Solid-Phase Synthesized Copolymers for the Assembly of pH-Sensitive Micelles Suitable for Drug Delivery Applications
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Determination of the Critical Micelle Concentration of the Three Copolymers
2.3. Assembly of Copolymers in Unloaded Micelles and DOX-Loaded Micelles
2.4. Encapsulation Efficiency (EE) and Drug Loading (DL) of DOX
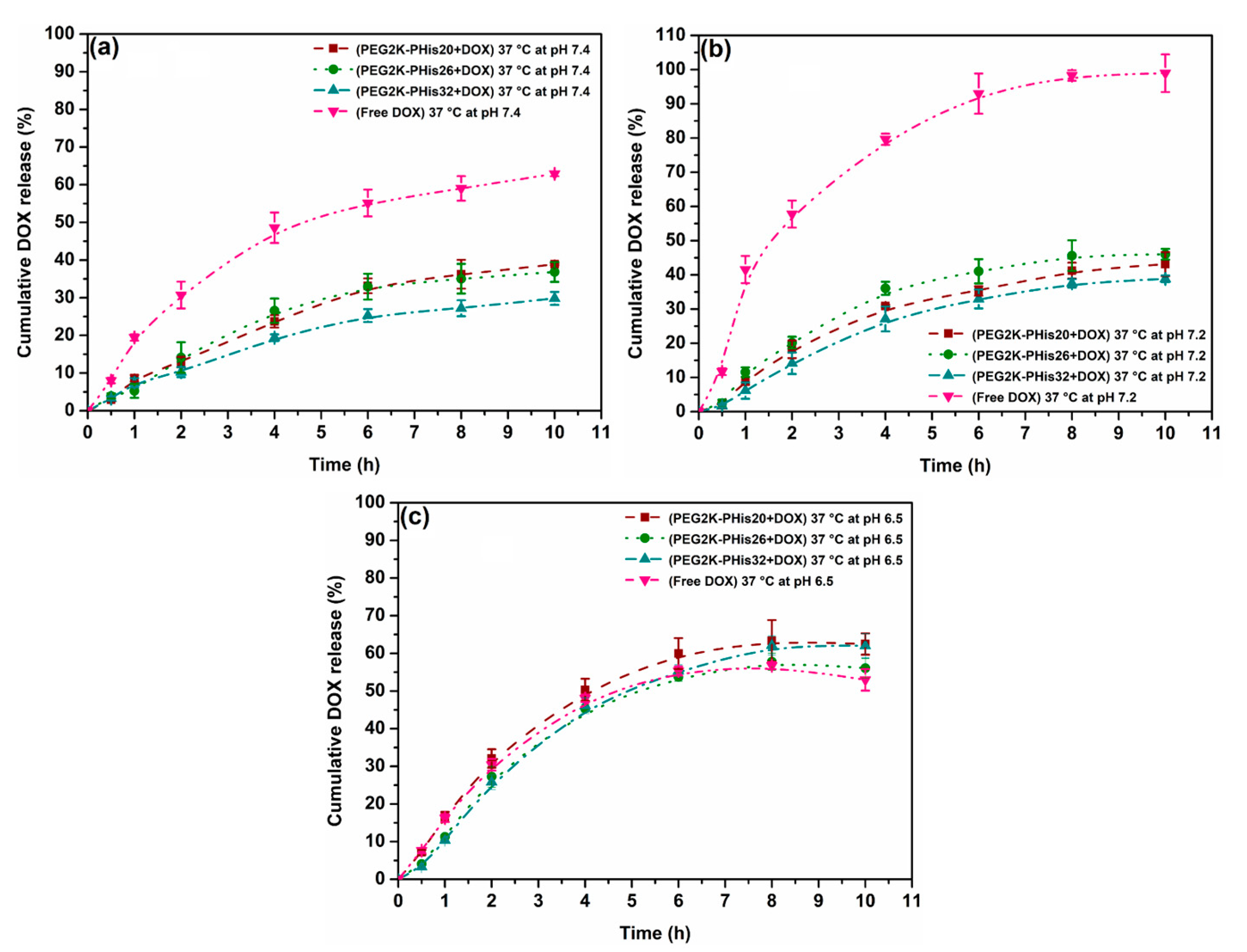
2.5. In Vitro pH-Dependent Release Studies
2.6. Characterization
2.7. Cell Culture
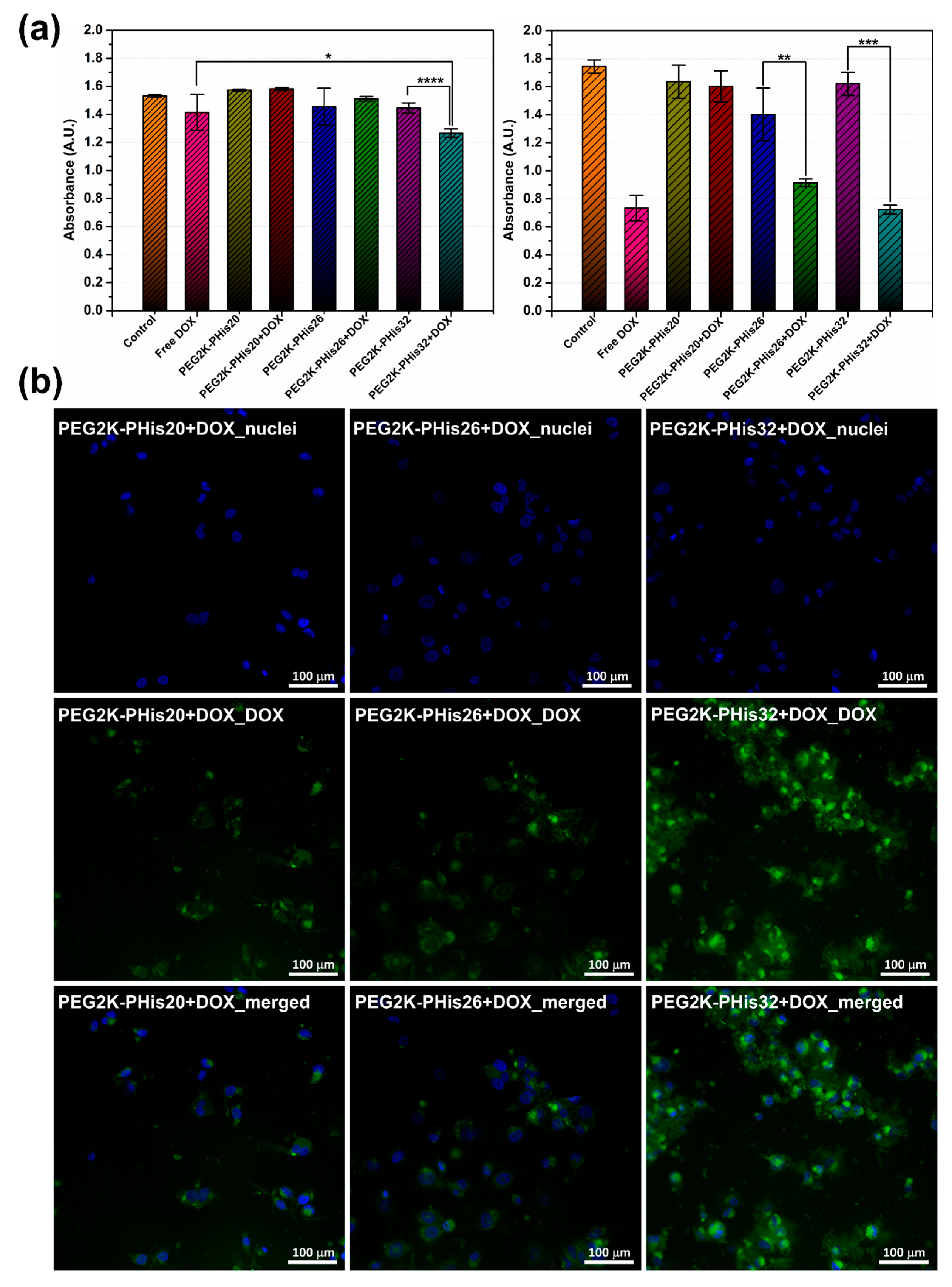
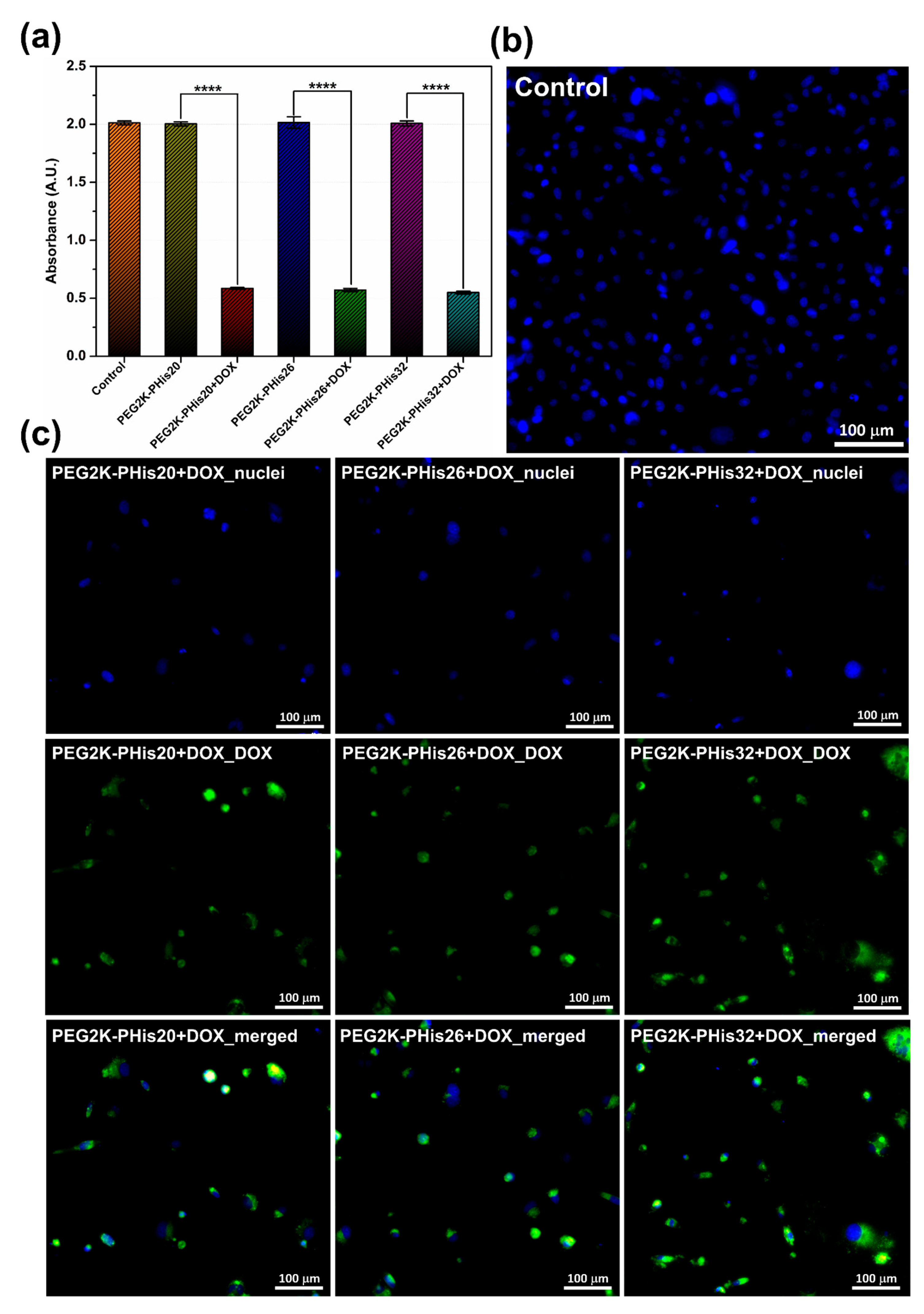
2.8. Cell Viability
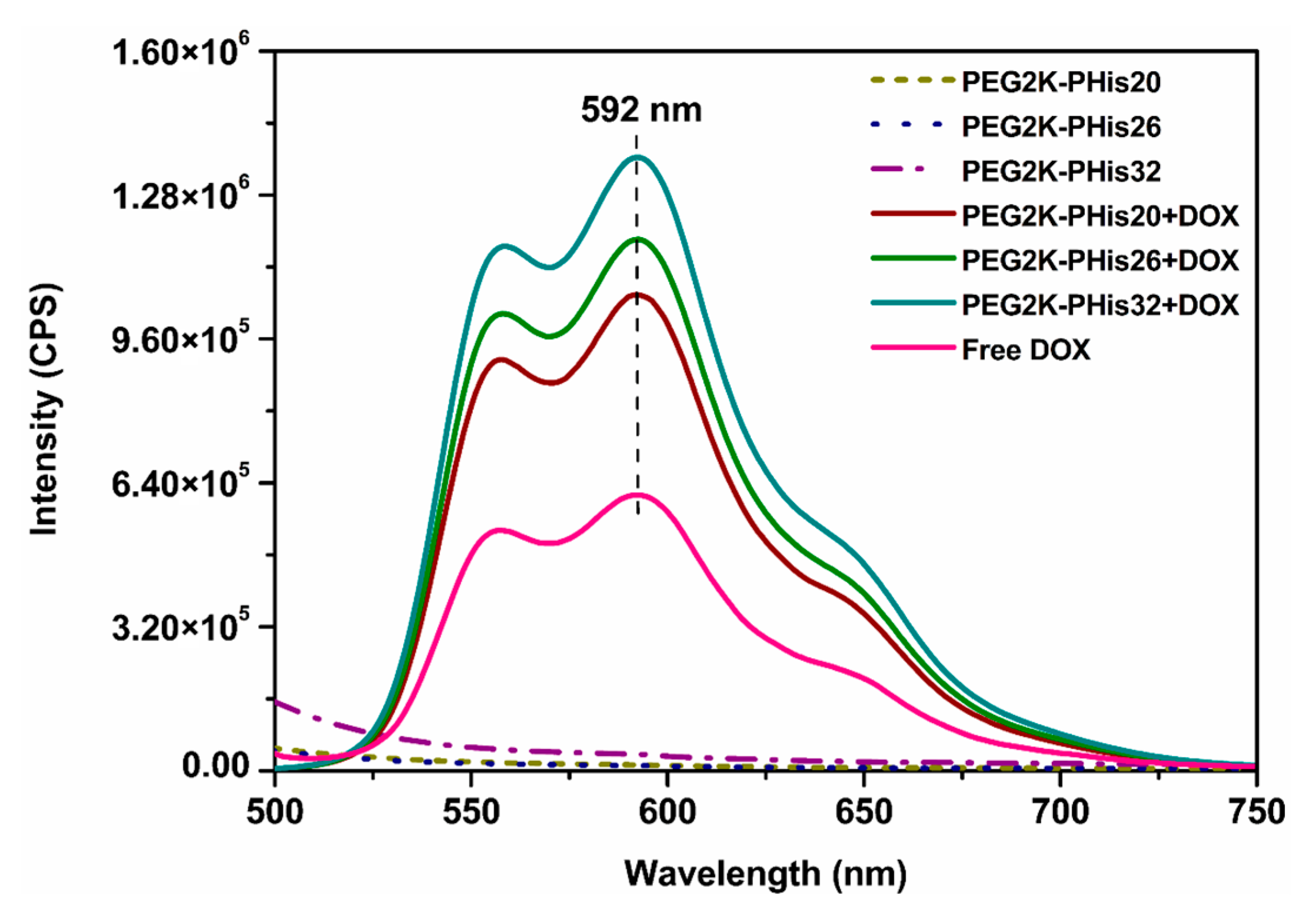
2.9. Fluorescence Measurements
2.10. Statistical Analysis
3. Results and Discussion
3.1. Synthesis of Copolymers
3.2. Critical Micelle Concentration (CMC)
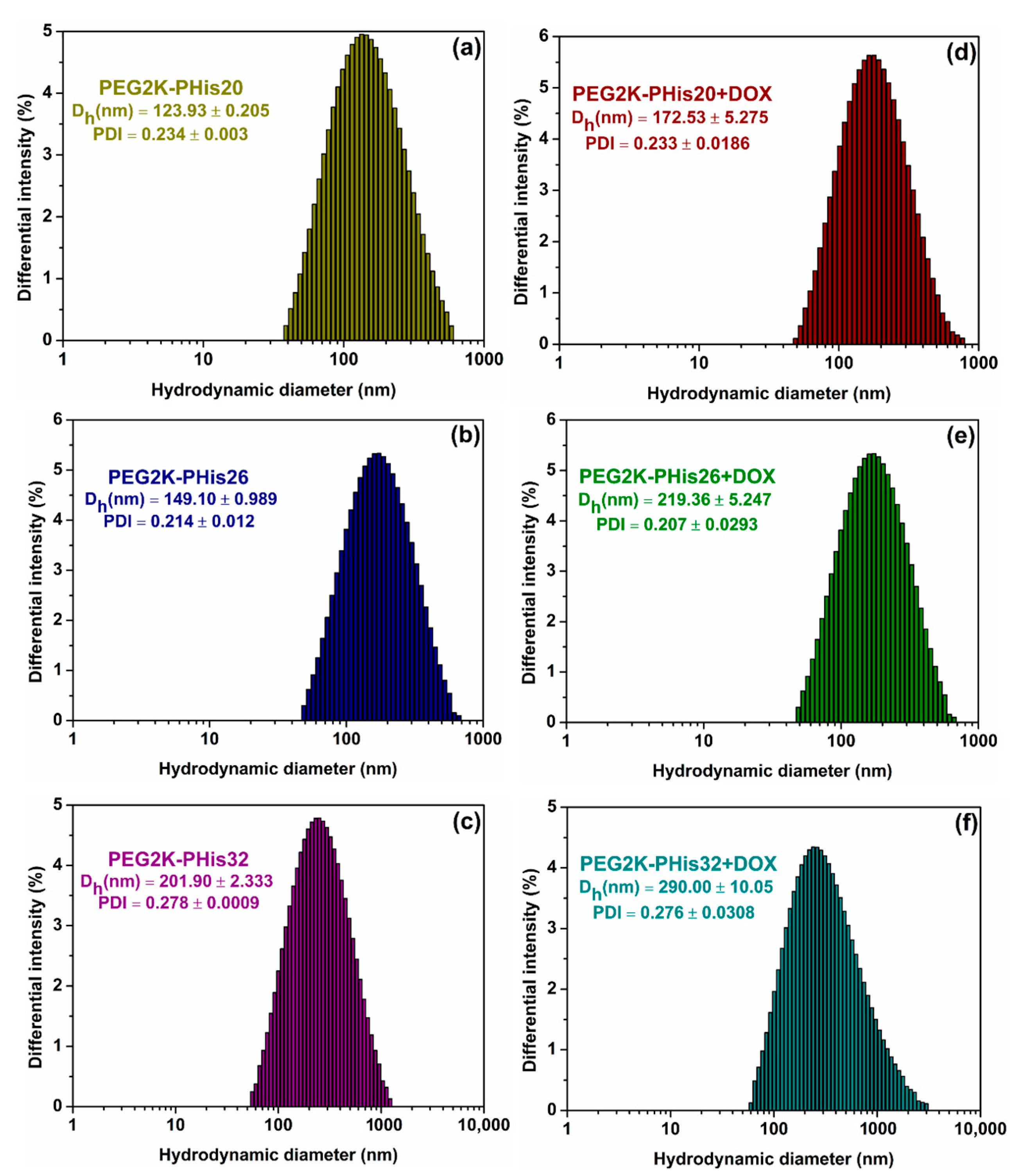
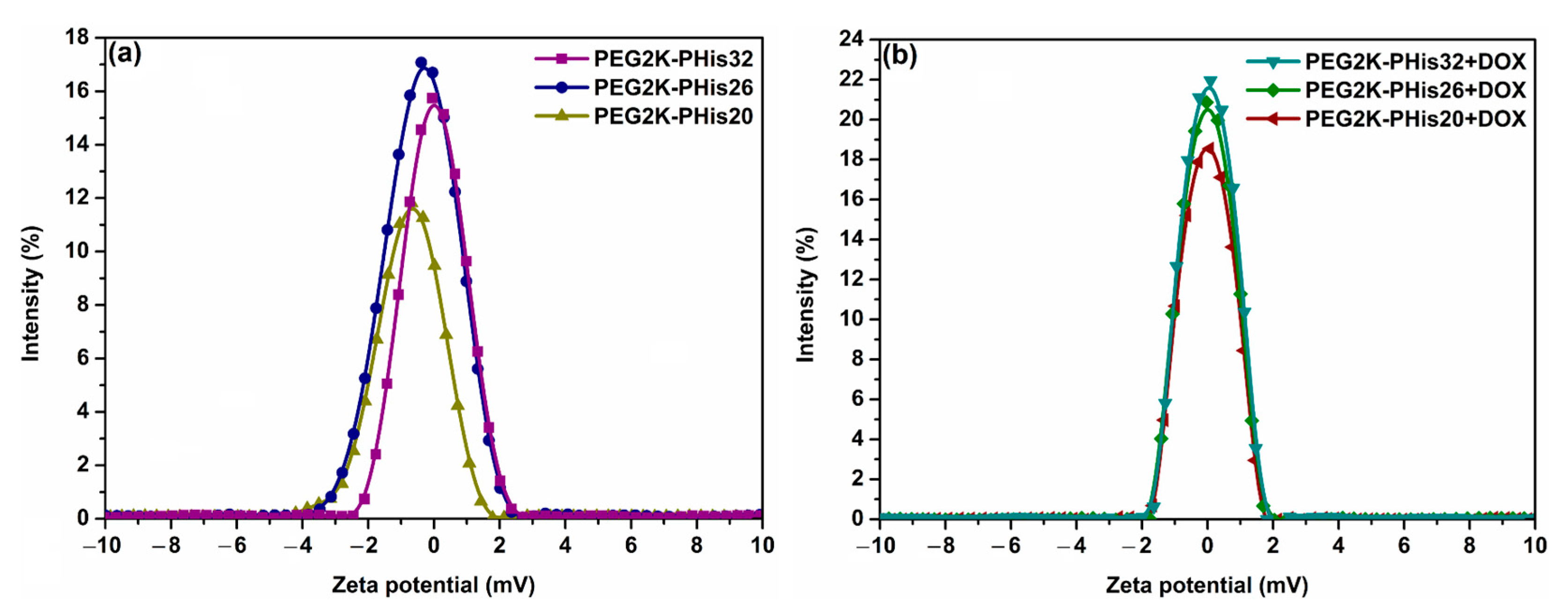
3.3. Dynamic Light Scattering (DLS) and Zeta Potential (ζ)
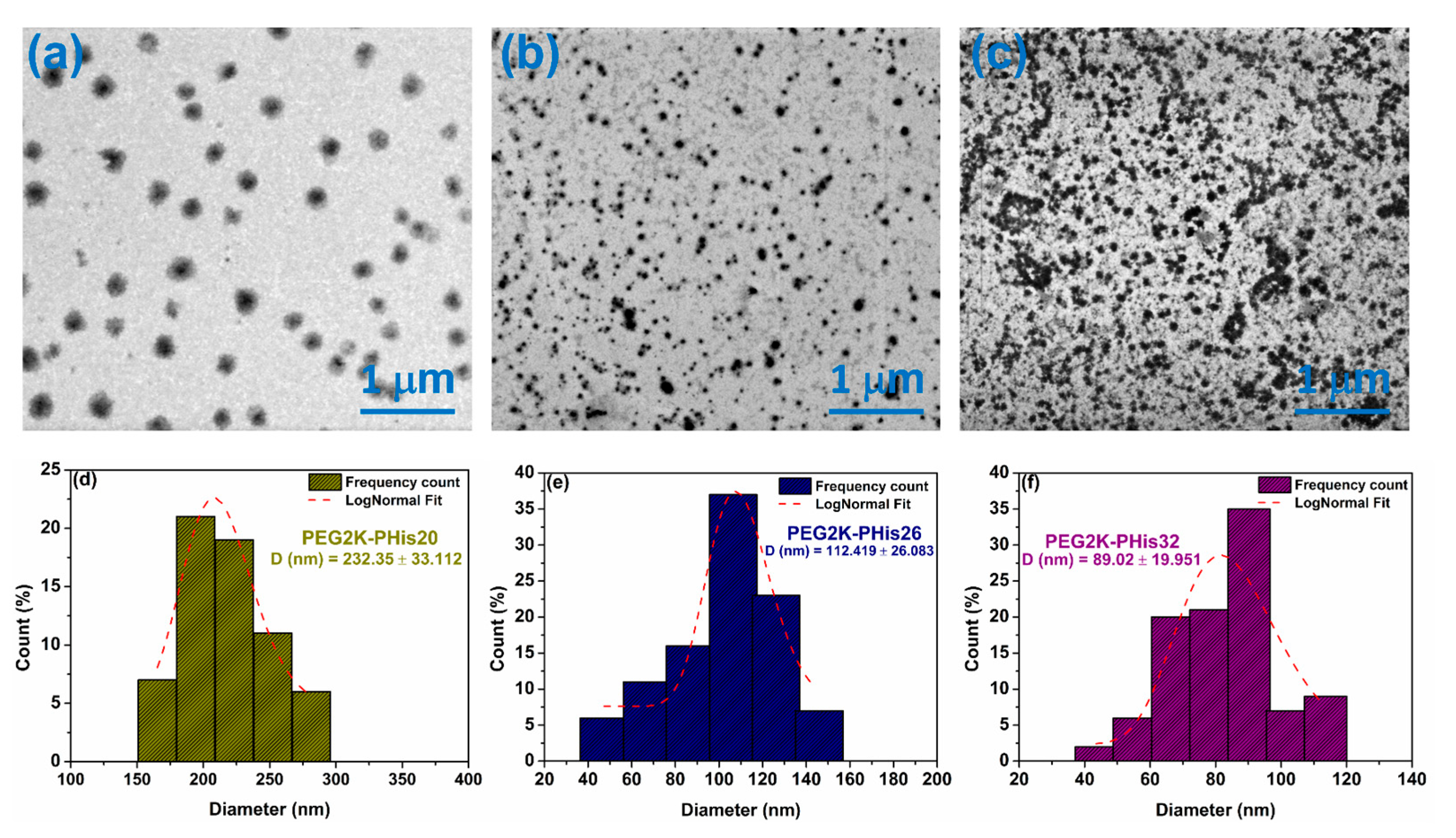
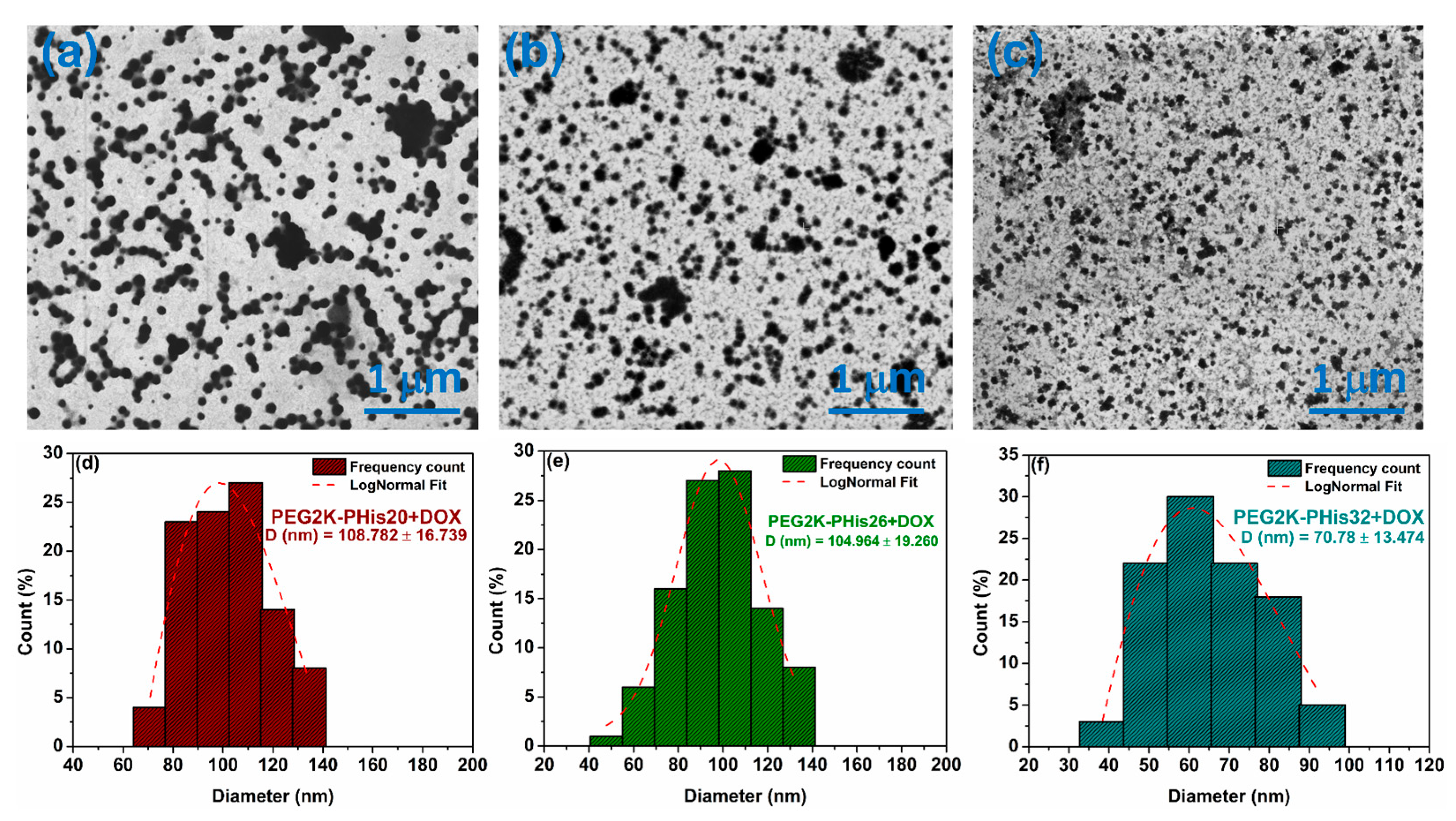
3.4. Scanning Transmission Electron Microscopy (STEM) of Micelles
3.5. Encapsulation Efficiency (EE) and Drug Loading (DL) of DOX in Micelles
3.6. pH-Triggered Doxorubicin Release Study
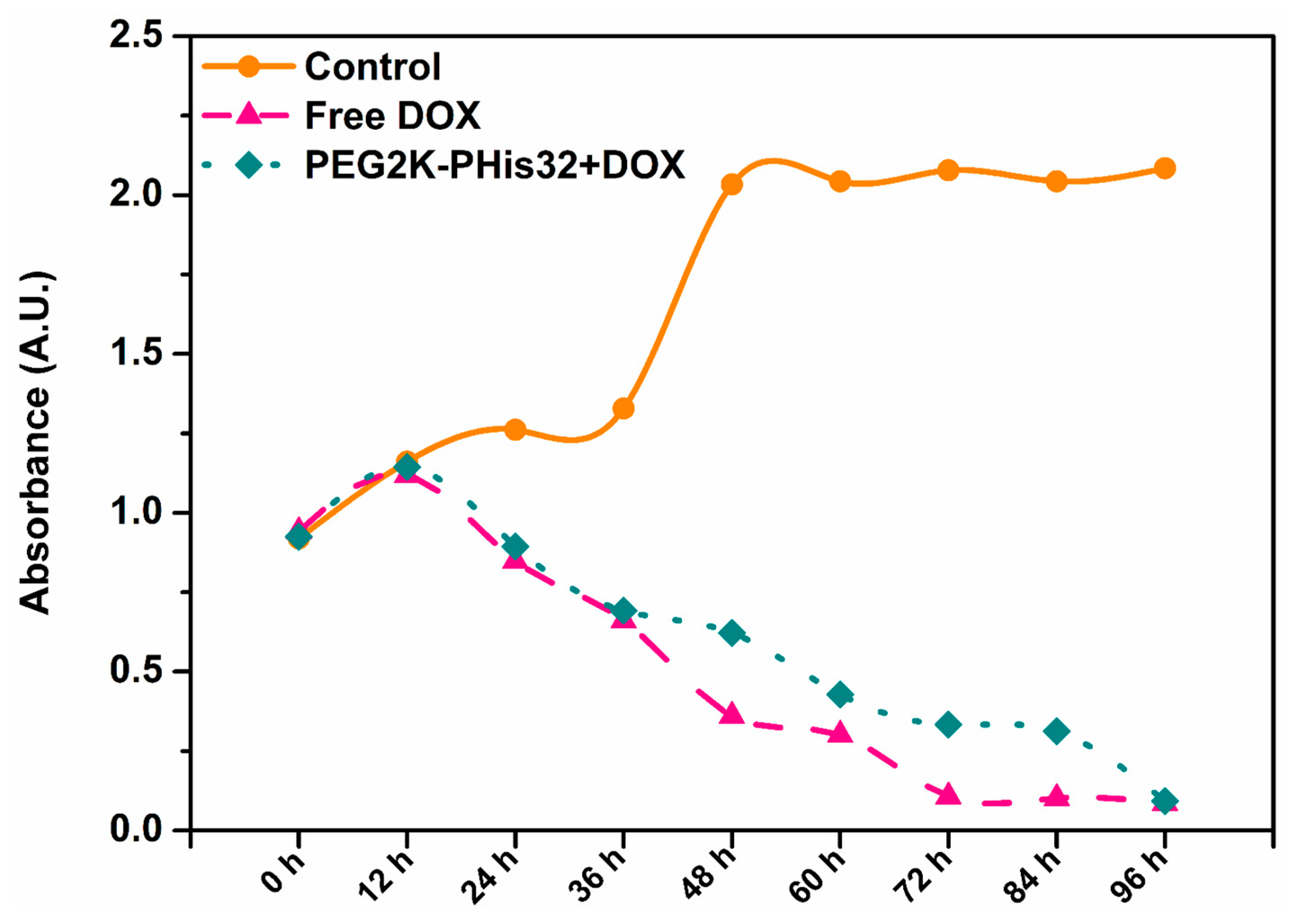
3.7. In Vitro Cell Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ge, Z.; Liu, S. Functional Block Copolymer Assemblies Responsive to Tumor and Intracellular Microenvironments for Site-Specific Drug Delivery and Enhanced Imaging Performance. Chem. Soc. Rev. 2013, 42, 7289–7325. [Google Scholar] [CrossRef] [PubMed]
- Tong, R.; Tang, L.; Ma, L.; Tu, C.; Baumgartner, R.; Cheng, J. Smart Chemistry in Polymeric Nanomedicine. Chem. Soc. Rev. 2014, 43, 6982–7012. [Google Scholar] [CrossRef] [PubMed]
- Ganta, S.; Devalapally, H.; Shahiwala, A.; Amiji, M. A Review of Stimuli-Responsive Nanocarriers for Drug and Gene Delivery. J. Control. Release 2008, 126, 187–204. [Google Scholar] [CrossRef] [PubMed]
- Kelley, E.G.; Albert, J.N.L.; Sullivan, M.O.; Epps, T.H. Stimuli-Responsive Copolymer Solution and Surface Assemblies for Biomedical Applications. Chem. Soc. Rev. 2013, 42, 7057–7071. [Google Scholar] [CrossRef]
- Colson, Y.L.; Grinstaff, M.W. Biologically Responsive Polymeric Nanoparticles for Drug Delivery. Adv. Mater. 2012, 24, 3878–3886. [Google Scholar] [CrossRef]
- Li, T.; Li, J.; Morozov, K.I.; Wu, Z.; Xu, T.; Rozen, I.; Leshansky, A.M.; Li, L.; Wang, J. Highly Efficient Freestyle Magnetic Nanoswimmer. Nano Lett. 2017, 17, 5092–5098. [Google Scholar] [CrossRef]
- Li, T.; Chang, X.; Wu, Z.; Li, J.; Shao, G.; Deng, X.; Qiu, J.; Guo, B.; Zhang, G.; He, Q.; et al. Autonomous Collision-Free Navigation of Microvehicles in Complex and Dynamically Changing Environments. ACS Nano 2017, 11, 9268–9275. [Google Scholar] [CrossRef]
- Ji, F.; Li, T.; Yu, S.; Wu, Z.; Zhang, L. Propulsion Gait Analysis and Fluidic Trapping of Swinging Flexible Nanomotors. ACS Nano 2021, 15, 5118–5128. [Google Scholar] [CrossRef]
- Yu, S.; Li, T.; Ji, F.; Zhao, S.; Liu, K.; Zhang, Z.; Zhang, W.; Mei, Y. Trimer-like Microrobots with Multimodal Locomotion and Reconfigurable Capabilities. Mater. Today Adv. 2022, 14, 100231. [Google Scholar] [CrossRef]
- Yu, S.; Ma, N.; Yu, H.; Sun, H.; Chang, X.; Wu, Z.; Deng, J.; Zhao, S.; Wang, W.; Zhang, G.; et al. Self-Propelled Janus Microdimer Swimmers under a Rotating Magnetic Field. Nanomaterials 2019, 9, 1672. [Google Scholar] [CrossRef]
- Liao, Z.S.; Huang, S.Y.; Huang, J.J.; Chen, J.K.; Lee, A.W.; Lai, J.Y.; Lee, D.J.; Cheng, C.C. Self-Assembled PH-Responsive Polymeric Micelles for Highly Efficient, Noncytotoxic Delivery of Doxorubicin Chemotherapy to Inhibit Macrophage Activation: In Vitro Investigation. Biomacromolecules 2018, 19, 2772–2781. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Huang, Y.; Kumar, A.; Tan, A.; Jin, S.; Mozhi, A.; Liang, X.J. PH-Sensitive Nano-Systems for Drug Delivery in Cancer Therapy. Biotechnol. Adv. 2014, 32, 693–710. [Google Scholar] [CrossRef] [PubMed]
- Meng, F.; Zhong, Y.; Cheng, R.; Deng, C.; Zhong, Z. PH-Sensitive Polymeric Nanoparticles for Tumor-Targeting Doxorubicin Delivery: Concept and Recent Advances. Nanomedicine 2014, 9, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Yang, Q.; Shi, K.; Xiao, Y.; Wei, X.; Qian, Z. Intratumoral Fate of Functional Nanoparticles in Response to Microenvironment Factor: Implications on Cancer Diagnosis and Therapy. Adv. Drug Deliv. Rev. 2019, 143, 37–67. [Google Scholar] [CrossRef]
- Ghiarasim, R.; Simionescu, N.; Coroaba, A.; Uritu, C.M.; Marangoci, N.L.; Ibanescu, S.-A.; Pinteala, M. SI-ATRP Decoration of Magnetic Nanoparticles with PHEMA and Post-Polymerization Modification with Folic Acid for Tumor Cells’ Specific Targeting. Int. J. Mol. Sci. 2022, 23, 155. [Google Scholar] [CrossRef]
- Li, J.; Dirisala, A.; Ge, Z.; Wang, Y.; Yin, W.; Ke, W.; Toh, K.; Xie, J.; Matsumoto, Y.; Anraku, Y.; et al. Therapeutic Vesicular Nanoreactors with Tumor-Specific Activation and Self-Destruction for Synergistic Tumor Ablation. Angew. Chem. Int. Ed. 2017, 56, 14025–14030. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Wang, Y.; Ke, W.; Chen, W.; Wang, W.; Ge, Z. Polymer Prodrug-Based Nanoreactors Activated by Tumor Acidity for Orchestrated Oxidation/Chemotherapy. Nano Lett. 2017, 17, 6983–6990. [Google Scholar] [CrossRef]
- Zhuo, S.; Zhang, F.; Yu, J.; Zhang, X.; Yang, G.; Liu, X. PH-Sensitive Biomaterials for Drug Deliv. Molecules 2020, 25, 5649. [Google Scholar] [CrossRef]
- Sang, W.; Zhang, Z.; Dai, Y.; Chen, X. Recent Advances in Nanomaterial-Based Synergistic Combination Cancer Immunotherapy. Chem. Soc. Rev. 2019, 48, 3771–3810. [Google Scholar] [CrossRef]
- Chen, W.; Meng, F.; Cheng, R.; Zhong, Z. PH-Sensitive Degradable Polymersomes for Triggered Release of Anticancer Drugs: A Comparative Study with Micelles. J. Control. Release 2010, 142, 40–46. [Google Scholar] [CrossRef]
- Petersen, L.K.; Sackett, C.K.; Narasimhan, B. High-Throughput Analysis of Protein Stability in Polyanhydride Nanoparticles. Acta Biomater. 2010, 6, 3873–3881. [Google Scholar] [CrossRef]
- Lee, S.; Saito, K.; Lee, H.R.; Lee, M.J.; Shibasaki, Y.; Oishi, Y.; Kim, B.S. Hyperbranched Double Hydrophilic Block Copolymer Micelles of Poly(Ethylene Oxide) and Polyglycerol for PH-Responsive Drug Delivery. Biomacromolecules 2012, 13, 1190–1196. [Google Scholar] [CrossRef] [PubMed]
- Fleige, E.; Quadir, M.A.; Haag, R. Stimuli-Responsive Polymeric Nanocarriers for the Controlled Transport of Active Compounds: Concepts and Applications. Adv. Drug Deliv. Rev. 2012, 64, 866–884. [Google Scholar] [CrossRef] [PubMed]
- Sim, T.; Lim, C.; Hoang, N.H.; Oh, K.T. Recent Advance of PH-Sensitive Nanocarriers Targeting Solid Tumors. J. Pharm. Investig. 2017, 47, 383–394. [Google Scholar] [CrossRef]
- Wang, Y.; Li, P.; Chen, F.; Jia, L.; Xu, Q.; Gai, X.; Yu, Y.; Di, Y.; Zhu, Z.; Liang, Y.; et al. A Novel PH-Sensitive Carrier for the Delivery of Antitumor Drugs: Histidine-Modified Auricularia Auricular Polysaccharide Nano-Micelles. Sci. Rep. 2017, 7, 4751. [Google Scholar] [CrossRef]
- Liu, Y.; Qiao, L.; Zhang, S.; Wan, G.; Chen, B.; Zhou, P.; Zhang, N.; Wang, Y. Dual PH-Responsive Multifunctional Nanoparticles for Targeted Treatment of Breast Cancer by Combining Immunotherapy and Chemotherapy. Acta Biomater. 2018, 66, 310–324. [Google Scholar] [CrossRef]
- Razzano, V.; Paolino, M.; Reale, A.; Giuliani, G.; Donati, A.; Giorgi, G.; Artusi, R.; Caselli, G.; Visintin, M.; Makovec, F.; et al. Poly-Histidine Grafting Leading to Fishbone-like Architectures. RSC Adv. 2018, 8, 8638–8656. [Google Scholar] [CrossRef]
- Oh, N.M.; Kwag, D.S.; Oh, K.T.; Youn, Y.S.; Lee, E.S. Electrostatic Charge Conversion Processes in Engineered Tumor-Identifying Polypeptides for Targeted Chemotherapy. Biomaterials 2012, 33, 1884–1893. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, D.; Ba, S.; Zhu, J.; Zhang, J.; Hong, W.; Zhao, X.; Hu, H.; Qiao, M. Poly(l-Histidine) Based Triblock Copolymers: PH Induced Reassembly of Copolymer Micelles and Mechanism Underlying Endolysosomal Escape for Intracellular Delivery. Biomacromolecules 2014, 15, 4032–4045. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, D.; Ba, S.; Chang, J.; Zhou, J.; Zhao, H.; Zhu, J.; Zhao, X.; Hu, H.; Qiao, M. Poly(l-Histidine) Based Copolymers: Effect of the Chemically Substituted l-Histidine on the Physio-Chemical Properties of the Micelles and In Vivo Biodistribution. Colloids Surf. B Biointerfaces 2016, 140, 176–184. [Google Scholar] [CrossRef]
- John, J.V.; Uthaman, S.; Augustine, R.; Manickavasagam Lekshmi, K.; Park, I.K.; Kim, I. Biomimetic PH/Redox Dual Stimuli-Responsive Zwitterionic Polymer Block Poly(L-Histidine) Micelles for Intracellular Delivery of Doxorubicin into Tumor Cells. J. Polym. Sci. Part A Polym. Chem. 2017, 55, 2061–2070. [Google Scholar] [CrossRef]
- Wu, H.; Zhu, L.; Torchilin, V.P. PH-Sensitive Poly(Histidine)-PEG/DSPE-PEG Co-Polymer Micelles for Cytosolic Drug Delivery. Biomaterials 2013, 34, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.S.; Gao, Z.; Bae, Y.H. Recent Progress in Tumor PH Targeting Nanotechnology. J. Control. Release 2008, 132, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.S.; Shin, H.J.; Na, K.; Bae, Y.H. Poly(L-Histidine)-PEG Block Copolymer Micelles and PH-Induced Destabilization. J. Control. Release 2003, 90, 363–374. [Google Scholar] [CrossRef]
- Lee, E.S.; Na, K.; Bae, Y.M. Super PH-Sensitive Multifunctional Polymeric Micelle. Nano Lett. 2005, 5, 325–329. [Google Scholar] [CrossRef]
- Lee, E.S.; Oh, K.T.; Kim, D.; Youn, Y.S.; Bae, Y.H. Tumor PH-Responsive Flower-like Micelles of Poly(l-Lactic Acid)-b-Poly(Ethylene Glycol)-b-Poly(l-Histidine). J. Control. Release 2007, 123, 19–26. [Google Scholar] [CrossRef]
- Kim, D.; Lee, E.S.; Park, K.; Kwon, I.C.; Bae, Y.H. Doxorubicin Loaded PH-Sensitive Micelle: Antitumoral Efficacy against Ovarian A2780/DOXR Tumor. Pharm. Res. 2008, 25, 2074–2082. [Google Scholar] [CrossRef]
- Kim, K.S.; Park, W.; Hu, J.; Bae, Y.H.; Na, K. A Cancer-Recognizable MRI Contrast Agents Using PH-Responsive Polymeric Micelle. Biomaterials 2014, 35, 337–343. [Google Scholar] [CrossRef]
- Li, Z.; Chen, Q.; Qi, Y.; Liu, Z.; Hao, T.; Sun, X.; Qiao, M.; Ma, X.; Xu, T.; Zhao, X.; et al. Rational Design of Multifunctional Polymeric Nanoparticles Based on Poly(l-Histidine) and d-α-Vitamin e Succinate for Reversing Tumor Multidrug Resistance. Biomacromolecules 2018, 19, 2595–2609. [Google Scholar] [CrossRef]
- Li, Z.; Qiu, L.; Chen, Q.; Hao, T.; Qiao, M.; Zhao, H.; Zhang, J.; Hu, H.; Zhao, X.; Chen, D.; et al. PH-Sensitive Nanoparticles of Poly(L-Histidine)-Poly(Lactide-Co-Glycolide)-Tocopheryl Polyethylene Glycol Succinate for Anti-Tumor Drug Delivery. Acta Biomater. 2015, 11, 137–150. [Google Scholar] [CrossRef]
- Behrendt, R.; White, P.; Offer, J. Advances in Fmoc Solid-Phase Peptide Synthesis. J. Pept. Sci. 2016, 22, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Bondalapati, S.; Jbara, M.; Brik, A. Expanding the Chemical Toolbox for the Synthesis of Large and Uniquely Modified Proteins. Nat. Chem. 2016, 8, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Geno Samaritoni, J.; Martynow, J.G.; O’Donnell, M.J.; Scott, W.L. Preparation and Use of a General Solid-Phase Intermediate to Biomimetic Scaffolds and Peptide Condensations. Molecules 2018, 23, 1762. [Google Scholar] [CrossRef] [PubMed]
- El-Faham, A.; Albericio, F. Peptide Coupling Reagents, More than a Letter Soup. Chem. Rev. 2011, 111, 6557–6602. [Google Scholar] [CrossRef]
- Nanda, J.S.; Lorsch, J.R. Labeling a Protein with Fluorophores Using NHS Ester Derivitization. Methods Enzymol. 2014, 536, 87–94. [Google Scholar] [CrossRef]
- Tacar, O.; Sriamornsak, P.; Dass, C.R. Doxorubicin: An Update on Anticancer Molecular Action, Toxicity and Novel Drug Delivery Systems. J. Pharm. Pharmacol. 2013, 65, 157–170. [Google Scholar] [CrossRef]
- Motlagh, N.S.H.; Parvin, P.; Ghasemi, F.; Atyabi, F. Fluorescence Properties of Several Chemotherapy Drugs: Doxorubicin, Paclitaxel and Bleomycin. Biomed. Opt. Express 2016, 7, 2400–2406. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, S.; Ying, X.; Wang, Y.; Geng, P.; Deng, A.; Yu, Z. Doxorubicin-Loaded Redox-Responsive Micelles Based on Dextran and Indomethacin for Resistant Breast Cancer. Int. J. Nanomed. 2017, 12, 6153–6168. [Google Scholar] [CrossRef]
- Ilhami, F.B.; Peng, K.C.; Chang, Y.S.; Alemayehu, Y.A.; Tsai, H.C.; Lai, J.Y.; Chiao, Y.H.; Kao, C.Y.; Cheng, C.C. Photo-Responsive Supramolecular Micelles for Controlled Drug Release and Improved Chemotherapy. Int. J. Mol. Sci. 2021, 22, 154. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Available online: http://www.chempeptide.com/ (accessed on 7 May 2021).
- Norouzy, A.; Lazar, A.I.; Karimi-Jafari, M.H.; Firouzi, R.; Nau, W.M. Electrostatically Induced PK a Shifts in Oligopeptides: The Upshot of Neighboring Side Chains. Amino Acids 2022, 54, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Lu, J.; Wang, J.; Hao, P.; Li, C.; Qi, L.; Yang, L.; He, B.; Zhong, Z.; Hao, N. Redox-Sensitive Polymeric Micelles with Aggregation-Induced Emission for Bioimaging and Delivery of Anticancer Drugs. J. Nanobiotechnol. 2021, 19, 1–15. [Google Scholar] [CrossRef]
- Zhou, X.X.; Jin, L.; Qi, R.Q.; Ma, T. Ph-Responsive Polymeric Micelles Self-Assembled from Amphiphilic Copolymer Modified with Lipid Used as Doxorubicin Delivery Carriers. R. Soc. Open Sci. 2018, 5, 171654. [Google Scholar] [CrossRef] [PubMed]
- Bloemen, M. Immunomagnetic Separation of Bacteria by Iron Oxide Nanoparticles. Ph.D. Thesis, KU Leuven, Leuven, Belgium, May 2015. [Google Scholar]
- Haghi, A.; Raissi, H.; Hashemzadeh, H.; Farzad, F. Development of the Poly(L-Histidine) Grafted Carbon Nanotube as a Possible Smart Drug Delivery Vehicle. Comput. Biol. Med. 2022, 143, 105336. [Google Scholar] [CrossRef]
- Cipolla, D.; Wu, H.; Salentinig, S.; Boyd, B.; Rades, T.; Vanhecke, D.; Petri-Fink, A.; Rothin-Rutishauser, B.; Eastman, S.; Redelmeier, T.; et al. Formation of Drug Nanocrystals under Nanoconfinement Afforded by Liposomes. RSC Adv. 2016, 6, 6223–6233. [Google Scholar] [CrossRef][Green Version]
- Wu, X.R.; Zhang, J.; Zhang, J.H.; Xiao, Y.P.; He, X.; Liu, Y.H.; Yu, X.Q. Amino Acid-Linked Low Molecular Weight Polyethylenimine for Improved Gene Delivery and Biocompatibility. Molecules 2020, 25, 975. [Google Scholar] [CrossRef]
- Chen, M.; Chen, C.; Shen, Z.; Zhang, X.; Chen, Y.; Lin, F.; Ma, X.; Zhuang, C.; Mao, Y.; Gan, H.; et al. Extracellular PH Is a Biomarker Enabling Detection of Breast Cancer and Liver Cancer Using CEST MRI. Oncotarget 2017, 8, 45759–45767. [Google Scholar] [CrossRef]
- Yamada, Y. Dimerization of Doxorubicin Causes Its Precipitation. ACS Omega 2020, 5, 33235–33241. [Google Scholar] [CrossRef]

| Sample Name | EE (%) | DL (%) |
|---|---|---|
| PEG2K-PHis20 + DOX | 52.98 | 8.11 |
| PEG2K-PHis26 + DOX | 60.51 | 9.16 |
| PEG2K-PHis32 + DOX | 71.31 | 10.62 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghiarasim, R.; Tiron, C.E.; Tiron, A.; Dimofte, M.-G.; Pinteala, M.; Rotaru, A. Solid-Phase Synthesized Copolymers for the Assembly of pH-Sensitive Micelles Suitable for Drug Delivery Applications. Nanomaterials 2022, 12, 1798. https://doi.org/10.3390/nano12111798
Ghiarasim R, Tiron CE, Tiron A, Dimofte M-G, Pinteala M, Rotaru A. Solid-Phase Synthesized Copolymers for the Assembly of pH-Sensitive Micelles Suitable for Drug Delivery Applications. Nanomaterials. 2022; 12(11):1798. https://doi.org/10.3390/nano12111798
Chicago/Turabian StyleGhiarasim, Razvan, Crina Elena Tiron, Adrian Tiron, Mihail-Gabriel Dimofte, Mariana Pinteala, and Alexandru Rotaru. 2022. "Solid-Phase Synthesized Copolymers for the Assembly of pH-Sensitive Micelles Suitable for Drug Delivery Applications" Nanomaterials 12, no. 11: 1798. https://doi.org/10.3390/nano12111798
APA StyleGhiarasim, R., Tiron, C. E., Tiron, A., Dimofte, M.-G., Pinteala, M., & Rotaru, A. (2022). Solid-Phase Synthesized Copolymers for the Assembly of pH-Sensitive Micelles Suitable for Drug Delivery Applications. Nanomaterials, 12(11), 1798. https://doi.org/10.3390/nano12111798







