Isolation and Characterization of Cell Envelope Fragments Comprising Archaeal S-Layer Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cultivation
2.2. Extraction of SSO Ghosts and Production of SSO Sacculi and Fragments
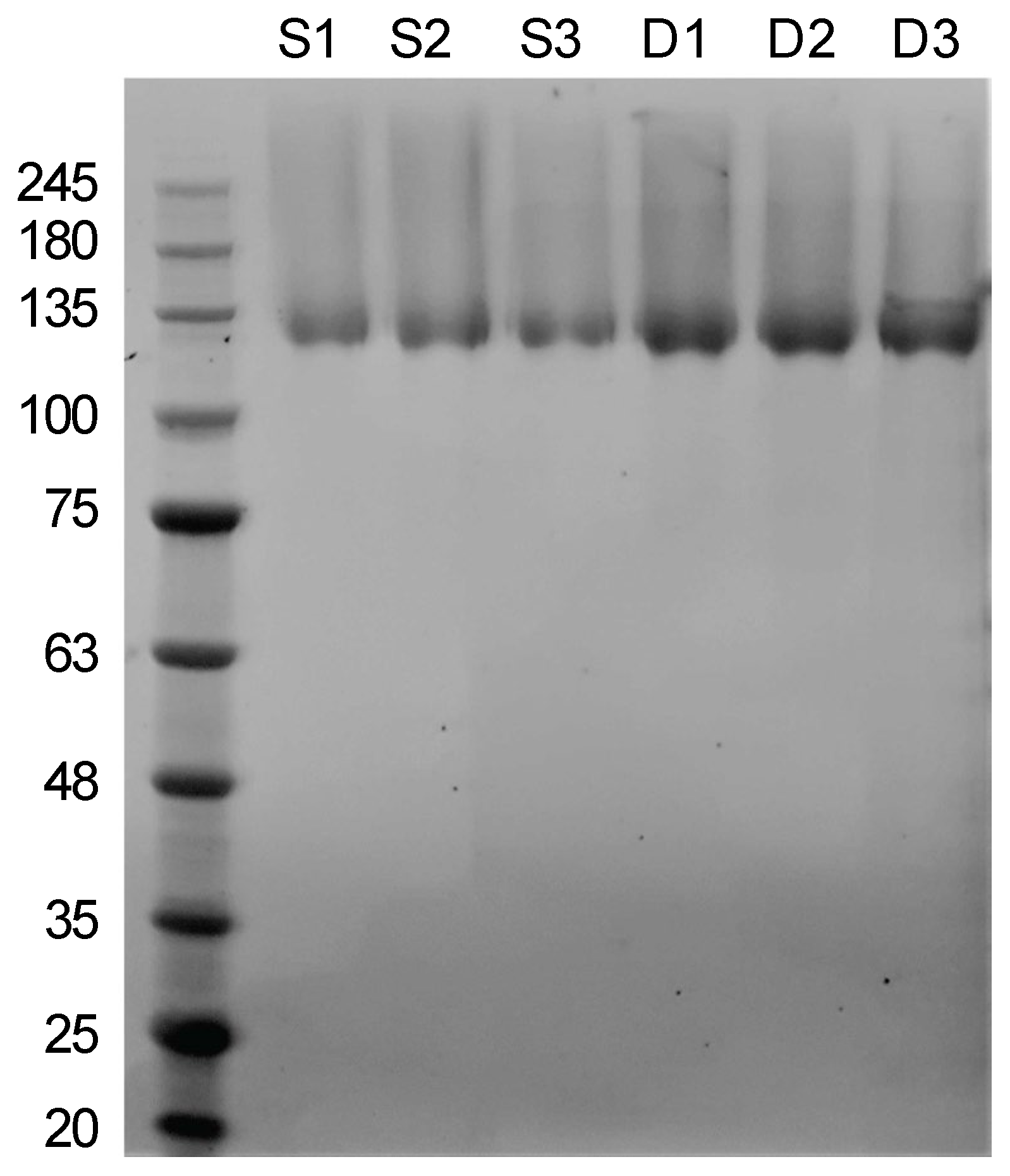
2.3. SDS Polyacrylamide Gel Electrophoresis (PAGE)
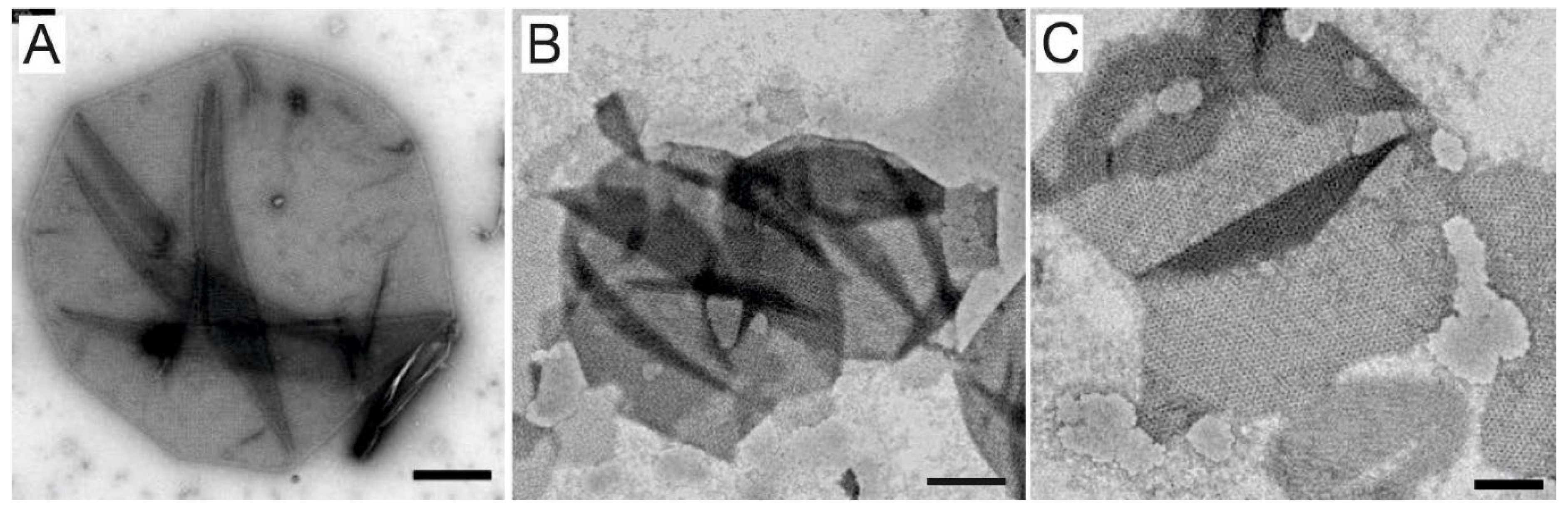
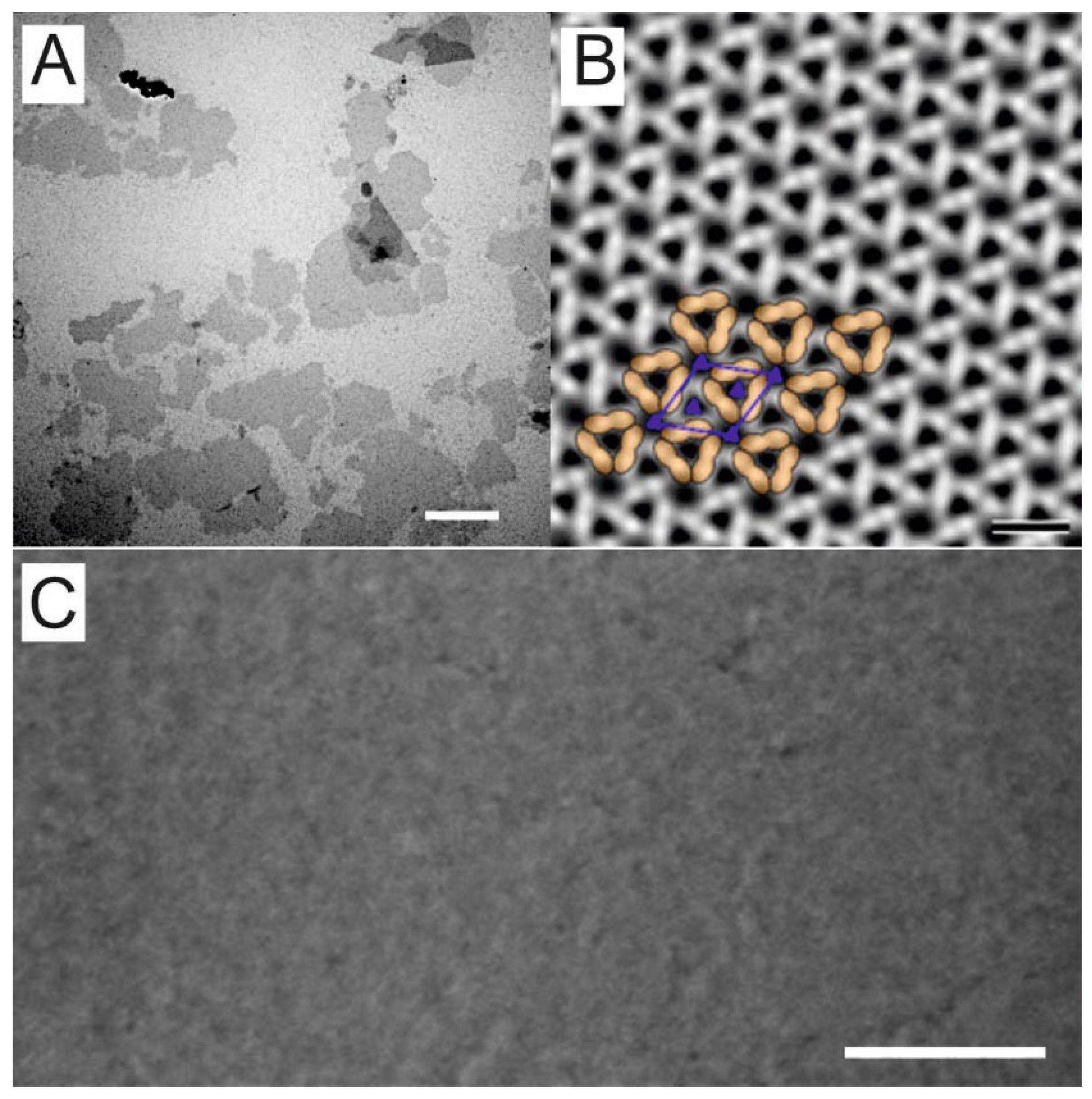
2.4. Electron Microscopy
2.5. Deposition of SSO Fragments on Microfilters
2.6. Investigation of Retention Efficiency against Proteins by SSO Fragments
3. Results and Discussion
3.1. A Modified and Cost-Effective Extraction Method for SSO Ghosts and Production of SSO Sacculi Comprising SlaA
3.2. Characterization of SSO Fragments on Microfilters
4. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| Abbreviation | Meaning |
| DEG | DNase-based extraction of ghosts |
| MF1 | microfilter 1 |
| MF2 | microfilter 2 |
| SDS | sodium dodecylsulfate |
| SDS-PAGE | sodium dodecylsulfate polyacrylamide gel electrophoresis |
| SEG | sonication-based extraction of ghosts |
| SEM | scanning electron microscopy |
| SlaA | surface layer large protein from Saccharolobus solfataricus P2 |
| SlaB | surface layer small protein from Saccharolobus solfataricus P2 |
| S-layer | surface-layer |
| SLP | surface layer protein and glycoprotein |
| SSO | Saccharolobus solfataricus P2 |
| SSOMF1 | cross-linked SSO fragments on microfilter 1 |
| SSOMF2 | cross-linked SSO fragments on microfilter 2 |
| SUM | S-layer ultrafiltration membranes |
| TEM | transmission electron microscopy |
References
- Houwink, A.L. A macromolecular monolayer in the cell wall of Spirillum spec. Biochim. Biophys. Acta 1953, 10, 360–366. [Google Scholar] [CrossRef]
- Salton, M.R.J.; Horne, R.W. Studies of the bacterial cell wall I. Electron microscopical observation on heated bacteria. Biochim. Biophys. Acta 1951, 7, 19–42. [Google Scholar] [CrossRef]
- Remsen, C.C.; Watson, S.W.; Waterbury, J.B.; Trüper, H.G. Fine structure of Ectothiorhodospira mobilis Pelsh. J. Bacteriol. 1968, 95, 2374–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sleytr, U.B.; Adam, H.; Klaushofer, H. Die Feinstruktur der Zellwandoberfläche von zwei thermophilen Clostridienarten, dargestellt mit Hilfe der Gefrierätztechnik. Mikroskopie 1968, 23, 1–10. [Google Scholar]
- Sleytr, U.B.; Messner, P.; Pum, D.; Sara, M. Crystalline Bacterial Cell Surface Layers; Springer: Berlin/Heidelberg, Germany, 1988; pp. 13–31. [Google Scholar] [CrossRef]
- Sleytr, U.B.; Schuster, B.; Egelseer, E.M.; Pum, D. S-layers: Principles and applications. FEMS Microbiol. Rev. 2014, 38, 823–864. [Google Scholar] [CrossRef]
- Albers, S.V.; Meyer, B.H. The archaeal cell envelope. Nat. Rev. Microbiol. 2011, 9, 414–426. [Google Scholar] [CrossRef]
- Lanzoni-Mangutchi, P.; Banerji, O.; Wilson, J.; Barwinska-Sendra, A.; Kirk, J.A.; Vaz, F.; O’Beirne, S.; Baslé, A.; El Omari, K.; Wagner, A.; et al. Structure and assembly of the S-layer in C. difficile. Nat. Commun. 2022, 13, 970. [Google Scholar] [CrossRef]
- Madhurantakam, C.; Howorka, S.; Remaut, H. S-layer Structure in Bacteria and Archaea. In Nanomicrobiology; Barton, L., Bazylinski, D., Xu, H., Eds.; Springer: New York, NY, USA, 2014; pp. 11–37. [Google Scholar] [CrossRef]
- Baumeister, W.; Barth, M.; Hegerl, R.; Guckenberger, R.; Hahn, M.; Saxton, W.O. Three-dimensional structure of the regular surface layer (HPI layer) of Deinococcus radiodurans. J. Mol. Biol. 1986, 187, 241–250. [Google Scholar] [CrossRef]
- Lister, T.E.; Pinhero, P.J. In vivo atomic force microscopy of surface proteins on Deinococcus radiodurans. Langmuir 2001, 17, 2624–2628. [Google Scholar] [CrossRef]
- Farci, D.; Kereïche, S.; Pangeni, S.; Haniewicz, P.; Bodrenko, I.V.; Ceccarelli, M.; Winterhalter, M.; Piano, D. Structural analysis of the architecture and in situ localization of the main S-layer complex in Deinococcus radiodurans. Structure 2021, 29, 1279–1285.e3. [Google Scholar] [CrossRef]
- Farci, D.; Haniewicz, P.; de Sanctis, D.; Iesu, L.; Kereïche, S.; Winterhalter, M.; Piano, D. The cryo-EM structure of the S-layer deinoxanthin-binding complex of Deinococcus radiodurans informs properties of its environmental interactions. J. Biol. Chem. 2022, 298, 102031. [Google Scholar] [CrossRef] [PubMed]
- Farci, D.; Aksoyoglu, M.A.; Farci, S.F.; Bafna, J.A.; Bodrenko, I.; Ceccarelli, M.; Kirkpatrick, J.; Winterhalter, M.; Kereïche, S.; Piano, D. Structural insights into the main S-layer unit of Deinococcus radiodurans reveal a massive protein complex with porin-like features. J. Biol. Chem. 2020, 295, 4224–4236. [Google Scholar] [CrossRef] [PubMed]
- Fagan, R.P.; Fairweather, N.F. Biogenesis and functions of bacterial S-layers. Nat. Rev. Microbiol. 2014, 12, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues-Oliveira, T.; Belmok, A.; Vasconcellos, D.; Schuster, B.; Kyaw, C.M. Archaeal S-Layers: Overview and Current State of the Art. Front. Microbiol. 2017, 8, 2597. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Wipfler, R.L.; Li, Y.; Wang, Z.; Hallett, E.N.; Whitaker, R.J. Cell Structure Changes in the Hyperthermophilic Crenarchaeon Sulfolobus islandicus Lacking the S-Layer. mBio 2019, 10, e01589-19. [Google Scholar] [CrossRef] [Green Version]
- Zink, I.A.; Pfeifer, K.; Wimmer, E.; Sleytr, U.B.; Schuster, B.; Schleper, C. CRISPR-mediated gene silencing reveals involvement of the archaeal S-layer in cell division and virus infection. Nat. Commun. 2019, 10, 2540. [Google Scholar] [CrossRef] [Green Version]
- Valentine, D.L. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat. Rev. Microbiol. 2007, 5, 316–323. [Google Scholar] [CrossRef]
- Sara, M.; Sleytr, U.B. S-layer proteins. J. Bacteriol. 2000, 182, 859–868. [Google Scholar] [CrossRef] [Green Version]
- Schuster, B.; Sleytr, U.B. S-Layer Ultrafiltration Membranes. Membranes 2021, 11, 275. [Google Scholar] [CrossRef]
- Pum, D.; Sleytr, U.B. Reassembly of S-layer proteins. Nanotechnology 2014, 25, 312001. [Google Scholar] [CrossRef]
- Pum, D.; Toca-Herrera, J.L.; Sleytr, U.B. S-layer protein self-assembly. Int. J. Mol. Sci. 2013, 14, 2484–2501. [Google Scholar] [CrossRef]
- Sleytr, U.B.; Huber, C.; Ilk, N.; Pum, D.; Schuster, B.; Egelseer, E.M. S-layers as a tool kit for nanobiotechnological applications. FEMS Microbiol. Lett. 2007, 267, 131–144. [Google Scholar] [CrossRef] [Green Version]
- Sleytr, U.B.; Sára, M. Ultrafiltration membranes with uniform pores from crystalline bacterial cell envelope layers. Appl. Microbiol. Biotechnol. 1986, 25, 83–90. [Google Scholar] [CrossRef]
- Pfeifer, K.; Ergal, İ.; Koller, M.; Basen, M.; Schuster, B.; Rittmann, S.K.-M.R. Archaea Biotechnology. Biotechnol. Adv. 2021, 47, 107668. [Google Scholar] [CrossRef]
- Ücisik, M.H.; Küpcü, S.; Debreczeny, M.; Schuster, B.; Sleytr, U.B. S-layer coated emulsomes as potential nanocarriers. Small 2013, 9, 2895–2904. [Google Scholar] [CrossRef]
- Damiati, S.; Schuster, B. Electrochemical Biosensors Based on S-Layer Proteins. Sensors 2020, 20, 1721. [Google Scholar] [CrossRef] [Green Version]
- Tornabene, T.G.; Langworthy, T.A.; Holzer, G.; Oró, J. Squalenes, Phytanes and Other Isoprenoids as Major Neutral Lipids of Methanogenic and Thermoacidophilic “Archaebacteria”. J. Mol. Evol. 1979, 13, 73–83. [Google Scholar] [CrossRef]
- Rastädter, K.; Wurm, D.J.; Spadiut, O.; Quehenberger, J. The Cell Membrane of Sulfolobus spp.—Homeoviscous Adaption and Biotechnological Applications. Int. J. Mol. Sci. 2020, 21, 3935. [Google Scholar] [CrossRef]
- Gambelli, L.; Meyer, B.H.; McLaren, M.; Sanders, K.; Quax, T.E.F.; Gold, V.A.M.; Albers, S.-V.; Daum, B. Architecture and modular assembly of Sulfolobus S-layers revealed by electron cryotomography. Proc. Nat. Acad. Sci. USA 2019, 116, 25278–25286. [Google Scholar] [CrossRef] [Green Version]
- Schuster, B.; Sleytr, U.B. Relevance of glycosylation of S-layer proteins for cell surface properties. Acta Biomater. 2015, 19, 149–157. [Google Scholar] [CrossRef] [Green Version]
- Grogan, D.W. Phenotypic characterization of the archaebacterial genus Sulfolobus: Comparison of five wild-type strains. J. Bacteriol. 1989, 171, 6710–6719. [Google Scholar] [CrossRef] [Green Version]
- Michel, H.; Neugebauer, D.-C.; Oesterhelt, D. The 2-D Crystalline Cell Wall of Sulfolobus acidocaldarius: Structure, Solubilization, and Reassembly. In Electron Microscopy at Molecular Dimensions, Proceedings in Life Sciences; Baumeister, W., Vogel, W., Eds.; Springer: Berlin/Heidelberg, Germany, 1980; pp. 27–35. [Google Scholar] [CrossRef]
- Grogan, D.W. Isolation and fractionation of cell envelope from the extreme thermo-acidophile Sulfolobus acidocaldarius. J. Microbiol. Methods 1996, 26, 35–43. [Google Scholar] [CrossRef]
- Veith, A.; Klingl, A.; Zolghadr, B.; Lauber, K.; Mentele, R.; Lottspeich, F.; Rachel, R.; Albers, S.-V.; Kletzin, A. Acidianus, Sulfolobus and Metallosphaera surface layers: Structure, composition, and gene expression. Mol. Microbiol. 2009, 73, 58–72. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Sára, M.; Sleytr, U.B. Production and characteristics of ultrafiltration membranes with uniform pores from two-dimensional arrays of proteins. J. Membr. Sci. 1987, 33, 27–49. [Google Scholar] [CrossRef]
- Sára, M.; Manigley, C.; Wolf, G.; Sleytr, U.B. Isoporous ultrafiltration membranes from bacterial cell envelope layers. J. Membr. Sci. 1988, 36, 179–186. [Google Scholar] [CrossRef]
- Sára, M.; Küpcü, S.; Sleytr, U.B. Crystalline bacterial cell surface layers used as ultrafiltration membranes and immobilization matrix. Gen. Eng. Biotechnol. 1990, 10, 10–13. [Google Scholar]
- Weigert, S.; Sára, M. Surface modification of an ultrafiltration membrane with crystalline structure and studies on interactions with selected protein molecules. J. Membr. Sci. 1995, 106, 147–159. [Google Scholar] [CrossRef]
- Sára, M.; Sleytr, U.B. Molecular sieving through S-layers of Bacillus stearothermophilus strains. J. Bacteriol. 1987, 169, 4092–4098. [Google Scholar] [CrossRef] [Green Version]
- Picher, M.M.; Küpcü, S.; Dostalek, J.; Pum, D.; Sleytr, U.B.; Ertl, P. Nanobiotechnology advanced antifouling surfaces for the continuous electrochemical monitoring of glucose in whole blood using a lab-on-a-chip. Lab Chip 2013, 13, 1780–1789. [Google Scholar] [CrossRef]
- Weiner, C.; Sára, M.; Sleytr, U.B. Novel protein A affinity matrix prepared from two-dimensional protein crystals. Biotechnol. Bioeng. 1994, 43, 321–330. [Google Scholar] [CrossRef]
- Küpcü, S.; Sara, M.; Sleytr, U.B. Influence of covalent attachment of low molecular weight substances on the rejection and adsorption properties of crystalline proteinaceous ultrafiltration membranes. Desalination 1993, 90, 65–76. [Google Scholar] [CrossRef]
- Egelseer, E.M.; Ilk, N.; Pum, D.; Messner, P.; Schäffer, C.; Schuster, B.; Sleytr, U.B. S-Layers, Microbial, Biotechnological Applications. In Encyclopedia of Industrial Biotechnology: Bioprocess, Bioseparation, and Cell Technology; Flickinger, M.C., Ed.; Wiley-VCH Verlag: Weinheim, Germany, 2010; pp. 4424–4448. [Google Scholar]
- Breitwieser, A.; Küpcü, S.; Howorka, S.; Weigert, S.; Langer, C.; Hoffmann-Sommergruber, K.; Scheiner, O.; Sleytr, U.B.; Sára, M. 2-D Protein Crystals as an Immobilization Matrix for Producing Reaction Zones in Dipstick-Style Immunoassays. BioTechniques 1996, 21, 918–925. [Google Scholar] [CrossRef] [Green Version]
- Schuster, B.; Pum, D.; Sára, M.; Braha, O.; Bayley, H.; Sleytr, U.B. S-layer ultrafiltration membranes: A new support for stabilizing functionalized lipid membranes. Langmuir 2001, 17, 499–503. [Google Scholar] [CrossRef]
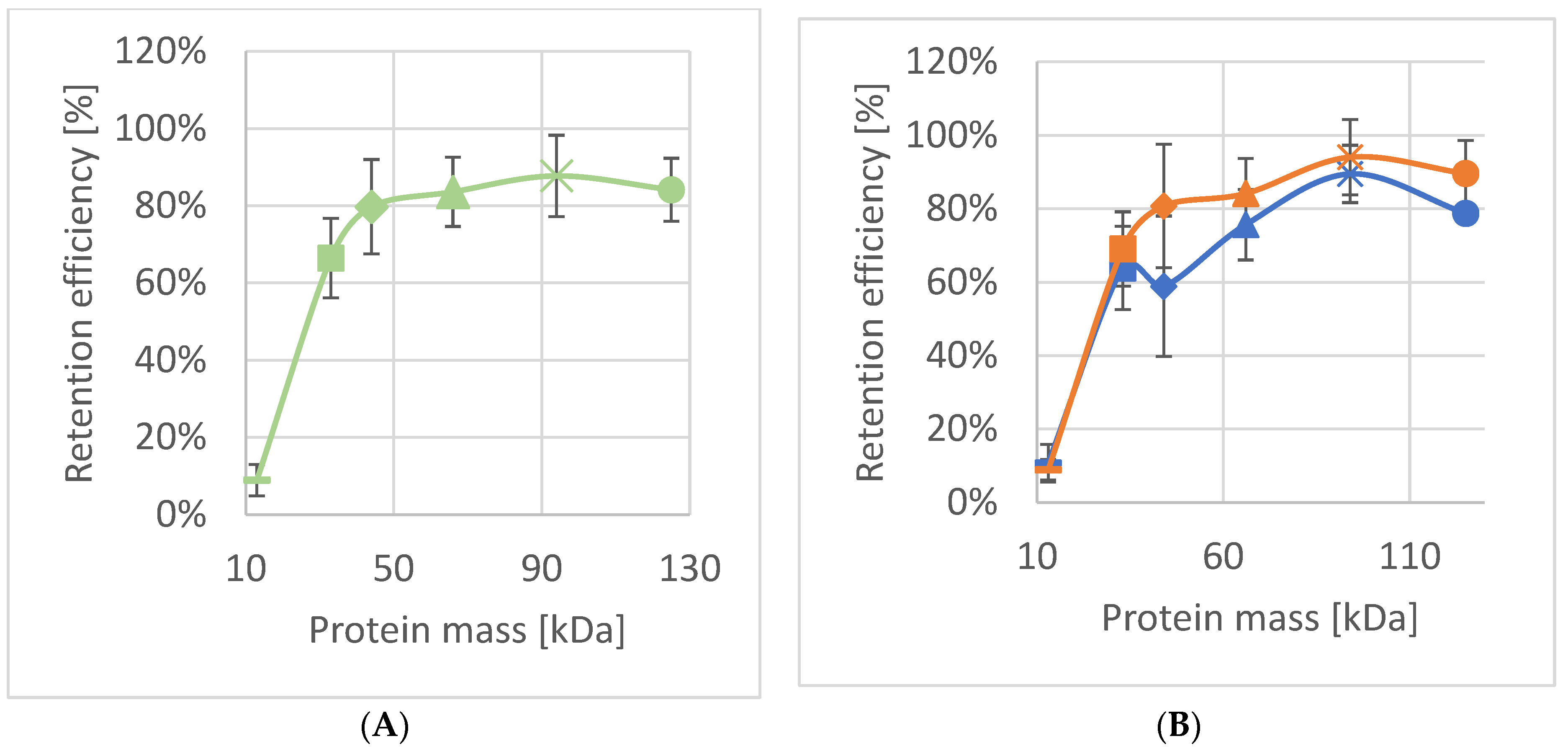

| Protein | Molec. Mass Mr (kDa) | Molecular Size (nm) | Retention Efficiency SSOMF1 | Retention Efficiency SSOMF2 |
|---|---|---|---|---|
| Myoglobin | 17 | 4.4 × 4.4 × 2.5 | 9% ± 4%; n = 9 | 12% ± 7%; n = 4 |
| Carbonic anhydrase | 31 | 4.1 × 4.1 × 4.7 | 66% ± 10%; n = 9 | 63% ± 3%; n = 4 |
| Horseradish peroxidase | 44 | 4.0 × 6.7 × 11.7 | 80% ± 12%; n = 6 | 70% ± 5%; n = 5 |
| Bovine serum albumin | 66 | 4.0 × 4.0 × 14.0 | 84% ± 9%; n = 7 | 97% ± 3%; n = 4 |
| Amylo-glucosidase | 97 | 5.7 × 7.3 × 10.7 | 88% ± 11%; n = 9 | 98% ± 2%; n = 4 |
| γ-globulin | 125 | 4.5 × 8.4 × 14.5 | 84% ± 8%; n = 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pfeifer, K.; Ehmoser, E.-K.; Rittmann, S.K.-M.R.; Schleper, C.; Pum, D.; Sleytr, U.B.; Schuster, B. Isolation and Characterization of Cell Envelope Fragments Comprising Archaeal S-Layer Proteins. Nanomaterials 2022, 12, 2502. https://doi.org/10.3390/nano12142502
Pfeifer K, Ehmoser E-K, Rittmann SK-MR, Schleper C, Pum D, Sleytr UB, Schuster B. Isolation and Characterization of Cell Envelope Fragments Comprising Archaeal S-Layer Proteins. Nanomaterials. 2022; 12(14):2502. https://doi.org/10.3390/nano12142502
Chicago/Turabian StylePfeifer, Kevin, Eva-Kathrin Ehmoser, Simon K.-M. R. Rittmann, Christa Schleper, Dietmar Pum, Uwe B. Sleytr, and Bernhard Schuster. 2022. "Isolation and Characterization of Cell Envelope Fragments Comprising Archaeal S-Layer Proteins" Nanomaterials 12, no. 14: 2502. https://doi.org/10.3390/nano12142502
APA StylePfeifer, K., Ehmoser, E.-K., Rittmann, S. K.-M. R., Schleper, C., Pum, D., Sleytr, U. B., & Schuster, B. (2022). Isolation and Characterization of Cell Envelope Fragments Comprising Archaeal S-Layer Proteins. Nanomaterials, 12(14), 2502. https://doi.org/10.3390/nano12142502







