicron hybrid variants: BA(x), XE, XD, XF more than just alphabets. Int. J. Surg. 2022, 104, 106727. [Google Scholar] [CrossRef] [PubMed]
- Thakur, V.; Bhola, S.; Thakur, P.; Patel, S.; Kulshrestha, S.; Ratho, R.K.; Kumar, P. Waves and variants of SARS-CoV-2: Understanding the causes and effect of the COVID-19 catastrophe. Infection 2022, 50, 309–325. [Google Scholar] [CrossRef]
- Kumar, N.; Kumbhat, S. Unique Properties. In Essentials in Nanoscience and Nanotechnology; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Tiloke, C.; Phulukdaree, A.; Chuturgoon, A.A. The Chemotherapeutic Potential of Gold Nanoparticles against Human Carcinomas: A Review. In Nanoarchitectonics for Smart Delivery and Drug Targeting, 1st ed.; Holban, A.M., Grumezescu, A.M., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 783–811. [Google Scholar]
- Thakur, N.; Kumar, A.K.; Kumar, A. Effect of (Ag, Zn) co-doping on structural, optical and bactericidal properties of CuO nanoparticles synthesized by a microwave-assisted method. Dalton Trans. 2021, 50, 6188–6203. [Google Scholar] [CrossRef]
- Pelaz, B.; Alexiou, C.; Alvarez-Puebla, R.A.; Alves, F.; Andrews, A.M.; Ashraf, S.; Balogh, L.P.; Ballerini, L.; Bestetti, A.; Brendel, C.; et al. Diverse Applications of Nanomedicine. ACS Nano 2017, 11, 2313–2381. [Google Scholar] [CrossRef] [PubMed]
- Ielo, I.; Rando, G.; Giacobello, F.; Sfameni, S.; Castellano, A.; Galletta, M.; Drommi, D.; Rosace, G.; Plutino, M.R. Synthesis, Chemical-Physical Characterization, and Biomedical Applications of Functional Gold Nanoparticles: A Review. Molecules 2021, 26, 5823. [Google Scholar] [CrossRef]
- Davies, G.L.; O’Brien, J.; Gun’ko, Y.K. Rare Earth Doped Silica Nanoparticles via Thermolysis of a Single Source Metallasilsesquioxane Precursor. Sci. Rep. 2017, 7, 45862. [Google Scholar] [CrossRef]
- Yu, X.; Pham, J.T.; Subramani, C.; Creran, B.; Yeh, Y.C.; Du, K.; Patra, D.; Miranda, O.R.; Crosby, A.J.; Rotello, V.M. Direct patterning of engineered ionic gold nanoparticles via nanoimprint lithography. Adv. Mater. 2012, 24, 6330–6334. [Google Scholar] [CrossRef]
- Sportelli, M.C.; Izzi, M.; Volpe, A.; Clemente, M.; Picca, R.A.; Ancona, A.; Lugarà, P.M.; Palazzo, G.; Cioffi, N. The Pros and Cons of the Use of Laser Ablation Synthesis for the Production of Silver Nano-Antimicrobials. Antibiotics 2018, 7, 67. [Google Scholar] [CrossRef]
- Mirzaei, A.; Neri, G. Microwave-assisted synthesis of metal oxide nanostructures for gas sensing application: A review. Sens. Actuators B Chem. 2016, 237, 749–775. [Google Scholar] [CrossRef]
- Flores-Rojas, G.; López-Saucedo, F.; Bucio, E. Gamma-irradiation applied in the synthesis of metallic and organic nanoparticles: A short review. Radiat. Phys. Chem. 2020, 169, 107962. [Google Scholar] [CrossRef]
- Hatakeyama, Y.; Onishi, K.; Nishikawa, K. Effects of sputtering conditions on formation of gold nanoparticles in sputter deposition technique. RSC Adv. 2011, 1, 1815–1821. [Google Scholar] [CrossRef]
- Singh, J.; Dutta, T.; Kim, K.H.; Rawat, M.; Samddar, P.; Kumar, P. ‘Green’ synthesis of metals and their oxide nanoparticles: Applications for environmental remediation. J. Nanobiotechnol. 2018, 16, 84. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Annu, I.S.; Yudha, S.S. Biosynthesis of gold nanoparticles: A green approach. J. Photochem. Photobiol. B 2016, 161, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Salem, S.S.; Fouda, A. Synthesis of Metallic Nanoparticles and Their Prospective Biotechnological Applications: An Overview. Biol. Trace Elem. Res. 2021, 199, 344–370. [Google Scholar] [CrossRef]
- Kumar, S.; Lather, V.; Pandita, D. Green synthesis of therapeutic nanoparticles: An expanding horizon. Nanomedicine 2015, 10, 2451–2471. [Google Scholar] [CrossRef] [PubMed]
- Caires, A.J.; Mansur, H.S.; Mansur, A.; Carvalho, S.M.; Lobato, Z.; Dos Reis, J. Gold nanoparticle-carboxymethyl cellulose nanocolloids for detection of human immunodeficiency virus type-1 (HIV-1) using laser light scattering immunoassay. Colloids. Surf. B Biointerfaces 2019, 177, 377–388. [Google Scholar] [CrossRef]
- Liu, Z.; Shang, C.; Ma, H.; You, M. An upconversion nanoparticle-based photostable FRET system for long-chain DNA sequence detection. Nanotechnology 2020, 31, 235501. [Google Scholar] [CrossRef]
- Medhi, R.; Srinoi, P.; Ngo, N.; Tran, H.V.; Lee, T.R. Nanoparticle-Based Strategies to Combat COVID-19. ACS Appl. Nano Mater. 2020, 3, 8557–8580. [Google Scholar] [CrossRef]
- Asdaq, S.; Ikbal, A.; Sahu, R.K.; Bhattacharjee, B.; Paul, T.; Deka, B.; Fattepur, S.; Widyowati, R.; Vijaya, J.; Al Mohaini, M.; et al. Nanotechnology Integration for SARS-CoV-2 Diagnosis and Treatment: An Approach to Preventing Pandemic. Nanomaterials 2021, 11, 1841. [Google Scholar] [CrossRef]
- Rabaan, A.A.; Al-Ahmed, S.H.; Haque, S.; Sah, R.; Tiwari, R.; Malik, Y.S.; Dhama, K.; Yatoo, M.I.; Bonilla-Aldana, D.K.; Rodriguez-Morales, A.J. SARS-CoV-2, SARS-CoV, and MERS-CoV- a comparative overview. Le Infez. Med. 2020, 28, 174–184. [Google Scholar]
- Demmler, G.J.; Ligon, B.L. Severe Acute Respiratory Syndrome (SARS)- A Review of the History, Epidemiology, Prevention, and Concerns for the Future. Semin. Pediatr. Infect. Dis. 2003, 14, 240–244. [Google Scholar] [CrossRef]
- Coleman, C.M.; Frieman, M.B. Coronaviruses: Important emerging human pathogens. J. Virol. 2014, 88, 5209–5212. [Google Scholar] [CrossRef] [PubMed]
- Skowronski, D.M.; Astell, C.; Brunham, R.C.; Low, D.E.; Petric, M.; Roper, R.L.; Talbot, P.J.; Tam, T.; Babiuk, L. Severe acute respiratory syndrome (SARS): A year in review. Annu. Rev. Med. 2005, 56, 357–381. [Google Scholar] [CrossRef]
- Baharoon, S.; Memish, Z.A. MERS-CoV as an emerging respiratory illness: A review of prevention methods. Travel Med. Infect. Dis. 2019, 32, 101520. [Google Scholar] [CrossRef]
- Mackay, I.M.; Arden, K.E. MERS coronavirus: Diagnostics, epidemiology and transmission. Virol. J. 2015, 12, 222. [Google Scholar] [CrossRef]
- Khailany, R.A.; Safdar, M.; Ozaslan, M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020, 19, 100682. [Google Scholar] [CrossRef]
- Majumder, J.; Minko, T. Recent Developments on Therapeutic and Diagnostic Approaches for COVID-19. AAPS J. 2021, 23, 14. [Google Scholar] [CrossRef]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef]
- Yuki, K.; Fujiogi, M.; Koutsogiannaki, S. COVID-19 pathophysiology: A review. Clin. Immunol. 2020, 215, 108427. [Google Scholar] [CrossRef]
- Turner, A.J.; Hiscox, J.A.; Hooper, N.M. ACE2: From vasopeptidase to SARS virus receptor. Trends Pharm. Sci 2004, 25, 291–294. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Tipnis, S.R.; Hooper, N.M.; Hyde, R.; Karran, E.; Christie, G.; Turner, A.J. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 2000, 275, 33238–33243. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.Y.; Chang, S.C.; Wu, H.Y.; Yu, T.C.; Wei, W.C.; Lin, S.; Chien, C.L.; Chang, M.F. Upregulation of the chemokine (C-C motif) ligand 2 via a severe acute respiratory syndrome coronavirus spike-ACE2 signaling pathway. J. Virol. 2010, 84, 7703–7712. [Google Scholar] [CrossRef]
- Chu, H.; Yuen, K.Y. Pathogenicity of SARS-CoV-2 Omicron. Clin. Transl. Med. 2022, 12, e880. [Google Scholar] [CrossRef]
- World Health Organization. Classification of Omicron (B.1.1.529): SARS-CoV-2 Variant of Concern. Available online: https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern (accessed on 9 September 2022).
- Wang, Y.; Liu, C.; Zhang, C.; Wang, Y.; Hong, Q.; Xu, S.; Li, Z.; Yang, Y.; Huang, Z.; Cong, Y. Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies. Nat. Commun. 2022, 13, 871. [Google Scholar] [CrossRef]
- Hart, W.S.; Miller, E.; Andrews, N.J.; Waight, P.; Maini, P.K.; Funk, S.; Thompson, R.N. Generation time of the alpha and delta SARS-CoV-2 variants: An epidemiological analysis. Lancet Infect. Dis. 2022, 25, 603–610. [Google Scholar] [CrossRef]
- Carter, L.J.; Garner, L.V.; Smoot, J.W.; Li, Y.; Zhou, Q.; Saveson, C.J.; Sasso, J.M.; Gregg, A.C.; Soares, D.J.; Beskid, T.R.; et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent. Sci. 2020, 6, 591–605. [Google Scholar] [CrossRef]
- Yu, L.; Wu, S.; Hao, X.; Dong, X.; Mao, L.; Pelechano, V.; Chen, W.; Yin, X. Rapid detection of COVID-19 coronavirus using a reverse transcriptional loop-mediated isothermal amplification (RT-LAMP) diagnostic platform. Clin. Chem. 2020, 66, 975–977. [Google Scholar] [CrossRef]
- Haijema, B.J.; Volders, H.; Rottier, P.J. Live, attenuated coronavirus vaccines through the directed deletion of group-specific genes provide protection against feline infectious peritonitis. J. Virol. 2004, 78, 3863–3871. [Google Scholar] [CrossRef]
- Fukushi, S.; Fukuma, A.; Kurosu, T.; Watanabe, S.; Shimojima, M.; Shirato, K.; Iwata-Yoshikawa, N.; Nagata, N.; Ohnishi, K.; Ato, M.; et al. Characterization of novel monoclonal antibodies against the MERS-coronavirus spike protein and their application in species-independent antibody detection by competitive ELISA. J. Virol. Methods 2018, 251, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Tahamtan, A.; Ardebili, A. Real-time RT-PCR in COVID-19 detection: Issues affecting the results. Expert Rev. Mol. Diagn. 2020, 20, 453–454. [Google Scholar] [CrossRef] [PubMed]
- Bhalla, N.; Pan, Y.; Yang, Z.; Payam, A.F. Opportunities and Challenges for Biosensors and Nanoscale Analytical Tools for Pandemics: COVID-19. ACS Nano 2020, 14, 7783–7807. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Duong Bang, D.; Wolff, A. 2019 Novel Coronavirus Disease (COVID-19): Paving the Road for Rapid Detection and Point-of-Care Diagnostics. Micromachines 2020, 11, 306. [Google Scholar] [CrossRef] [PubMed]
- Palestino, G.; Garcia-Silva, I.; Gonzalez-Ortega, O.; Rosales-Mendoza, S. Can nanotechnology help in the fight against COVID-19? Expert Rev. Anti. Infect. 2020, 18, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Rabiee, N.; Bagherzadeh, M.; Ghasemi, A.; Zare, H.; Ahmadi, S.; Fatahi, Y.; Dinarvand, R.; Rabiee, M.; Ramakrishna, S.; Shokouhimehr, M.; et al. Point-of-Use Rapid Detection of SARS-CoV-2: Nanotechnology-Enabled Solutions for the COVID-19 Pandemic. Int. J. Mol. Sci. 2020, 21, 5126. [Google Scholar] [CrossRef]
- Li, Y.; Yao, L.; Li, J.; Chen, L.; Song, Y.; Cai, Z.; Yang, C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020, 92, 903–908. [Google Scholar] [CrossRef]
- Tang, Y.; Schmitz, J.; Persing, D.; CW, S. Laboratory Diagnosis of COVID-19- Current Issues and Challenges. J Clin Microbiol 2020, 58, e00512-20. [Google Scholar] [CrossRef]
- Al-Tawfiq, J.A.; Memish, Z.A. Diagnosis of SARS-CoV-2 infection based on CT scan vs RT-PCR: Reflecting on experience from MERS-CoV. J. Hosp. Infect. 2020, 105, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Han, X.; Jiang, N.; Cao, Y.; Alwalid, O.; Gu, J.; Fan, Y.; Zheng, C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: A descriptive study. Lancet Infect. Dis. 2020, 20, 425–434. [Google Scholar] [CrossRef]
- Ai, T.; Yang, Z.; Hou, H.; Zhan, C.; Chen, C.; Lv, W.; Tao, Q.; Sun, Z.; Xia, L. Correlation of Chest CT and RT-PCR Testing in Coronavirus Disease 2019 (COVID-19) in Chin—A Report of 1014 Cases. Radiology 2020, 296, E32–E40. [Google Scholar] [CrossRef]
- Peeling, R.W.; Olliaro, P.L.; Boeras, D.I.; Fongwen, N. Scaling up COVID-19 rapid antigen tests: Promises and challenges. Lancet Infect. Dis. 2021, 21, e290–e295. [Google Scholar] [CrossRef]
- Khandker, S.S.; Nik Hashim, N.; Deris, Z.Z.; Shueb, R.H.; Islam, M.A. Diagnostic Accuracy of Rapid Antigen Test Kits for Detecting SARS-CoV-2: A Systematic Review and Meta-Analysis of 17,171 Suspected COVID-19 Patients. J. Clin. Med. 2021, 10, 3493. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Liu, Y.; Fan, B.; Xiao, Y.; Tian, Q.; Chen, L.; Zhao, H.; Chen, W. Dynamic changes of serum SARS-coronavirus IgG, pulmonary function and radiography in patients recovering from SARS after hospital discharge. Respir. Res. 2005, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Wen, T.; Huang, C.; Shi, F.J.; Zeng, X.Y.; Lu, T.; Ding, S.N.; Jiao, Y.J. Development of a lateral flow immunoassay strip for rapid detection of IgG antibody against SARS-CoV-2 virus. Analyst 2020, 145, 5345–5352. [Google Scholar] [CrossRef] [PubMed]
- Oscorbin, I.P.; Belousova, E.A.; Zakabunin, A.I.; Boyarskikh, U.A.; Filipenko, M.L. Comparison of fluorescent intercalating dyes for quantitative loop-mediated isothermal amplification (qLAMP). Biotechniques 2016, 61, 20–25. [Google Scholar] [CrossRef]
- Goto, M.; Honda, E.; Ogura, A.; Nomoto, A.; Hanaki, K. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 2009, 46, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Park, G.S.; Ku, K.; Baek, S.H.; Kim, S.J.; Kim, S.I.; Kim, B.T.; Maeng, J.S. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 2020, 22, 729–735. [Google Scholar] [CrossRef]
- Sahoo, P.R.; Sethy, K.; Mohapatra, S.; Panda, D. Loop mediated isothermal amplification: An innovative gene amplification technique for animal diseases. Vet. World 2016, 9, 465–469. [Google Scholar] [CrossRef]
- Deguo, W.; Guicheng, H.; Fugui, W.; Yonggang, L.; Daxi, R. Drawback of loop-mediated isothermal amplification. Afr. J. Food Sci. 2008, 2, 83–86. [Google Scholar]
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627. [Google Scholar] [CrossRef]
- Nikaeen, G.; Abbaszadeh, S.; Yousefinejad, S. Application of nanomaterials in treatment, anti-infection and detection of coronaviruses. Nanomedicine 2020, 15, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Layqah, L.A.; Eissa, S. An electrochemical immunosensor for the corona virus associated with the Middle East respiratory syndrome using an array of gold nanoparticle-modified carbon electrodes. Mikrochim. Acta 2019, 186, 224. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Park, M.; Hwang, J.; Kim, J.H.; Chung, D.R.; Lee, K.S.; Kang, M. Development of Label-Free Colorimetric Assay for MERS-CoV Using Gold Nanoparticles. ACS Sens. 2019, 4, 1306–1312. [Google Scholar] [CrossRef]
- Huang, P.; Wang, H.; Cao, Z.; Jin, H.; Chi, H.; Zhao, J.; Yu, B.; Yan, F.; Hu, X.; Wu, F.; et al. A Rapid and Specific Assay for the Detection of MERS-CoV. Front. Microbiol. 2018, 9, 1101. [Google Scholar] [CrossRef] [PubMed]
- Teengam, P.; Siangproh, W.; Tuantranont, A.; Vilaivan, T.; Chailapakul, O.; Henry, C.S. Multiplex Paper-Based Colorimetric DNA Sensor Using Pyrrolidinyl Peptide Nucleic Acid-Induced AgNPs Aggregation for Detecting MERS-CoV, MTB, and HPV Oligonucleotides. Anal. Chem. 2017, 89, 5428–5435. [Google Scholar] [CrossRef]
- Qiao, J.; Li, Y.; Wei, C.; Yang, H.; Yu, J.; Wei, H. Rapid detection of viral antibodies based on multifunctional Staphylococcus aureus nanobioprobes. Enzym. Microb. Technol. 2016, 95, 94–99. [Google Scholar] [CrossRef]
- Park, T.J.; Lee, S.Y.; Lee, S.J.; Park, J.P.; Yang, S.K.; Lee, K.B.; Ko, S.; Park, J.B.; Kim, T.; Kim, S.K.; et al. Protein Nanopatterns and Biosensors Using Gold Binding Polypeptide as a Fusion Partner. Anal. Chem. 2006, 78, 7197–7205. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.C.; Chang, Y.F.; Chen, K.H.; Su, L.C.; Lee, C.W.; Chen, C.C.; Chen, Y.M.; Chou, C. Detection of severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein in human serum using a localized surface plasmon coupled fluorescence fiber-optic biosensor. Biosens. Bioelectron. 2009, 25, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Paredes, G.; González-García, M.B.; Costa-García, A. Genosensor for SARS Virus Detection Based on Gold Nanostructured Screen-Printed Carbon Electrodes. Electroanalysis 2009, 21, 379–385. [Google Scholar] [CrossRef]
- Ishikawa, F.N.; Curreli, M.; Olson, C.A.; Liao, H.I.; Sun, R.; Roberts, R.W.; Cote, R.J.; Thompson, M.E.; Zhou, C. Importance of Controlling Nanotube Density for Highly Sensitive and Reliable Biosensors Functional in Physiological Conditions. ACS Nano 2010, 4, 6914–6922. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.; He, X.; Wang, K.; Tan, W.; Xie, W.; Wu, P.; Li, H. Combination of functionalized nanoparticles and polymerase chain reaction-based method for SARS-CoV gene detection. Nanosci. Nanotechnol. 2008, 8, 293–300. [Google Scholar] [CrossRef]
- Kumar, V.; Mishra, S.; Sharma, R.; Agarwal, J.; Ghoshal, U.; Khanna, T.; Sharma, L.K.; Verma, S.K.; Mishra, P.; Tiwari, S. Development of RNA-based assay for rapid detection of SARS-CoV-2 in clinical samples. Intervirology 2022, 1–7. [Google Scholar] [CrossRef]
- Li, J.; Wu, D.; Yu, Y.; Li, T.; Li, K.; Xiao, M.M.; Li, Y.; Zhang, Z.Y.; Zhang, G.J. Rapid and unamplified identification of COVID-19 with morpholino-modified graphene field-effect transistor nanosensor. Biosens. Bioelectron. 2021, 183, 113206. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.; Gai, Z.; Tao, Y.; Schmitt, J.; Kullak-Ublick, G.A.; Wang, J. Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection. ACS Nano 2020, 14, 5268–5277. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, Z.; Zhai, X.; Li, Y.; Lin, L.; Zhao, H.; Bian, L.; Li, P.; Yu, L.; Wu, Y.; et al. Rapid and Sensitive Detection of anti-SARS-CoV-2 IgG, Using Lanthanide-Doped Nanoparticles-Based Lateral Flow Immunoassay. Anal. Chem. 2020, 92, 7226–7231. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wen, T.; Shi, F.J.; Zeng, X.Y.; Jiao, Y.J. Rapid Detection of IgM Antibodies against the SARS-CoV-2 Virus via Colloidal Gold Nanoparticle-Based Lateral-Flow Assay. ACS Omega 2020, 5, 12550–12556. [Google Scholar] [CrossRef]
- Li, Z.; Yi, Y.; Luo, X.; Xiong, N.; Liu, Y.; Li, S.; Sun, R.; Wang, Y.; Hu, B.; Chen, W.; et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J. Med. Virol. 2021, 92, 1518–1524. [Google Scholar] [CrossRef]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.H.; Choi, M.; Ku, K.B.; Lee, C.S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Rösch, E.L.; Viereck, T.; Schilling, M.; Ludwig, F. Toward Rapid and Sensitive Detection of SARS-CoV-2 with Functionalized Magnetic Nanoparticles. ACS Sens. 2021, 6, 976–984. [Google Scholar] [CrossRef] [PubMed]
- Hryniewicz, B.M.; Volpe, J.; Bach-Toledo, L.; Kurpel, K.C.; Deller, A.E.; Soares, A.L.; Nardin, J.M.; Marchesi, L.F.; Simas, F.F.; Oliveira, C.C.; et al. Development of polypyrrole (nano)structures decorated with gold nanoparticles toward immunosensing for COVID-19 serological diagnosis. Mater. Today Chem. 2022, 24, 100817. [Google Scholar] [CrossRef] [PubMed]
- Durmus, C.; Balaban Hanoglu, S.; Harmanci, D.; Moulahoum, H.; Tok, K.; Ghorbanizamani, F.; Sanli, S.; Zihnioglu, F.; Evran, S.; Cicek, C.; et al. Indiscriminate SARS-CoV-2 multivariant detection using magnetic nanoparticle-based electrochemical immunosensing. Talanta 2022, 243, 123356. [Google Scholar] [CrossRef] [PubMed]
- Ellipilli, S.; Wang, H.; Lee, W.J.; Shu, D.; Guo, P. Proof-of-concept for speedy development of rapid and simple at-home method for potential diagnosis of early COVID-19 mutant infections using nanogold and aptamer. Nanomedicine 2022, 45, 102590. [Google Scholar] [CrossRef]
- Blumenfeld, N.R.; Bolene, M.; Jaspan, M.; Ayers, A.G.; Zarrandikoetxea, S.; Freudman, J.; Shah, N.; Tolwani, A.M.; Hu, Y.; Chern, T.L.; et al. Multiplexed reverse-transcriptase quantitative polymerase chain reaction using plasmonic nanoparticles for point-of-care COVID-19 diagnosis. Nat. Nanotechnol. 2022, 17, 984–992. [Google Scholar] [CrossRef] [PubMed]
- Miripour, Z.S.; Sarrami-Forooshani, R.; Sanati, H.; Makarem, J.; Taheri, M.S.; Shojaeian, F.; Eskafi, A.H.; Abbasvandi, F.; Namdar, N.; Ghafari, H.; et al. Real-time diagnosis of reactive oxygen species (ROS) in fresh sputum by electrochemical tracing; correlation between COVID-19 and viral-induced ROS in lung/respiratory epithelium during this pandemic. Biosens. Bioelectron. 2020, 165, 112435. [Google Scholar] [CrossRef]
- Shan, B.; Broza, Y.Y.; Li, W.; Wang, Y.; Wu, S.; Liu, Z.; Wang, J.; Gui, S.; Wang, L.; Zhang, Z.; et al. Multiplexed Nanomaterial-Based Sensor Array for Detection of COVID-19 in Exhaled Breath. ACS Nano 2020, 14, 12125–12132. [Google Scholar] [CrossRef]


| Virus | Nanoparticle (NP) Type | Limit of Detection (LOD) | Mechanism of Detection | Qualities and Significance |
|---|---|---|---|---|
| MERS-CoV (79) | Gold NPs | 1.0 pg/mL | Electromechanical immunosensor: Detection of voltametric response using an array of carbon electrodes modified with a gold NP coating. | Rapid; 20 min for results Lower LOD compared to ELISA (1 ng/mL) |
| MERS-CoV (80) | Gold NPs | 1.0 pmol/µL | Colorimetric assay: in the absence of MERS-CoV targets, the gold NPs aggregate together and produce a color change | Rapid; 10 min for results Result is easy to interpret and can be seen with the naked eye Can detect virus with little-to-no amplification |
| MERS-CoV (82) | Silver NPs | 1.53 nM | Colorimetric sensor: silver NPs used as colorimetric agent where color change is associated with the aggregation of NPs | High selectivity for desired oligonucleotides Can detect virus with little-to-no amplification Low-cost and disposable |
| SARS-CoV (85) | Gold NPs | 1.0 pg/mL | Localized surface plasmon coupled fluorescence (LSPCF) fiber-optic biosensor: detection of fluorescence of a fluorophore-labeled viral target (N protein) | Quantitative: linear response seen between fluorescence signal and concentration of viral target (0.1 pg/mL to 1.0 ng/mL) LOD is 104-fold more sensitive than conventional ELISA Easy to use and disposable |
| SARS-CoV (86) | Gold NPs | 2.5 pmol/L | Electrochemical genosensor: enzymatic amplification of hybridization signal | Rapid; 20 min for results |
| SARS-CoV (88) | Silica-coated superparamagnetic NPs | 2.0 × 103 copies | NPs used to capture cDNA targets for amplification. Amplified cDNA (via PCR) was separated from PCR products using NPs. Functionalized NPs were subsequently used as signaling probes and fluorescence was measured | Able to isolate desired cDNA targets from samples that contain both target cDNA and non-target cDNA Can be used as an adjuvant to PCR |
| SARS-CoV-2 (77) | Gold NPs | 0.18 ng/µL | Colorimetric bioassay: in the presence of the N gene of SARS-CoV-2, the gold NP would aggregate, changing their surface plasmon resonance and precipitating | Rapid; 10 min for results Result can be seen with the naked eye Selective; no cross-reactivity with MERS-CoV Affordable and does not require sophisticated equipment |
| SARS-CoV-2 (90) | Gold NPs | 2.29 fM for throat swab 3.99 fM for serum | Graphene field-effect transistor sensor: by targeting the RdRp genes, electrical changes on the sensor chip can be monitored | Near-perfect agreeability with RT-PCR (92%) Extremely rapid; <2 min for results |
| SARS-CoV-2 (94) | Gold NPs | - | Lateral flow immunoassay: able to detect both IgM and IgG antibodies against SARS-CoV-2. Result is portrayed via detection lines on the strip | Rapid; 15 min for results High selectivity and sensitivity; tested 525 clinical samples Easy to use and does not need additional equipment |
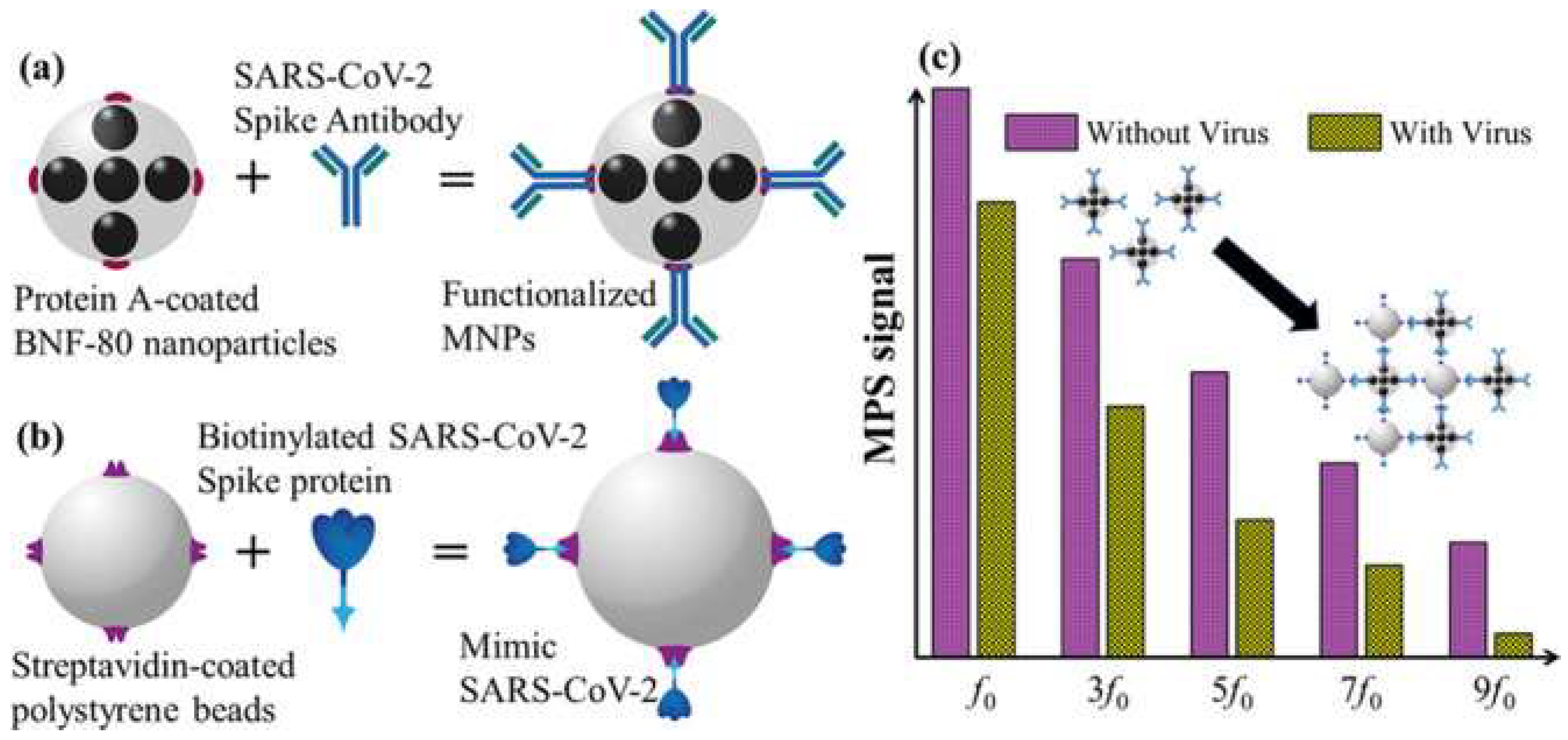
| SARS-CoV-2 (96) | Magnetic NPs (Iron NPs) | 0.084 nM | Magnetic particle spectroscopy: binding of mimic SARS-CoV-2 and functionalized NPs lead to a change in magnetic response within an AC magnetic field | Rapid; <15 min for results Sensitive and selective Low cost and easy to handle |
| SARS-CoV-2 (101) | Multi-wall carbon nanotubes | - | Electrochemical sensor: detection of reactive oxygen species in sputum samples. Changes in electrode potential was measured using cyclin voltammetry | High selectivity and sensitivity Extremely rapid; 30 s till results Effective screening tool: cannot independently diagnose as reactive oxygen species can be alleviated by other respiratory conditions (e.g., asthma, acute bacterial pneumonia, etc.) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Hindawi, A.; AlDallal, U.; Waly, Y.M.; Hussain, M.H.; Shelig, M.; Saleh ElMitwalli, O.S.M.M.; Deen, G.R.; Henari, F.Z. An Exploration of Nanoparticle-Based Diagnostic Approaches for Coronaviruses: SARS-CoV-2, SARS-CoV and MERS-CoV. Nanomaterials 2022, 12, 3550. https://doi.org/10.3390/nano12203550
Al-Hindawi A, AlDallal U, Waly YM, Hussain MH, Shelig M, Saleh ElMitwalli OSMM, Deen GR, Henari FZ. An Exploration of Nanoparticle-Based Diagnostic Approaches for Coronaviruses: SARS-CoV-2, SARS-CoV and MERS-CoV. Nanomaterials. 2022; 12(20):3550. https://doi.org/10.3390/nano12203550
Chicago/Turabian StyleAl-Hindawi, Ahmed, Usama AlDallal, Yousef Mostafa Waly, Muhammed Hesham Hussain, Mohamed Shelig, Omar Samir Mohamed Megahed Saleh ElMitwalli, G. Roshan Deen, and Fryad Z. Henari. 2022. "An Exploration of Nanoparticle-Based Diagnostic Approaches for Coronaviruses: SARS-CoV-2, SARS-CoV and MERS-CoV" Nanomaterials 12, no. 20: 3550. https://doi.org/10.3390/nano12203550
APA StyleAl-Hindawi, A., AlDallal, U., Waly, Y. M., Hussain, M. H., Shelig, M., Saleh ElMitwalli, O. S. M. M., Deen, G. R., & Henari, F. Z. (2022). An Exploration of Nanoparticle-Based Diagnostic Approaches for Coronaviruses: SARS-CoV-2, SARS-CoV and MERS-CoV. Nanomaterials, 12(20), 3550. https://doi.org/10.3390/nano12203550









