Silver Nanoparticles Formation by Jatropha integerrima and LC/MS-QTOF-Based Metabolite Profiling
Abstract
:1. Introduction
2. Results
2.1. Characterization of Biogenic J-AgNPs
2.2. Analytical RP-HPLC and LC–QTOF-MS Identified Compounds
2.3. Functional Groups Analyzed by FTIR
2.4. Antibacterial Screening
2.5. Amp-J-AgNPs Nanocomposites
2.6. Cytotoxicity Test
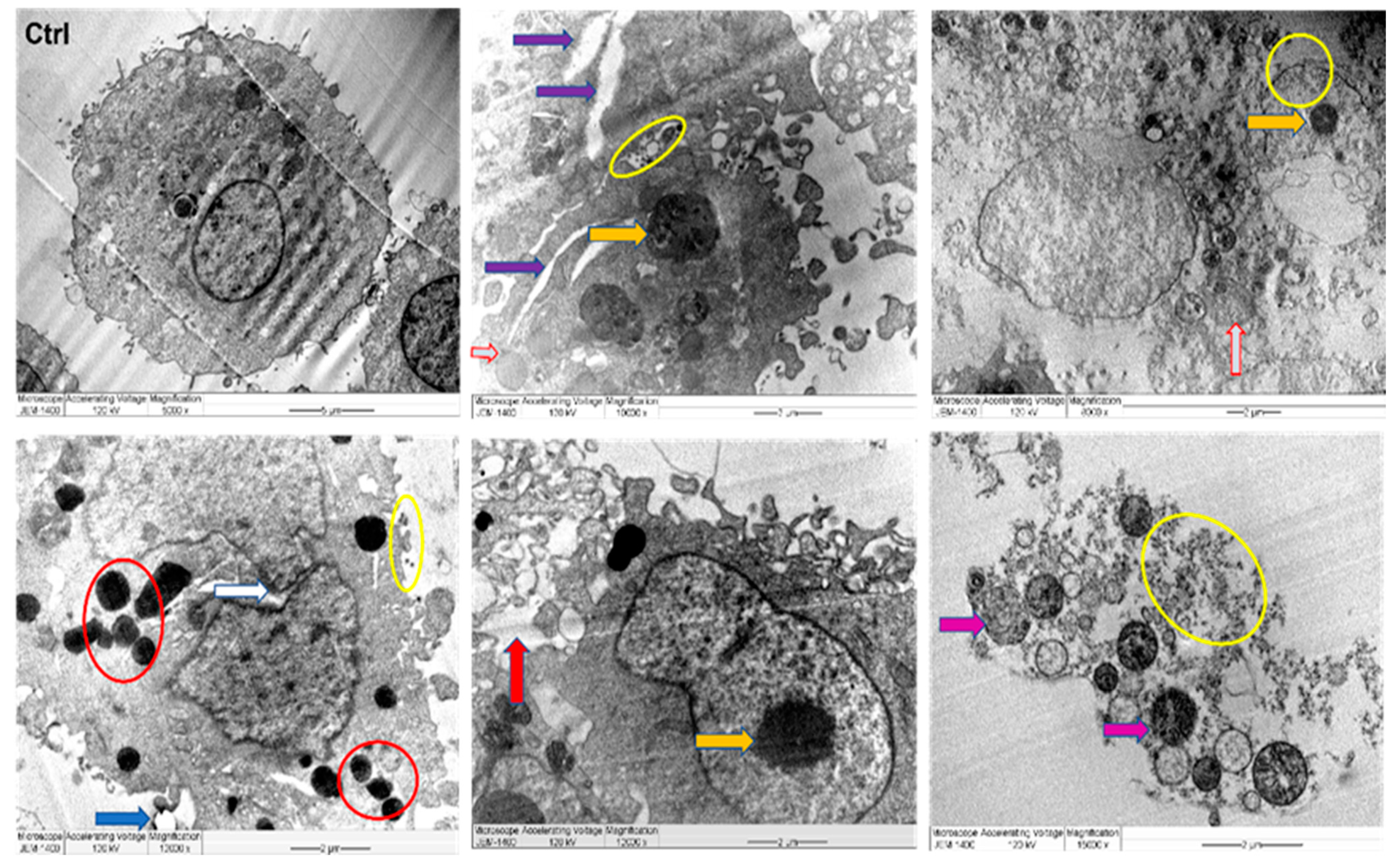
2.7. TEM and LSM Analysis of Cancer Cells Treated by J-AgNPs
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Aqueous Extracts for AgNPs Synthesis
4.3. Characterization of J-AgNPs
4.3.1. Ultraviolet-Visible Spectroscopy
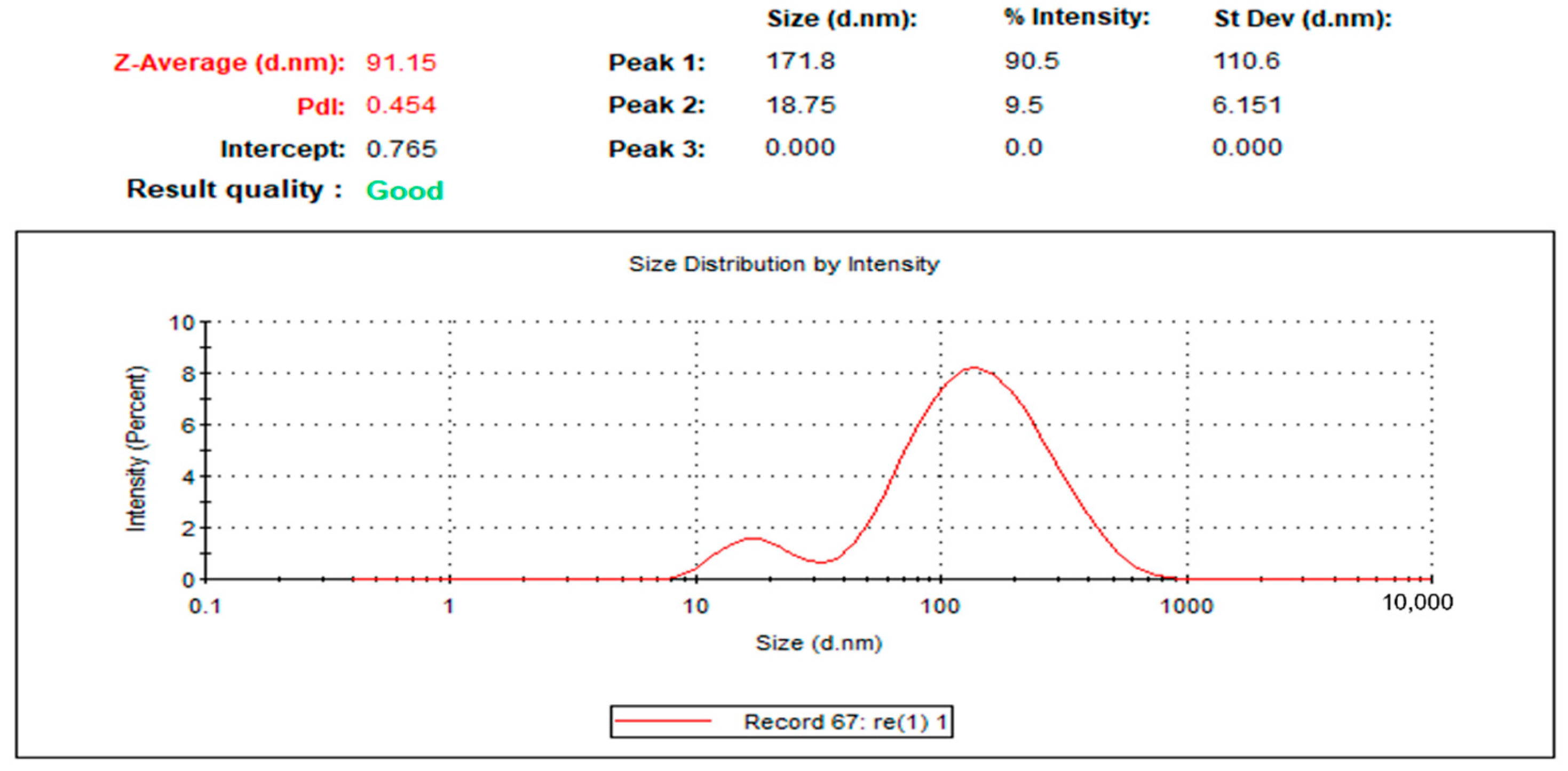
4.3.2. Hydrodynamic Size and Surface Area Analysis
4.3.3. Size Distribution and Morphology Analysis Using Transmission Electron Microscopy (TEM)
4.3.4. Energy-Dispersive X-ray Spectroscopy (EDS)
4.4. Analysis of the Plant Extracts by Analytical RP-HPLC Method
4.5. C–QTOF-MS Method
4.6. Analysis of Surface Functional Groups and Compounds
4.7. Antibacterial Screening
4.8. Minimum Inhibitory Concentration and Minimum Bactericidal Concentration
4.9. Ampicillin-Conjugated J-AgNPs (Amp-J-AgNPs) Synthesis, Analysis and Application
4.10. Anticancer Action and Apoptosis Induced by J-AgNPs
4.11. Cell Structural Changes by Transmission Electron Microscopy
4.12. Laser Scanning Microscopy (LSM)
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendation; Government of the United Kingdom: London, UK, 2016. [Google Scholar]
- Alqahtani, M.A.; Al Othman, M.R.; Mohammed, A.E. Bio fabrication of silver nanoparticles with antibacterial and cytotoxic abilities using lichens. Sci. Rep. 2020, 10, 16781. [Google Scholar] [CrossRef]
- Gour, A.; Jain, N.K. Advances in green synthesis of nanoparticles. Artif. Cells Nanomed. Biotechnol. 2019, 47, 844–851. [Google Scholar] [CrossRef] [Green Version]
- Dakal, T.C.; Kumar, A.; Majumdar, R.S.; Yadav, V. Mechanistic Basis of Antimicrobial Actions of Silver Nanoparticles. Front. Microbiol. 2016, 7, 1831. [Google Scholar] [CrossRef] [Green Version]
- Jena, P.; Bhattacharya, M.; Bhattacharjee, G.; Satpati, B.; Mukherjee, P.; Senapati, D.; Srinivasan, R. Bimetallic gold-silver nanoparticles mediate bacterial killing by disrupting the actin cytoskeleton MreB. Nanoscale 2020, 12, 3731–3749. [Google Scholar] [CrossRef] [PubMed]
- Sanpui, P.; Chattopadhyay, A.; Ghosh, S.S. Induction of apoptosis in cancer cells at low silver nanoparticle concentrations using chitosan nanocarrier. ACS Appl. Mater. Interfaces 2011, 3, 218–228. [Google Scholar] [CrossRef]
- Akintelu, S.A.; Bo, Y.; Folorunso, A.S. A review on synthesis, optimization, mechanism, characterization, and antibacterial application of silver nanoparticles synthesized from plants. J Chem. 2020, 2020, 3189043. [Google Scholar] [CrossRef]
- Dong, H.; Gao, Y.; Sinko, P.J.; Wu, Z.; Xu, J.; Jia, L. The nanotechnology race between China and the United States. Nano Today 2016, 11, 7–12. [Google Scholar] [CrossRef]
- Suresh, G.; Gunasekar, P.H.; Kokila, D.; Prabhu, D.; Dinesh, D.; Ravichandran, N.; Ramesh, B.; Koodalingam, A.; Vijaiyan Siva, G. Green synthesis of silver nanoparticles using Delphinium denudatum root extract exhibits antibacterial and mosquito larvicidal activities. Spectrochim. Acta Part A Mol. Biomol. Spectroscopy 2014, 127, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Ovais, M.; Nadhman, A.; Khalil, A.T.; Raza, A.; Khuda, F.; Sohail, M.F.; Islam, N.U.; Sarwar, H.S.; Shahnaz, G.; Ahmad, I.; et al. Biosynthesized colloidal silver and gold nanoparticles as emerging leishmanicidal agents: An insight. Nanomedicine 2017, 12, 2807–2819. [Google Scholar] [CrossRef]
- Ovais, M.; Khalil, A.T.; Raza, A.; Khan, M.A.; Ahmad, I.; Islam, N.U.; Saravanan, M.; Ubaid, M.F.; Ali, M.; Shinwari, Z.K. Green synthesis of silver nanoparticles via plant extracts: Beginning a new era in cancer theranostics. Nanomedicine 2016, 12, 3157–3177. [Google Scholar] [CrossRef]
- Mohammed, A.E.; Al-Qahtani, A.; Al-Mutairi, A.; Al-Shamri, B.; Aabed, K. Antibacterial and cytotoxic potential of biosynthesized silver nanoparticles by some plant extracts. Nanomaterials 2018, 8, 382. [Google Scholar] [CrossRef] [Green Version]
- Aabed, K.; Mohammed, A.E. Synergistic and Antagonistic Effects of Biogenic Silver Nanoparticles in Combination with Antibiotics Against Some Pathogenic Microbes. Front. Bioeng. Biotechnol. 2021, 9, 1–14. [Google Scholar] [CrossRef]
- Salayová, A.; Bedlovičová, Z.; Daneu, N.; Baláž, M.; Lukáčová Bujňáková, Z.; Balážová, L’.; Tkáčiková, L’. Green synthesis of silver nanoparticles with antibacterial activity using various medicinal plant extracts: Morphology and antibacterial efficacy. Nanomaterials 2021, 11, 1005. [Google Scholar]
- Mohammed, A.E.; Al-Megrin, W.A. Biological Potential of Silver Nanoparticles Mediated by Leucophyllum frutescens and Russelia equisetiformis Extracts. Nanomaterials 2021, 11, 2098. [Google Scholar] [CrossRef]
- Mikhailov, O.V.; Mikhailova, E.O. Elemental silver nanoparticles: Biosynthesis and bio applications. Materials 2019, 12, 3177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonanomi, G.; Vinale, F.; Scala, F. The role of natural products in plant-microbe interactions. In Plant-Derived Natural Products; Springer: Cham, Switzerland, 2009; pp. 301–320. [Google Scholar]
- Rahman, M.M.; Habib, M.R.; Hasan, S.R.; Sayeed, M.A.; Rana, M.S. Antibacterial, cytotoxic and antioxidant potential of methanolic extract of Phyllanthus acidus L. Int. J. Drug Dev. Res. 2011, 3, 154–161. [Google Scholar]
- Panzu, N.N.; Lengbiye, E.M.; Domondo, A.; Inkoto, C.L.; Muanyishay, C.L.; Gbolo, B.Z.; Ashande, C.M.; Mawi, C.F.; Tshibangu, D.S.T. A review on the Bioactivity and Phytochemistry of Jatropha podagrica Hook (Euphorbiaceae). Discov. Phytomed. 2020, 7, 186–194. [Google Scholar] [CrossRef]
- Ngbolua, K.N.; Lengbiye, E.M.; Likolo, J.B.; Djolu, R.D.; Masengo, C.A.; Mpiana, P.T. A mini-review on the pharmacognosy and phytochemistry of the tropical medicinal plant Annona senegalensis Persoon (Annonaceae). Tro. Plant Res. 2017, 4, 168–175. [Google Scholar] [CrossRef]
- Laviola, B.G.; Rodrigues, E.V.; Teodoro, P.E.; de Azevedo Peixoto, L.; Bhering, L.L. Biometric and biotechnology strategies in Jatropha genetic breeding for biodiesel production. Renew. Sustain. Energy Rev. 2017, 76, 894–904. [Google Scholar] [CrossRef]
- Sabandar, C.W.; Ahmat, N.; Jaafar, F.M.; Sahidin, I. Medicinal property, phytochemistry and pharmacology of several Jatropha species (Euphorbiaceae): A review. Phytochemistry 2013, 85, 7–29. [Google Scholar] [CrossRef]
- Can-Aké, R.; Erosa-Rejón, G.; May-Pat, F.; Peña-Rodríguez, L.; Peraza-Sánchez, S. Bioactive Terpenoids from Roots and Leaves of Jatropha gaumeri. Rev. Soc. Química México 2004, 48, 11–14. [Google Scholar]
- Bhaumik, J.; Thakur, N.S.; Aili, P.K.; Ghanghoriya, A.; Mittal, A.K.; Banerjee, U.C. Bioinspired Nanotheranostic Agents: Synthesis, Surface Functionalization, and Antioxidant Potential. ACS Biomater. Sci. Eng. 2015, 1, 382–392. [Google Scholar] [CrossRef] [PubMed]
- Banerji, J.; Das, B.; Bose, P.; Chakabarti, R.; Chaterjee, A. Traditional Medicine; Oxford and IBH Publishing Co. Pvt. Ltd.: New Dehli, India, 1993. [Google Scholar]
- Cavalcante, N.B.; Diego da, C.S.A.; Guedes da Silva Almeida, J.R. The genus Jatropha (Euphorbiaceae): A review on secondary chemical metabolites and biological aspects. Chem. Biol. Interac. 2020, 318, 108976. [Google Scholar] [CrossRef]
- Minh, T.N.; Xuan, T.D.; Tran, H.D.; Van, T.M.; Andriana, Y.; Khanh, T.D.; Van Quan, N.; Ahmad, A. Isolation and purification of bioactive compounds from the stem bark of Jatropha podagrica. Molecules 2019, 24, 889. [Google Scholar] [CrossRef] [Green Version]
- Mongkolvisut, W.; Sutthivaiyakit, S.; Leutbecher, H.; Mika, S.; Klaiber, I.; Möller, W.; Rösner, H.; Beifuss, U.; Conrad, J. Integerrimides A and B, cyclic heptapeptides from the latex of Jatropha integerrima. J. Nat. Prod. 2006, 69, 1435–1441. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.Y.; Lou, L.L.; Guo, Y.Q.; Li, W.; Guo, Y.H.; Bao, J.M.; Tang, G.H.; Bu, X.Z.; Yin, S. Natural thioredoxin reductase inhibitors from Jatropha integerrima. RSC Adv. 2015, 5, 47235–47243. [Google Scholar] [CrossRef]
- Li, A.N.; Li, S.; Li, H.B.; Xu, D.P.; Xu, X.R.; Chen, F. Total phenolic contents and antioxidant capacities of 51 edible and wild flowers. J. Funct. Foods 2014, 6, 319–330. [Google Scholar] [CrossRef]
- Nazeema, T.H.; Sugannya, P.K. Synthesis and characterization of silver nanoparticle from two medicinal plants and it anticancer property. Int. J. Eng. Res. Technol. 2014, 2, 49–56. [Google Scholar]
- Huang, J.D.; Wang, C.F.; Lian, C.L.; Huang, M.Y.; Zhang, C.; Liu, J.Q. Isolation and identification of five new diterpenoids from Jatropha curcas. Phytochem. Lett. 2020, 40, 37–41. [Google Scholar] [CrossRef]
- Xu, L.; Wang, Y.Y.; Huang, J.; Chen, C.Y.; Wang, Z.X.; Xie, H. Silver nanoparticles: Synthesis, medical applications and biosafety. Theranostics 2020, 10, 8996. [Google Scholar] [CrossRef]
- Bhattacharjee, S. DLS and zeta potential-What they are and what they are not? J. Control. Release 2016, 235, 337–351. [Google Scholar] [CrossRef]
- Sujitha, M.V.; Kannan, S. Green synthesis of gold nanoparticles using Citrus fruits (Citrus limon, Citrus reticulata and Citrus sinensis) aqueous extract and its characterization. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 102, 15–23. [Google Scholar] [CrossRef]
- El-Naggar, N.E.A.; Mohamedin, A.; Hamza, S.S.; Sherief, A.D. Extracellular biofabrication, characterization, and antimicrobial efficacy of silver nanoparticles loaded on cotton fabrics using newly isolated Streptomyces sp. SSHH-1E. J. Nanomater. 2016, 2016, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Lanzotti, V. Diterpenes for Therapeutic Use. In Natural Products; Ramawat, K., Mérillon, J.M., Eds.; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar] [CrossRef] [Green Version]
- Eshilokun, A.O.; Kasali, A.A.; Ogunwande, I.A.; Walker, T.M.; Setzer, W.N. Chemical Composition and Antimicrobial Studies of the Essential Oils of Jatropha integerrima Jacq (Leaf and Seeds). Nat. Prod. Commun. 2007, 2, 853–855. [Google Scholar] [CrossRef] [Green Version]
- Yuan, W.J.; Yang, G.P.; Zhang, J.H.; Zhang, Y.; Chen, D.Z.; Li, S.L.; Di, Y.T.; Hao, X.J. Three new diterpenes with cytotoxic activity from the roots of Euphorbia ebracteolata Hayata. Phytochem. Lett. 2016, 18, 176–179. [Google Scholar] [CrossRef] [Green Version]
- Khandel, P.; Shahi, S.K.; Kanwar, L.; Yadaw, R.K.; Soni, D.K. Biochemical profiling of microbes inhibiting silver nanoparticles using symbiotic organisms. Int. J. Pharm. Sci. Invent. 2018, 9, 273–285. [Google Scholar]
- Dasari, S.; Suresh, K.A.; Rajesh, M.; Reddy, S.; Samba, C.; Hemalatha, C.S.; Wudayagiri, R.; Valluru, L. Biosynthesis, characterization, antibacterial and antioxidant activity of silver nanoparticles produced by lichens. J. Bionanosci. 2013, 7, 237–244. [Google Scholar] [CrossRef]
- Sonbol, H.; Ameen, F.; AlYahya, S.; Almansob, A.; Alwakeel, S. Padina boryana mediated green synthesis of crystalline palladium nanoparticles as potential nanodrug against multidrug resistant bacteria and cancer cells. Sci. Rep. 2021, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Guidelli, E.J.; Ramos, A.P.; Zaniquelli, M.E.D.; Baffa, O. Green synthesis of colloidal silver nanoparticles using natural rubber latex extracted from Hevea brasiliensis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2011, 82, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Ali, K.; Ahmed, B.; Dwivedi, S.; Saquib, Q.; Al-Khedhairy, A.A.; Musarra, J. Microwave accelerated green synthesis of stable silver nanoparticles with Eucalyptus globulus leaf extract and their antibacterial and antibiofilm activity on clinical isolates. PLoS ONE 2015, 10, 15. [Google Scholar] [CrossRef]
- Aiyelaagbe, O.O.; Hamid, A.A.; Fattorusso, E.; Taglialatela-Scafati, O.; Schröder, H.C.; Werner, E.G.; Müller, W.E.G. Cytotoxic Activity of Crude Extracts as well as of Pure Components from Jatropha Species, Plants Used Extensively in African Traditional Medicine. Evid.-Based Complementary Altern. Med. 2011, 2011, 134954. [Google Scholar] [CrossRef] [Green Version]
- Chauhan, N.; Tyagi, A.K.; Kumar, P.; Malik, A. Antibacterial Potential of Jatropha curcas Synthesized Silver Nanoparticles against Food Borne Pathogens. Front. Microbiol. 2016, 7, 1748. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.M.; Arif, M.; Ahmed, Z. Antimicrobial activity in leaf, seed extract and seed oil of Jatropha curcas L. plant. J. Appl. Nat. Sci. 2011, 3, 102–105. [Google Scholar] [CrossRef] [Green Version]
- Gurunathan, S.; Qasim, M.; Park, C.; Yoo, H.; Choi, D.Y.; Song, H.; Park, C.; Kim, J.-H.; Hong, K. Cytotoxicity and transcriptomic analysis of silver nanoparticles in mouse embryonic fibroblast cells. Int. J. Mol. Sci. 2018, 19, 3618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, Y.; Pauer, A.C.; Gonzales, A.A.; Fenniri, H. Enhanced antibiotic activity of ampicillin conjugated to gold nanoparticles on PEGylated rosette nanotubes. Int. J. Nanomed. 2019, 14, 7281–7289. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masri, A.; Anwar, A.; Ahmed, D.; Siddiqui, R.B.; Shah, M.R.; Khan, N.A. Silver Nanoparticle Conjugation-Enhanced Antibacterial Efficacy of Clinically Approved Drugs Cephradine and Vildagliptin. Antibiotics 2018, 7, 100. [Google Scholar] [CrossRef] [Green Version]
- Payne, J.N.; Wahwani, H.K.; Connor, M.G.; Hamilton, L.; Tockstein, S.; Moolani, H.; Chavda, F.; Badwaik, V.; Lawrenz, M.B.; Dakshinamurthy, R. Novel Synthesis of Kanamycin Conjugated Gold Nanoparticles with Potent Antibacterial Activity. Front Microbiol. 2016, 7, 607. [Google Scholar] [CrossRef] [PubMed]
- Shahbandeh, M.; Eghdami, A.; Moghaddam, M.M.; Nadoushan, M.J.; Salimi, A.; Fasihi-Ramandi, M.; Mohammadi, S.; Mirzaei, M.; Mirnejad, R. Conjugation of imipenem to silver nanoparticles for enhancement of its antibacterial activity against multidrug-resistant isolates of Pseudomonas aeruginosa. J. Biosci. 2021, 46, 1–19. [Google Scholar] [CrossRef]
- Din, L.B.; Mie, R.; Samsudin, M.W.; Ahmad, A.; Ibrahim, N. Biomimetic synthesis of silver nanoparticles using the lichen Ramalina dumeticola and the antibacterial activity. Malay. J. Anal. Sci. 2015, 19, 369–376. [Google Scholar]
- Deng, H.; McShan, D.; Zhang, Y.; Sinha, S.S.; Arslan, Z.; Ray, P.C.; Yu, H. Mechanistic study of the synergistic antibacterial activity of combined silver nanoparticles and common antibiotics. Environ. Sci. Technol. 2016, 50, 8840–8848. [Google Scholar] [CrossRef] [Green Version]
- Oskoueian, E.; Abdullah, N.; Saad, W.Z.; Omar, A.R.; Ahmad, S.; Kuan, W.B.; Zolkifli, N.A.; Hendra, R.; Ho, Y.N. Antioxidant, anti-inflammatory and anticancer activities of methanolic extracts from Jatropha curcas Linn. J. Med. Plant Res. 2011, 5, 49–57. [Google Scholar]
- De Almeida, P.M.; Araújo, S.D.; Marin-Morales, M.A.; Benko-Iseppon, A.M.; Brasileiro-Vidal, A.C. Genotoxic potential of the latex from cotton-leaf physicnut (Jatropha gossypiifolia L.). Genet. Mol. Biol. 2015, 38, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Idrissa, N.; Adama, D.; Mamadou, B.; Rokhaya, S.G.; Yoro, T.; Alassane, W.; Djibril, F. Novel Cytotoxic Cycloheptapeptide from the Latex of Jatropha integerrima. J. Chem. Pharm. Res. 2016, 8, 135–139. [Google Scholar]
- Kepsutlu, B.; Wycisk, V.; Achazi, K.; Kapishnikov, S.; Pérez-Berná, A.J.; Guttmann, P.; Cossmer, A.; Pereiro, E.; Ewers, H.; Ballau, M.; et al. Cells Undergo Major Changes in the Quantity of Cytoplasmic Organelles after Uptake of Gold Nanoparticles with Biologically Relevant Surface Coatings. ACS Nano 2020, 14, 2248–2264. [Google Scholar] [CrossRef]
- Kodama, M.; Wang, Y.M.; Hutter, E.; Maysinger, D.; Stochaj, U. Off to the Organelles—Killing Cancer Cells with Targeted Gold Nanoparticles. Theranostics 2015, 5, 357–370. [Google Scholar]
- Manke, A.; Wang, L.; Rojanasakul, Y. Mechanisms of Nanoparticle-Induced Oxidative Stress and Toxicity. BioMed Res. Int. 2013, 2013, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrader, M.; Fahimi, H.D. Mammalian peroxisomes and reactive oxygen species. Histochem. Cell Biol. 2004, 122, 383–393.–393. [Google Scholar] [CrossRef] [PubMed]
- Vieira, L.F.D.A.; Lins, M.P.; Viana, I.M.M.N.; Dos Santos, J.E.; Smaniotto, S.; Reis, M.D.D.S. Metallic nanoparticles reduce the migration of human fibroblasts in vitro. Nanoscale Res. Lett. 2017, 12, 200. [Google Scholar] [CrossRef] [Green Version]
- El-Boubbou, K.; Ali, R.; Bahhari, H.M.; Boudjelal, M. Magnetic nanocarriers enhance drug delivery selectively to human leukemic cells. J. Nanomed. Nanotechnol. 2017, 8, 1–7. [Google Scholar]
- Kajta, M.; Wo’jtowicz, A.K.; Mac´kowiak, M.; Lason´, W. Aryl hydrocarbon receptor-mediated apoptosis of neuronal cells: A possible interaction with estrogen receptor signaling. Neuroscience 2009, 158, 811–822. [Google Scholar] [CrossRef]
- Palvai, S.; Kuman, M.M.; Sengupta, P.; Basu, S. Hyaluronic acid layered chimeric nanoparticles: Targeting MAPK-PI3K signaling hub in colon cancer cells. ACS Omega 2017, 2, 7868–7880. [Google Scholar] [CrossRef]
- Skonieczna, M.; Hudy, D. Biological activity of silver nanoparticles and their applications in anticancer therapy. In Silver Nanoparticles—Fabrication, Characterization and Applications; IntechOpen: London, UK, 2018; p. 131. [Google Scholar]
- Siddiqi, K.S.; Rashid, M.; Rahman, A.; Husen, A.; Rehman, S. Biogenic fabrication and characterization of silver nanoparticles using aqueous-ethanolic extract of lichen (Usnea longissima) and their antimicrobial activity. Biomater. Res. 2018, 22, 1–9. [Google Scholar] [CrossRef]
- Mie, R.; Samsudin, M.W.; Din, L.B.; Ahmad, A.; Ibrahim, N.; Adnan, S.N.A. Synthesis of silver nanoparticles with antibacterial activity using the lichen Parmotrema praesorediosum. Int. J. Nanomed. 2014, 9, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, S.; Singh, A.R.J.; Sasikumar, C.S. Green synthesis of Bio-Silver Nanoparticles by Parmelia perlata, Ganoderma lucidum and Phellinus igniarius & Their Fields of Application. Indian J. Res. Pharm. Biotech. 2015, 5674, 100–110. [Google Scholar]
- Wayne, P.A. Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing: Report No. 1-56238-838-X 20th. CLSI M100. 2018. Available online: https://clsi.org/media/1930/m100ed28_sample.pdf (accessed on 1 June 2021).
- Ali, R.; Samman, N.; Al Zahrani, H.; Nehdi, A.; Rahman, S.; Khan, A.L.; Al Balwi, M.; Alriyees, L.A.; Alzaid, M.; Al Askar, A.; et al. Isolation and characterization of a new naturally immortalized human breast carcinoma cell line, KAIMRC1. BMC Cancer 2017, 17, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammed, A.E.; Sonbol, H.; Alwakeel, S.S.; Alotaibi, M.O.; Alotaibi, S.; Alothman, N.; Ali, R. Investigation of biological activity of soil fungal extracts and LC/MS-QTOF based metabolite profiling. Sci. Rep. 2021, 11, 1–17. [Google Scholar]






 ) and Amp-AgNPs (
) and Amp-AgNPs ( ) against 4 different bacteria. Two-way ANOVA with multiple comparisons was performed to identify differences between groups. p < 0.01 (**), p < 0.001 (****).
) against 4 different bacteria. Two-way ANOVA with multiple comparisons was performed to identify differences between groups. p < 0.01 (**), p < 0.001 (****).
 ) and Amp-AgNPs (
) and Amp-AgNPs ( ) against 4 different bacteria. Two-way ANOVA with multiple comparisons was performed to identify differences between groups. p < 0.01 (**), p < 0.001 (****).
) against 4 different bacteria. Two-way ANOVA with multiple comparisons was performed to identify differences between groups. p < 0.01 (**), p < 0.001 (****).

 ) and MDA MB 231 (
) and MDA MB 231 ( ) and a normal cell line; MCF 10A (
) and a normal cell line; MCF 10A ( ). Two-way ANOVA p < 0.0001 (****) (A). Log dose-response relationship of J-AgNPs on the normalized viability of two human cancer cell lines; HCT116 (
). Two-way ANOVA p < 0.0001 (****) (A). Log dose-response relationship of J-AgNPs on the normalized viability of two human cancer cell lines; HCT116 ( ) and MDA MB 231 (
) and MDA MB 231 ( ) and one normal cell line MCF 10A (
) and one normal cell line MCF 10A ( ). IC50 values of J-AgNPs on each cell line were calculated using log viability vs. normalized response–variable slope (four parameters) (B).
). IC50 values of J-AgNPs on each cell line were calculated using log viability vs. normalized response–variable slope (four parameters) (B).
 ) and MDA MB 231 (
) and MDA MB 231 ( ) and a normal cell line; MCF 10A (
) and a normal cell line; MCF 10A ( ). Two-way ANOVA p < 0.0001 (****) (A). Log dose-response relationship of J-AgNPs on the normalized viability of two human cancer cell lines; HCT116 (
). Two-way ANOVA p < 0.0001 (****) (A). Log dose-response relationship of J-AgNPs on the normalized viability of two human cancer cell lines; HCT116 ( ) and MDA MB 231 (
) and MDA MB 231 ( ) and one normal cell line MCF 10A (
) and one normal cell line MCF 10A ( ). IC50 values of J-AgNPs on each cell line were calculated using log viability vs. normalized response–variable slope (four parameters) (B).
). IC50 values of J-AgNPs on each cell line were calculated using log viability vs. normalized response–variable slope (four parameters) (B).

| Microbes | MIC (mg/mL) | MBC (mg/mL) | MIC/MBC |
|---|---|---|---|
| S. aureus | 0.8 | 1.1 | 0.72 |
| S. mutans | 0.8 | 1.1 | 0.72 |
| E. coli | 1.1 | 1.4 | 0.78 |
| K. pneumoniae | 1.1 | 1.4 | 0.78 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammed, A.E.; Al-Keridis, L.A.; Rahman, I.; Alotaibi, M.O.; Suliman, R.S.; Alrajhi, A.M.; Elobeid, M.M.; Alothman, M.R.; Alhomaidi, E.A.; Korany, S.M. Silver Nanoparticles Formation by Jatropha integerrima and LC/MS-QTOF-Based Metabolite Profiling. Nanomaterials 2021, 11, 2400. https://doi.org/10.3390/nano11092400
Mohammed AE, Al-Keridis LA, Rahman I, Alotaibi MO, Suliman RS, Alrajhi AM, Elobeid MM, Alothman MR, Alhomaidi EA, Korany SM. Silver Nanoparticles Formation by Jatropha integerrima and LC/MS-QTOF-Based Metabolite Profiling. Nanomaterials. 2021; 11(9):2400. https://doi.org/10.3390/nano11092400
Chicago/Turabian StyleMohammed, Afrah E., Lamya Ahmed Al-Keridis, Ishrat Rahman, Modhi O. Alotaibi, Rasha Saad Suliman, Aisha Mohammed Alrajhi, Mudawi M. Elobeid, Monerah R. Alothman, Eman A. Alhomaidi, and Shereen M. Korany. 2021. "Silver Nanoparticles Formation by Jatropha integerrima and LC/MS-QTOF-Based Metabolite Profiling" Nanomaterials 11, no. 9: 2400. https://doi.org/10.3390/nano11092400
APA StyleMohammed, A. E., Al-Keridis, L. A., Rahman, I., Alotaibi, M. O., Suliman, R. S., Alrajhi, A. M., Elobeid, M. M., Alothman, M. R., Alhomaidi, E. A., & Korany, S. M. (2021). Silver Nanoparticles Formation by Jatropha integerrima and LC/MS-QTOF-Based Metabolite Profiling. Nanomaterials, 11(9), 2400. https://doi.org/10.3390/nano11092400







