Shrinkable Hydrogel-Enhanced Biomarker Detection with X-ray Fluorescent Nanoparticles
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
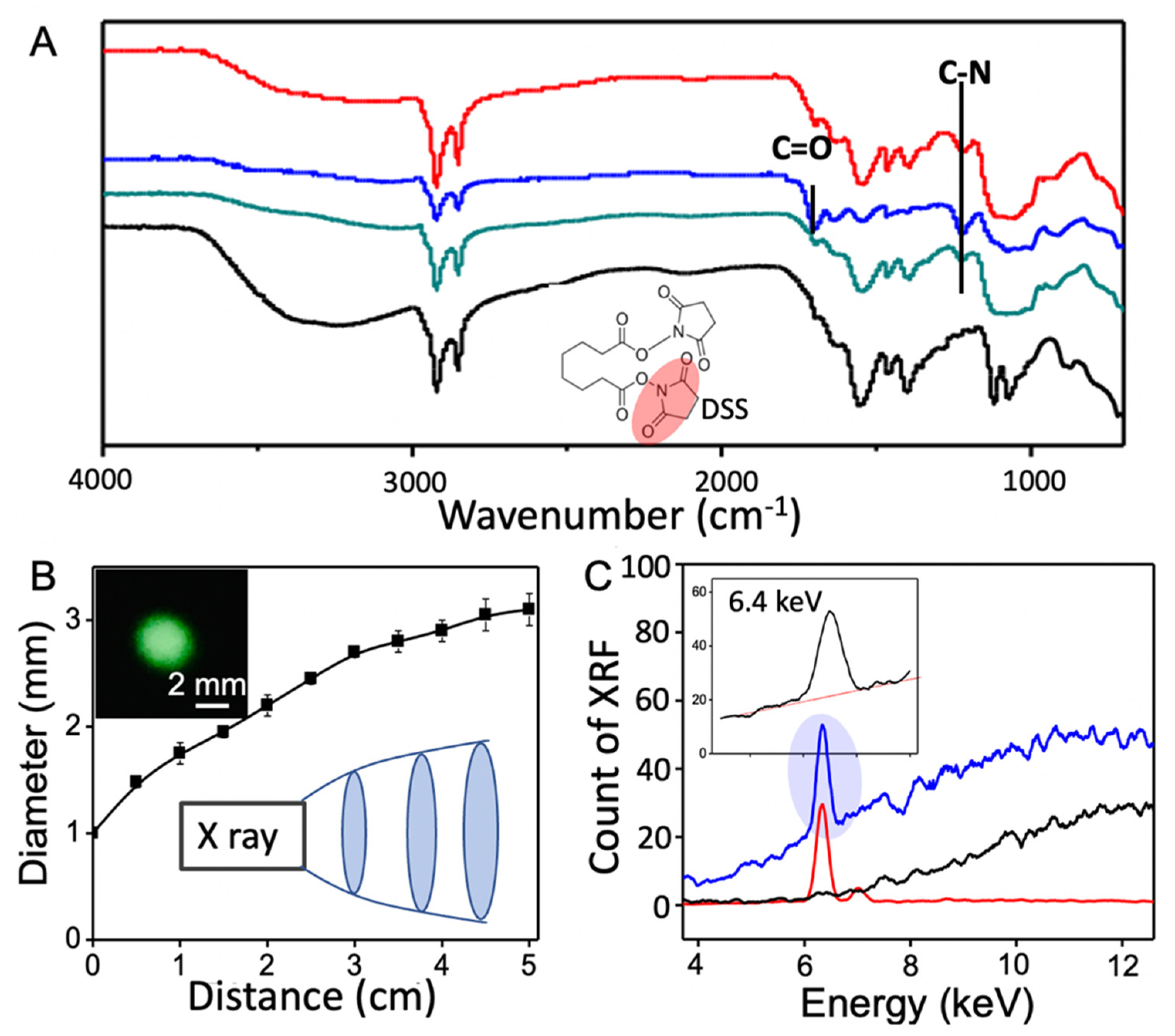
2.2. Chemical Modification of Agarose
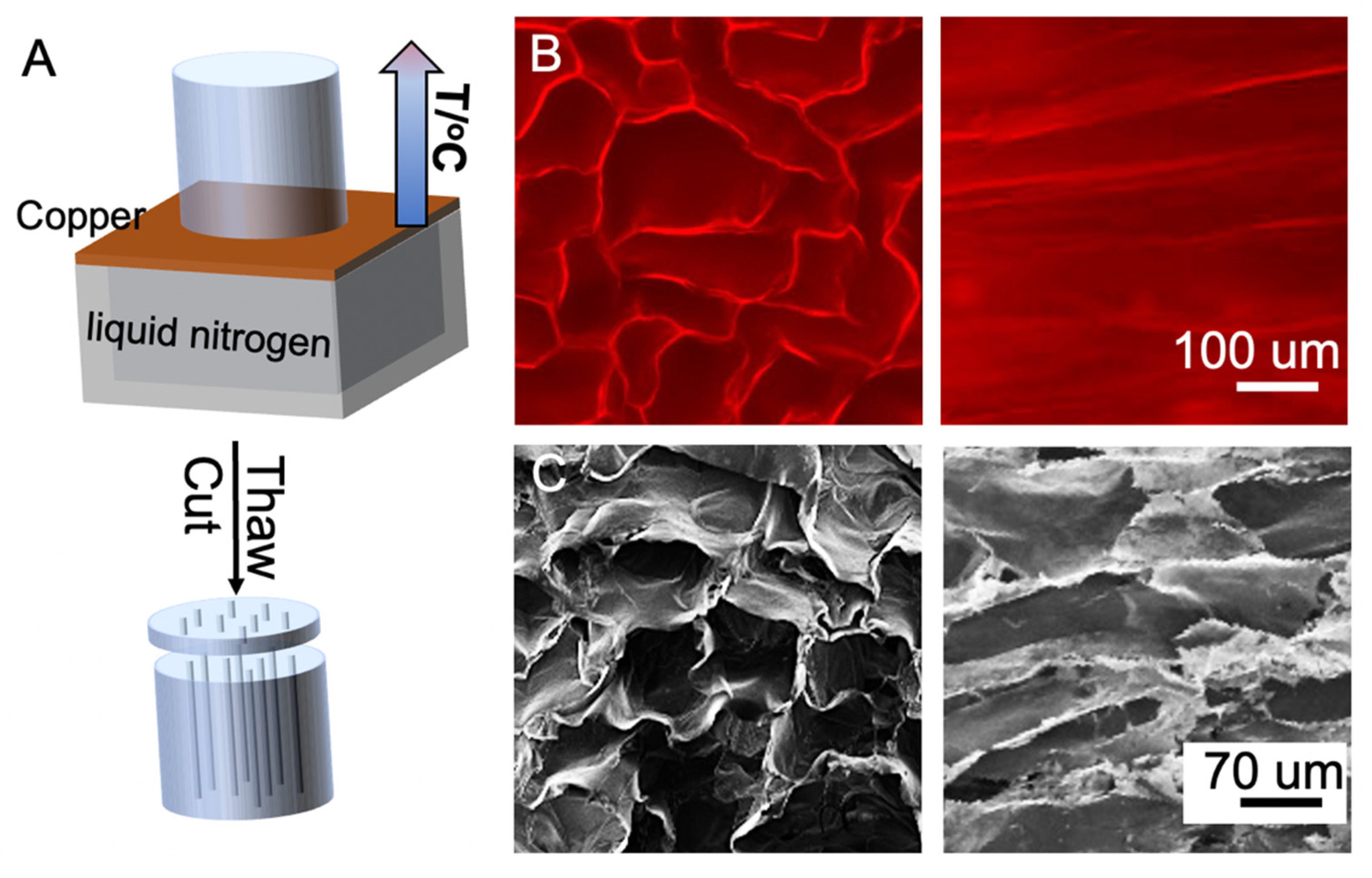
2.3. Preparation of Hydrogels with Aligned Channels
2.4. Biotin-Conjugation on Iron Oxide Nanoparticles
2.5. Protein Detection with X-ray Fluorescence
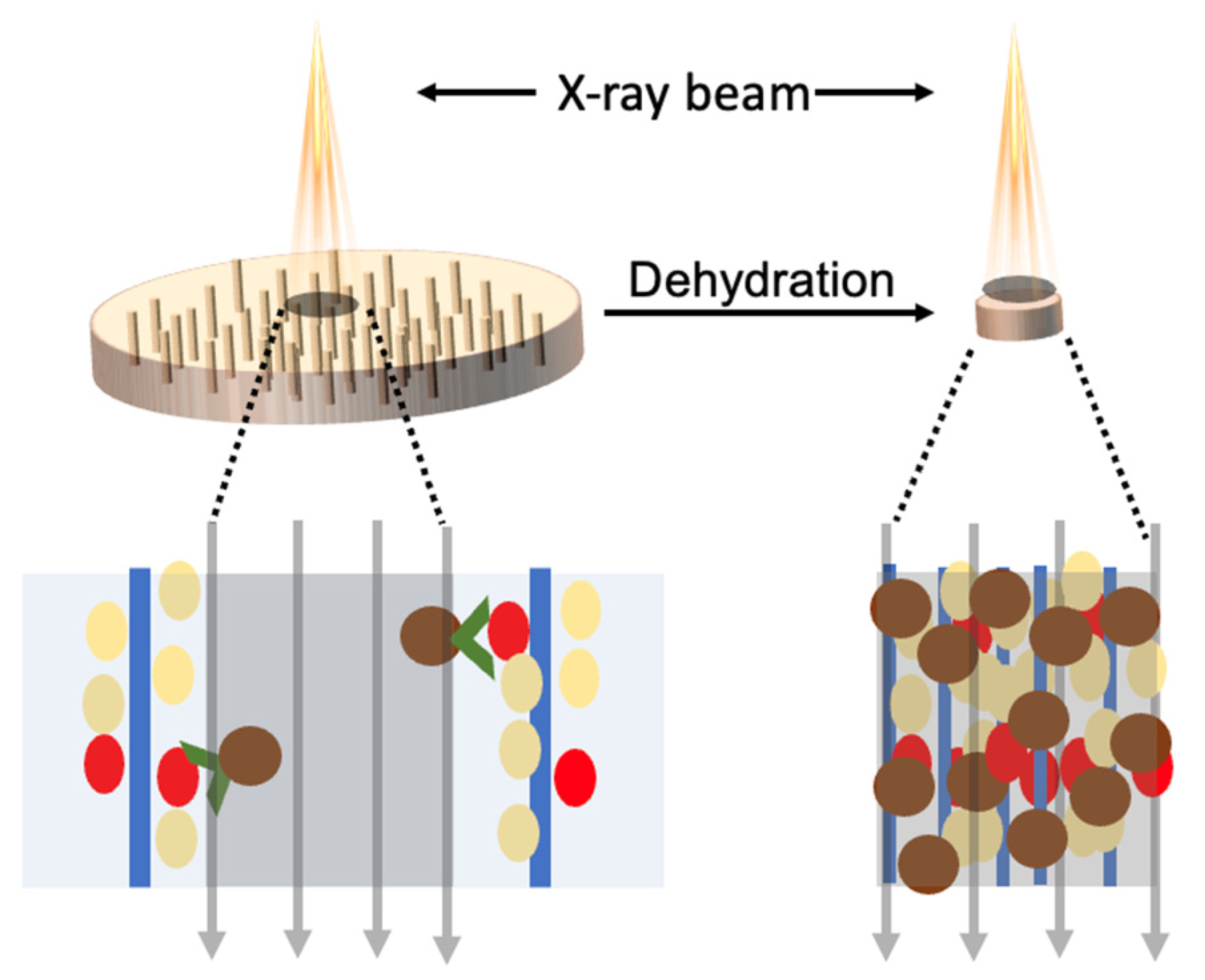
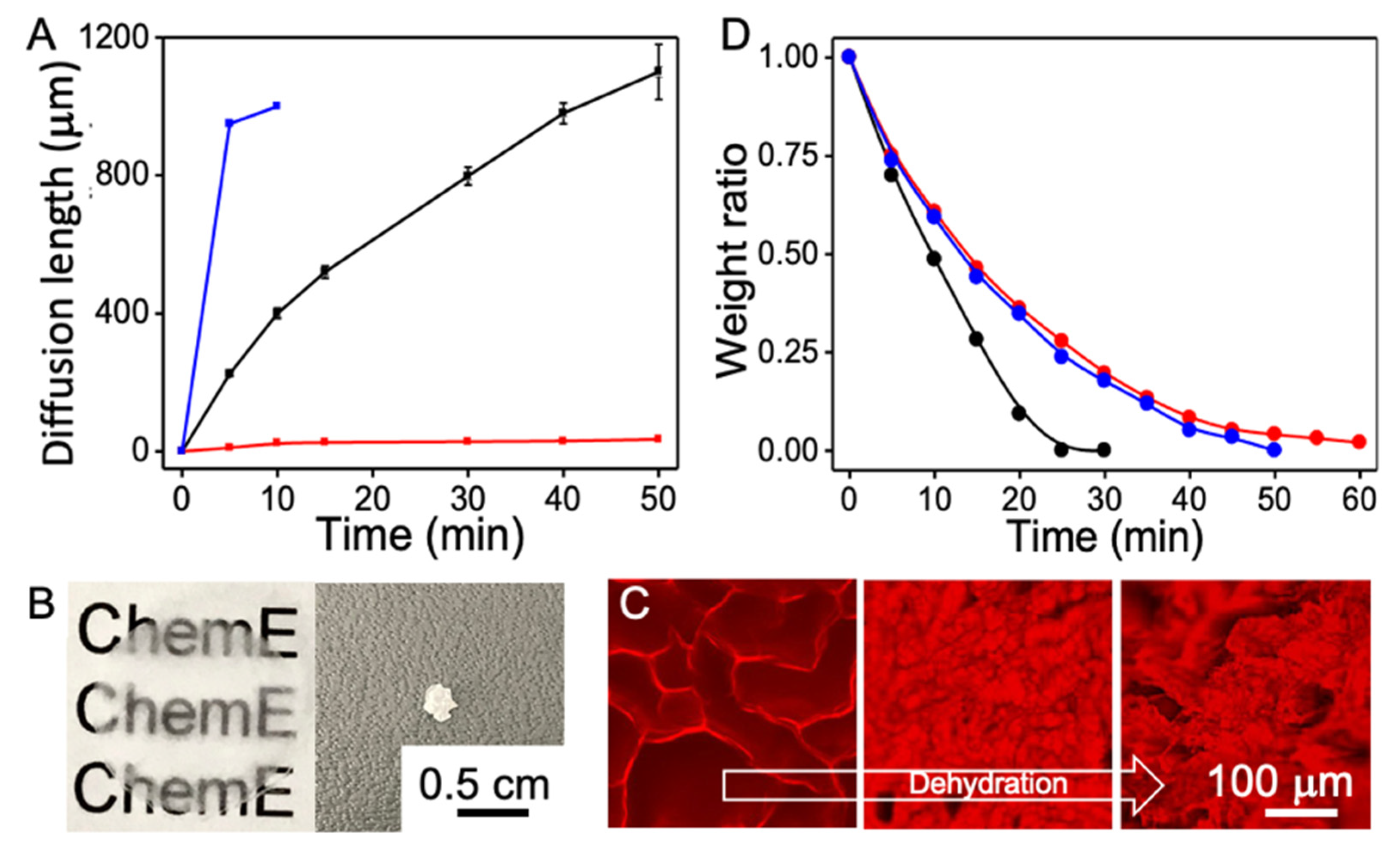
3. Results and Discussions
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Chinen, A.B.; Guan, C.M.; Ferrer, J.R.; Barnaby, S.N.; Merkel, T.J.; Mirkin, C.A. Nanoparticle Probes for the Detection of Cancer Biomarkers, Cells, and Tissues by Fluorescence. Chem. Rev. 2015, 115, 10530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swierczewska, M.; Liu, G.; Lee, S.; Chen, X. High-Sensitivity Nanosensors for Biomarker Detection. Chem. Soc. Rev. 2012, 41, 2641. [Google Scholar] [CrossRef] [PubMed]
- Thaxton, C.S.; Georganopoulou, D.G.; Mirkin, C.A. Gold Nanoparticle Probes for the Detection of Nucleic Acid Targets. Clin. Chim. Acta 2006, 363, 120. [Google Scholar] [CrossRef] [PubMed]
- Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Nanoparticles for Nucleic-Acid-Based Biosensing: Opportunities, Challenges, and Prospects. Anal. Bioanal. Chem. 2019, 411, 1791. [Google Scholar] [CrossRef]
- Perfézou, M.; Turner, A.; Merkoçi, A. Cancer Detection Using Nanoparticle-Based Sensors. Chem. Soc. Rev. 2012, 41, 2606. [Google Scholar] [CrossRef]
- Luo, X.; Morrin, A.; Killard, A.J.; Smyth, M.R. Application of Nanoparticles in Electrochemical Sensors and Biosensors. Electroanalysis 2006, 18, 319. [Google Scholar] [CrossRef] [Green Version]
- Milyutin, Y.; Abud-Hawa, M.; Kloper-Weidenfeld, V.; Mansour, E.; Broza, Y.Y.; Shani, G.; Haick, H. Fabricating and Printing Chemiresistors Based on Monolayer-Capped Metal Nanoparticles. Nat. Protoc. 2021, 16, 2968. [Google Scholar] [CrossRef]
- Kim, D.-S.; Kang, E.-S.; Baek, S.; Choo, S.-S.; Chung, Y.-H.; Lee, D.; Min, J.; Kim, T.-H. Electrochemical Detection of Dopamine Using Periodic Cylindrical Gold Nanoelectrode Arrays. Sci. Rep. 2018, 8, 14049. [Google Scholar] [CrossRef] [Green Version]
- Xia, F.; White, R.J.; Zuo, X.; Patterson, A.; Xiao, Y.; Kang, D.; Gong, X.; Plaxco, K.W.; Heeger, A.J. An Electrochemical Supersandwich Assay for Sensitive and Selective DNA Detection in Complex Matrices. J. Am. Chem. Soc. 2010, 132, 14346. [Google Scholar] [CrossRef] [Green Version]
- Sikes, H.D.; Hansen, R.R.; Johnson, L.M.; Jenison, R.; Birks, J.W.; Rowlen, K.L.; Bowman, C.N. Using Polymeric Materials to Generate an Amplified Response to Molecular Recognition Events. Nat. Mater. 2008, 7, 52. [Google Scholar] [CrossRef] [Green Version]
- Appleyard, D.C.; Chapin, S.C.; Srinivas, R.L.; Doyle, P.S. Bar-Coded Hydrogel Microparticles for Protein Detection: Synthesis, Assay and Scanning. Nat. Protoc. 2011, 6, 1761. [Google Scholar] [CrossRef] [PubMed]
- Savina, I.N.; Ingavle, G.C.; Cundy, A.B.; Mikhalovsky, S.V. A Simple Method for the Production of Large Volume 3d Macroporous Hydrogels for Advanced Biotechnological, Medical and Environmental Applications. Sci. Rep. 2016, 6, 21154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meiring, J.E.; Schmid, M.J.; Grayson, S.M.; Rathsack, B.M.; Johnson, D.M.; Kirby, R.; Kannappan, R.; Manthiram, K.; Hsia, B.; Hogan, Z.L.; et al. Hydrogel Biosensor Array Platform Indexed by Shape. Chem. Mater. 2004, 16, 5574. [Google Scholar] [CrossRef]
- Zubtsov, D.A.; Savvateeva, E.N.; Rubina, A.Y.; Pan’kov, S.V.; Konovalova, E.V.; Moiseeva, O.V.; Chechetkin, V.R.; Zasedatelev, A.S. Comparison of Surface and Hydrogel-Based Protein Microchips. Anal. Biochem. 2007, 368, 205. [Google Scholar] [CrossRef] [PubMed]
- Burdick, J.A.; Prestwich, G.D. Hyaluronic Acid Hydrogels for Biomedical Applications. Adv. Mater. 2011, 23, H41. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.L.; Moshe, M.; Greener, J.; Therien-Aubin, H.; Nie, Z.; Sharon, E.; Kumacheva, E. Three-Dimensional Shape Transformations of Hydrogel Sheets Induced by Small-Scale Modulation of Internal Stresses. Nat. Commun. 2013, 4, 1586. [Google Scholar] [CrossRef]
- Kudo, K.; Ishida, J.; Syuu, G.; Sekine, Y.; Ikeda-Fukazawa, T. Structural Changes of Water in Poly(Vinyl Alcohol) Hydrogel During Dehydration. J. Chem. Phys. 2014, 140, 44909. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, Q.; Wang, C.; Su, M. Enhanced Turn-on Fluorescence Detection of Aqueous Lead Ions with Size-Shrinkable Hydrogels. ACS Omega 2021, 6, 11897. [Google Scholar] [CrossRef]
- Guschin, D.; Yershov, G.; Zaslavsky, A.; Gemmell, A.; Shick, V.; Proudnikov, D.; Arenkov, P.; Mirzabekov, A. Manual Manufacturing of Oligonucleotide, DNA, and Protein Microchips. Anal. Biochem. 1997, 250, 203. [Google Scholar] [CrossRef] [Green Version]
- Buenger, D.; Topuz, F.; Groll, J. Hydrogels in Sensing Applications. Prog. Polym. Sci. 2012, 37, 1678. [Google Scholar] [CrossRef]
- Sandrin, D.; Wagner, D.; Sitta, C.E.; Thoma, R.; Felekyan, S.; Hermes, H.E.; Janiak, C.; de Sousa Amadeu, N.; Kühnemuth, R.; Löwen, H.; et al. Diffusion of Macromolecules in a Polymer Hydrogel: From Microscopic to Macroscopic Scales. Phys. Chem. Chem. Phys. 2016, 18, 12860. [Google Scholar] [CrossRef] [PubMed]
- Axpe, E.; Chan, D.; Offeddu, G.S.; Chang, Y.; Merida, D.; Hernandez, H.L.; Appel, E.A. A Multiscale Model for Solute Diffusion in Hydrogels. Macromolecules 2019, 52, 6889. [Google Scholar] [CrossRef] [Green Version]
- Stokols, S.; Tuszynski, M.H. Freeze-Dried Agarose Scaffolds with Uniaxial Channels Stimulate and Guide Linear Axonal Growth Following Spinal Cord Injury. Biomaterials 2006, 27, 443. [Google Scholar] [CrossRef] [PubMed]
- Hua, M.; Wu, S.; Ma, Y.; Zhao, Y.; Chen, Z.; Frenkel, I.; Strzalka, J.; Zhou, H.; Zhu, X.; He, X. Strong Tough Hydrogels Via the Synergy of Freeze-Casting and Salting Out. Nature 2021, 590, 594. [Google Scholar] [CrossRef] [PubMed]
- Manohar, N.; Reynoso, F.J.; Diagaradjane, P.; Krishnan, S.; Cho, S.H. Quantitative Imaging of Gold Nanoparticle Distribution in a Tumor-Bearing Mouse Using Benchtop X-Ray Fluorescence Computed Tomography. Sci. Rep. 2016, 6, 22079. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Li, Y.; Wong, M.D.; Liu, H. A Method of Measuring Gold Nanoparticle Concentrations by X-ray Fluorescence for Biomedical Applications. Med. Phys. 2013, 40, 51901. [Google Scholar] [CrossRef]
- Hossain, M.; Wang, C.; Su, M. Multiplexed Biomarker Detection Using X-Ray Fluorescence of Composition-Encoded Nanoparticles. Appl. Phys. Lett. 2010, 97, 263704. [Google Scholar] [CrossRef] [Green Version]
- Landmark, K.J.; DiMaggio, S.; Ward, J.; Kelly, C.; Vogt, S.; Hong, S.; Kotlyar, A.; Myc, A.; Thomas, T.P.; Penner-Hahn, J.E.; et al. Synthesis, Characterization, and in Vitro Testing of Superparamagnetic Iron Oxide Nanoparticles Targeted Using Folic Acid-Conjugated Dendrimers. ACS Nano 2008, 2, 773. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Huo, R.; Su, M. Shrinkable Hydrogel-Enhanced Biomarker Detection with X-ray Fluorescent Nanoparticles. Nanomaterials 2022, 12, 2412. https://doi.org/10.3390/nano12142412
Zheng Y, Huo R, Su M. Shrinkable Hydrogel-Enhanced Biomarker Detection with X-ray Fluorescent Nanoparticles. Nanomaterials. 2022; 12(14):2412. https://doi.org/10.3390/nano12142412
Chicago/Turabian StyleZheng, Yiting, Ruiqing Huo, and Ming Su. 2022. "Shrinkable Hydrogel-Enhanced Biomarker Detection with X-ray Fluorescent Nanoparticles" Nanomaterials 12, no. 14: 2412. https://doi.org/10.3390/nano12142412
APA StyleZheng, Y., Huo, R., & Su, M. (2022). Shrinkable Hydrogel-Enhanced Biomarker Detection with X-ray Fluorescent Nanoparticles. Nanomaterials, 12(14), 2412. https://doi.org/10.3390/nano12142412




