Lipoperoxide Nanoemulsion as Adjuvant in Cisplatin Cancer Therapy: In Vitro Study on Human Colon Adenocarcinoma DLD-1 Cells
Abstract
:1. Introduction
2. Material and Methods
2.1. Cell Cultures
2.2. NK600 Preparations
2.3. N-K600 Size and Electrophoretic Measurements
2.4. N-K600 Stability
2.5. Cisplatin-CCDP
2.6. Experimental Conditions
2.7. MTT Assay
2.8. DNA Assay
2.9. qPCR
2.10. Western Blotting
2.10.1. Nuclear/Cytosol Fractionation
2.10.2. Whole Lysates collection
2.10.3. Immunoblotting
2.11. GSH Measurement
2.12. Statistical Analysis
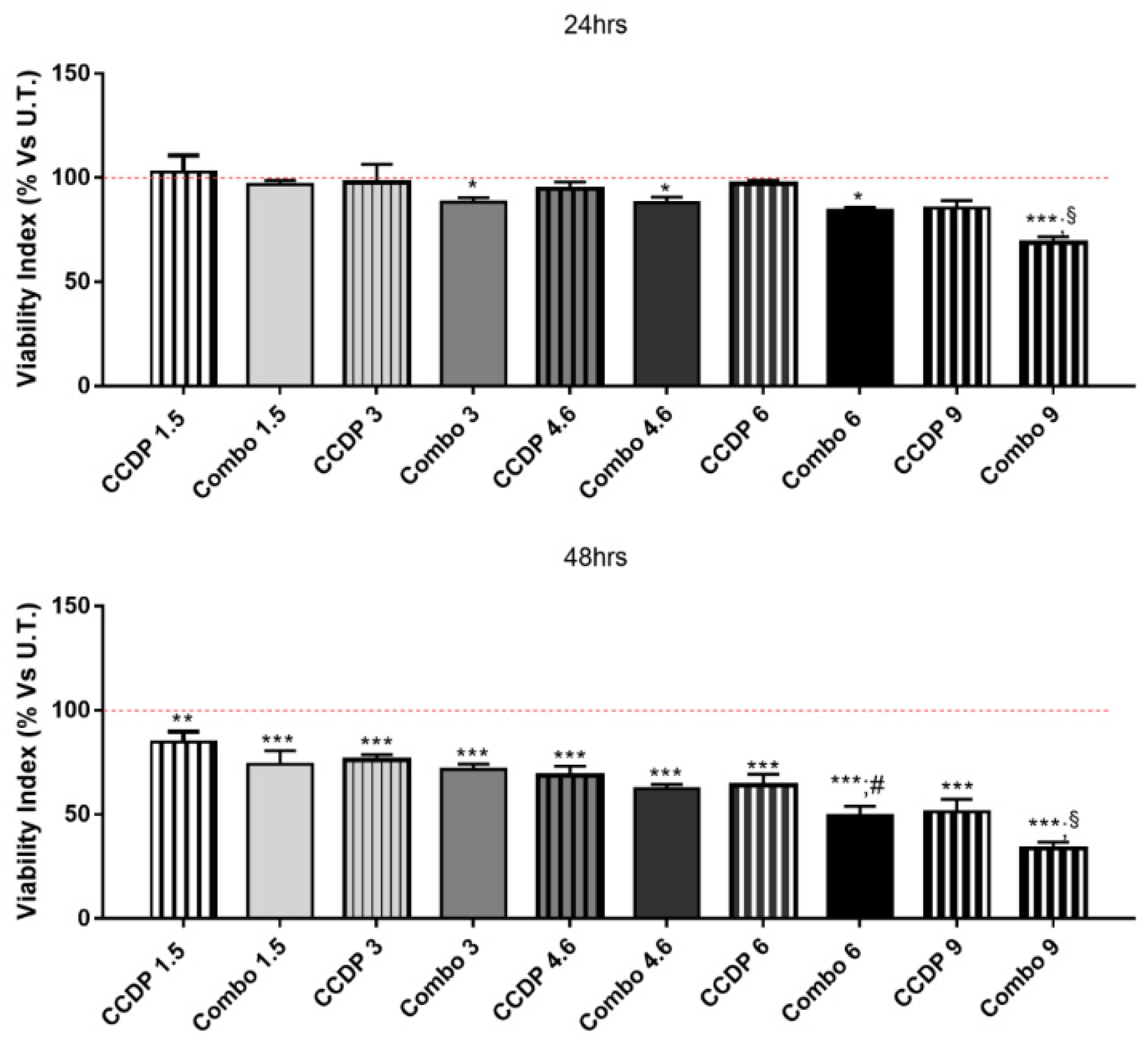
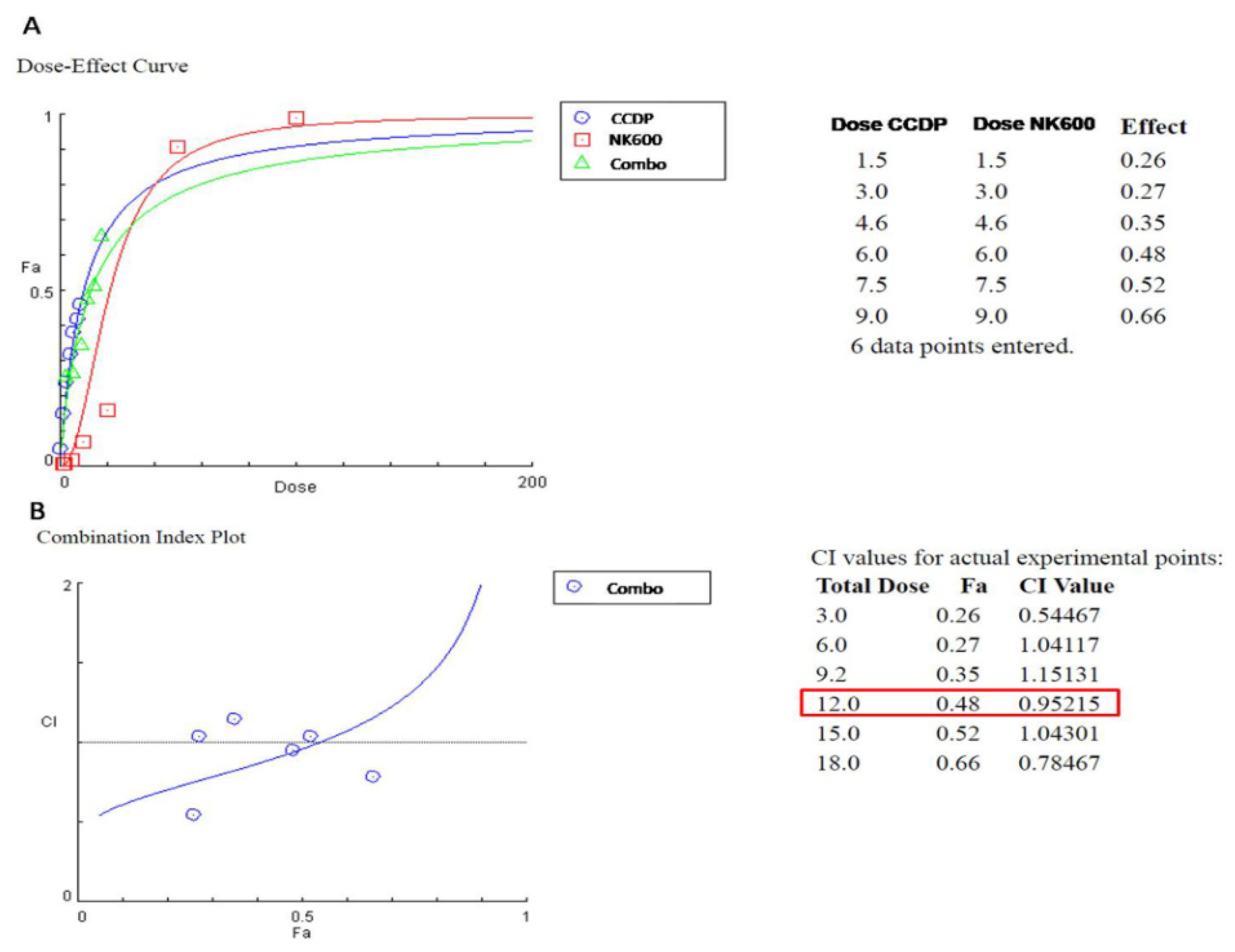
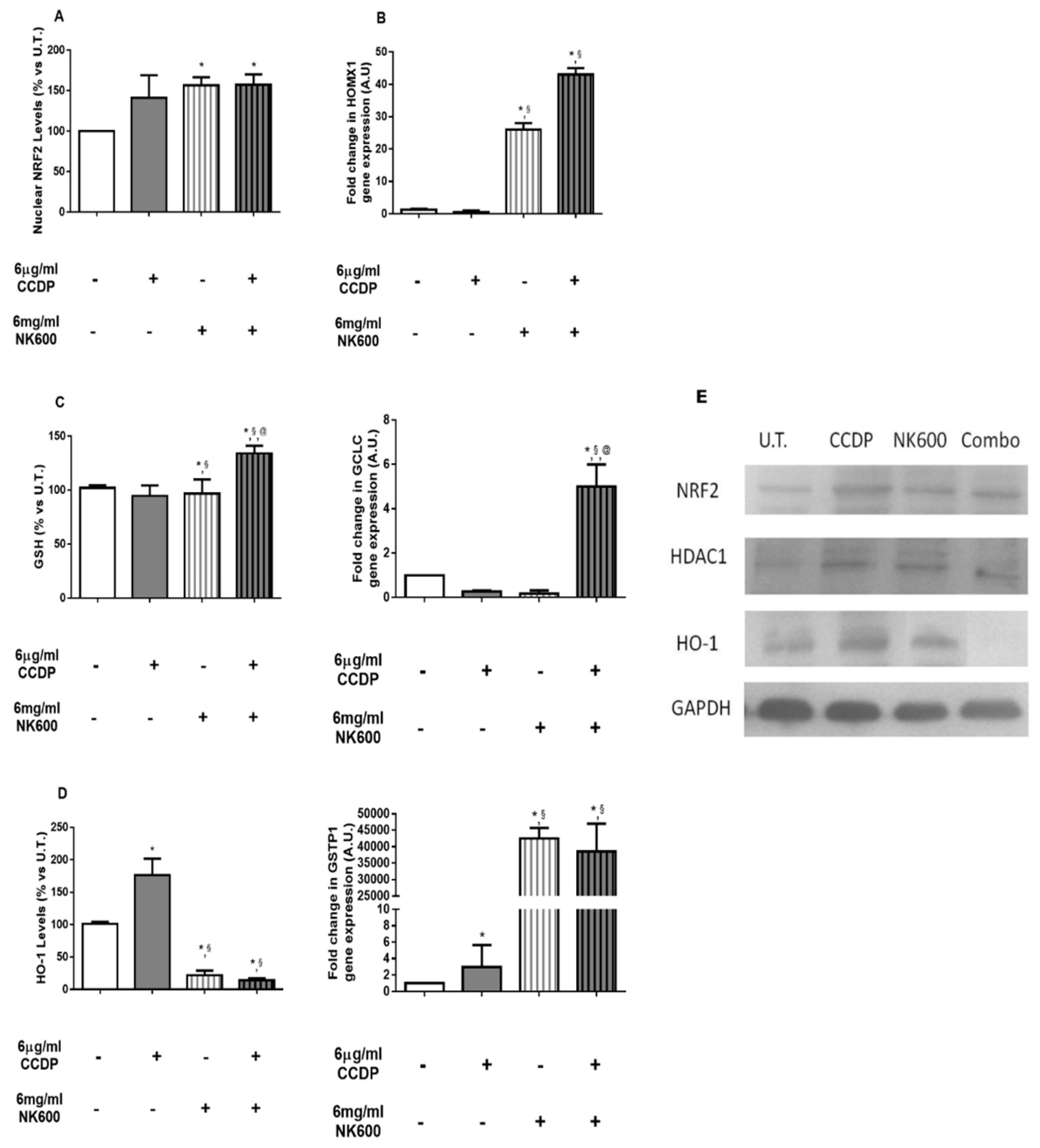
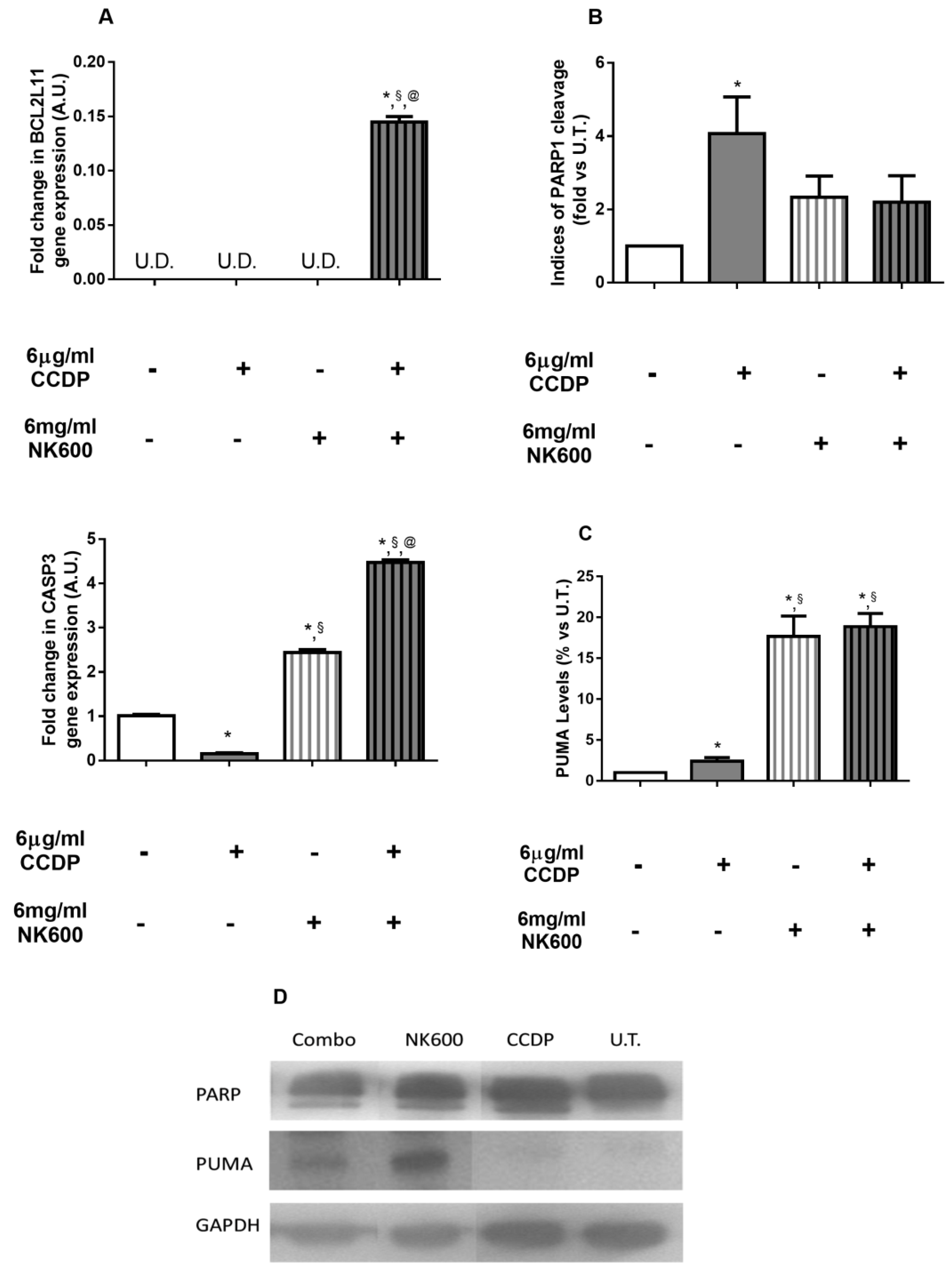
3. Results
3.1. NK600 Characterization
3.2. Synergic Effect of NK600-CCDP Combination
3.3. Basal Nuclear Levels of NRF2 and Gene/Protein Involved in the NRF2 Pathway on DLD1 Cells Treated with CCDP, NK600, and Combo 6
3.4. P53-Dependent Pathway Interferes with Antioxidant Responses
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siddik, Z.H. Cisplatin: Mode of Cytotoxic Action and Molecular Basis of Resistance. Oncogene 2003, 22, 7265–7279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borst, P.; Evers, R.; Kool, M.; Wijnholds, J. A Family of Drug Transporters: The Multidrug Resistance-Associated Proteins. J. Natl. Cancer Inst. 2000, 92, 1295–1302. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.-C.; Chien, C.-Y.; Chiang, W.-F.; Lin, C.-S.; Hour, T.-C.; Chen, H.-R.; Wang, L.-F.; Ko, J.-Y.; Chang, C.-H.; Chen, J.Y.-F. P22phox Confers Resistance to Cisplatin, by Blocking Its Entry into the Nucleus. Oncotarget 2015, 6, 4110–4125. [Google Scholar] [CrossRef]
- Wijdeven, R.H.; Pang, B.; Assaraf, Y.G.; Neefjes, J. Old Drugs, Novel Ways out: Drug Resistance toward Cytotoxic Chemotherapeutics. Drug Resist. Updates. 2016, 28, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Omran, Z.; Scaife, P.; Stewart, S.; Rauch, C. Physical and Biological Characteristics of Multi Drug Resistance (MDR): An Integral Approach Considering PH and Drug Resistance in Cancer. Semin. Cancer Biol. 2017, 43, 42–48. [Google Scholar] [CrossRef]
- Xu, X.; Xie, K.; Zhang, X.-Q.; Pridgen, E.M.; Park, G.Y.; Cui, D.S.; Shi, J.; Wu, J.; Kantoff, P.W.; Lippard, S.J.; et al. Enhancing Tumor Cell Response to Chemotherapy through Nanoparticle-Mediated Codelivery of SiRNA and Cisplatin Prodrug. Proc. Natl. Acad. Sci. USA 2013, 110, 18638–18643. [Google Scholar] [CrossRef] [Green Version]
- Iyer, A.K.; Singh, A.; Ganta, S.; Amiji, M.M. Role of Integrated Cancer Nanomedicine in Overcoming Drug Resistance. Adv. Drug Deliv. Rev. 2013, 65, 1784–1802. [Google Scholar] [CrossRef] [PubMed]
- Pelicano, H.; Carney, D.; Huang, P. ROS Stress in Cancer Cells and Therapeutic Implications. Drug Resist. Updat. 2004, 7, 97–110. [Google Scholar] [CrossRef]
- Schumacker, P.T. Reactive Oxygen Species in Cancer Cells: Live by the Sword, Die by the Sword. Cancer Cell 2006, 10, 175–176. [Google Scholar] [CrossRef] [Green Version]
- Trachootham, D.; Alexandre, J.; Huang, P. Targeting Cancer Cells by ROS-Mediated Mechanisms: A Radical Therapeutic Approach? Nat. Rev. Drug Discov. 2009, 8, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Trachootham, D.; Zhou, Y.; Zhang, H.; Demizu, Y.; Chen, Z.; Pelicano, H.; Chiao, P.J.; Achanta, G.; Arlinghaus, R.B.; Liu, J.; et al. Selective Killing of Oncogenically Transformed Cells through a ROS-Mediated Mechanism by Beta-Phenylethyl Isothiocyanate. Cancer Cell 2006, 10, 241–252. [Google Scholar] [CrossRef] [Green Version]
- Finocchiaro, C.; Segre, O.; Fadda, M.; Monge, T.; Scigliano, M.; Schena, M.; Tinivella, M.; Tiozzo, E.; Catalano, M.G.; Pugliese, M.; et al. Effect of N-3 Fatty Acids on Patients with Advanced Lung Cancer: A Double-Blind, Placebo-Controlled Study. Br. J. Nutr. 2012, 108, 327–333. [Google Scholar] [CrossRef] [Green Version]
- Miccadei, S.; Masella, R.; Mileo, A.M.; Gessani, S. Ω3 Polyunsaturated Fatty Acids as Immunomodulators in Colorectal Cancer: New Potential Role in Adjuvant Therapies. Front. Immunol. 2016, 7, 486. [Google Scholar] [CrossRef] [Green Version]
- Sturlan, S.; Baumgartner, M.; Roth, E.; Bachleitner-Hofmann, T. Docosahexaenoic Acid Enhances Arsenic Trioxide-Mediated Apoptosis in Arsenic Trioxide-Resistant HL-60 Cells. Blood 2003, 101, 4990–4997. [Google Scholar] [CrossRef] [Green Version]
- Baumgartner, M.; Sturlan, S.; Roth, E.; Wessner, B.; Bachleitner-Hofmann, T. Enhancement of Arsenic Trioxide-Mediated Apoptosis Using Docosahexaenoic Acid in Arsenic Trioxide-Resistant Solid Tumor Cells. Int. J. Cancer 2004, 112, 707–712. [Google Scholar] [CrossRef] [PubMed]
- Pettazzoni, P.; Pizzimenti, S.; Toaldo, C.; Sotomayor, P.; Tagliavacca, L.; Liu, S.; Wang, D.; Minelli, R.; Ellis, L.; Atadja, P.; et al. Induction of Cell Cycle Arrest and DNA Damage by the HDAC Inhibitor Panobinostat (LBH589) and the Lipid Peroxidation End Product 4-Hydroxynonenal in Prostate Cancer Cells. Free Radic. Biol. Med. 2011, 50, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Kanner, J. Dietary Advanced Lipid Oxidation Endproducts Are Risk Factors to Human Health. Mol. Nutr. Food Res. 2007, 51, 1094–1101. [Google Scholar] [CrossRef] [PubMed]
- Ganta, S.; Amiji, M. Coadministration of Paclitaxel and Curcumin in Nanoemulsion Formulations To Overcome Multidrug Resistance in Tumor Cells. Mol. Pharm. 2009, 6, 928–939. [Google Scholar] [CrossRef]
- Zheng, H.; Li, J.; Ning, F.; Wijaya, W.; Chen, Y.; Xiao, J.; Cao, Y.; Huang, Q. Improving in Vitro Bioaccessibility and Bioactivity of Carnosic Acid Using a Lecithin-Based Nanoemulsion System. Food Funct. 2021. [Google Scholar] [CrossRef]
- Desai, A.; Vyas, T.; Amiji, M. Cytotoxicity and Apoptosis Enhancement in Brain Tumor Cells Upon Coadministration of Paclitaxel and Ceramide in Nanoemulsion Formulations. J. Pharm. Sci. 2008, 97, 2745–2756. [Google Scholar] [CrossRef]
- Mosmann, T. Rapid Colorimetric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Bijnsdorp, I.V.; Giovannetti, E.; Peters, G.J. Analysis of Drug Interactions. In Cancer Cell Culture; Cree, I.A., Ed.; Humana Press: Totowa, NJ, Canada, 2011; Volume 731, pp. 421–434. ISBN 978-1-61779-079-9. [Google Scholar]
- Chou, T.-C. Drug Combination Studies and Their Synergy Quantification Using the Chou-Talalay Method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rao, J.; Otto, W.R. Fluorimetric DNA Assay for Cell Growth Estimation. Anal. Biochem. 1992, 207, 186–192. [Google Scholar] [CrossRef]
- Hissin, P.J.; Hilf, R. A Fluorometric Method for Determination of Oxidized and Reduced Glutathione in Tissues. Anal. Biochem. 1976, 74, 214–226. [Google Scholar] [CrossRef]
- Kaul, N.; Choi, J.; Forman, H.J. Transmembrane Redox Signaling Activates NF-ΚB in Macrophages. Free Radic. Biol. Med. 1998, 24, 202–207. [Google Scholar] [CrossRef]
- Heo, W.; Kim, J.H.; Pan, J.H.; Kim, Y.J. Lecithin-Based Nano-Emulsification Improves the Bioavailability of Conjugated Linoleic Acid. J. Agric. Food Chem. 2016, 64, 1355–1360. [Google Scholar] [CrossRef] [PubMed]
- Zweers, M.L.T.; Grijpma, D.W.; Engbers, G.H.M.; Feijen, J. The Preparation of Monodisperse Biodegradable Polyester Nanoparticles with a Controlled Size. J. Biomed. Mater. Res. 2003, 66B, 559–566. [Google Scholar] [CrossRef]
- Yu, X.; Kensler, T. Nrf2 as a Target for Cancer Chemoprevention. Mutat. Res. 2005, 591, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Furfaro, A.L.; Traverso, N.; Domenicotti, C.; Piras, S.; Moretta, L.; Marinari, U.M.; Pronzato, M.A.; Nitti, M. The Nrf2/HO-1 Axis in Cancer Cell Growth and Chemoresistance. Oxidative Med. Cell. Longev. 2016, 2016, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Silva, M.M.; Rocha, C.R.R.; Kinker, G.S.; Pelegrini, A.L.; Menck, C.F.M. The Balance between NRF2/GSH Antioxidant Mediated Pathway and DNA Repair Modulates Cisplatin Resistance in Lung Cancer Cells. Sci. Rep. 2019, 9, 17639. [Google Scholar] [CrossRef] [PubMed]
- Twentyman, P.; Bagrij, T. The Influence of Glutathione Metabolism on Multidrug Resistance in MRP-Overexpressing Cells. Drug Resist. Updat. 1998, 1, 121–127. [Google Scholar] [CrossRef]
- Franklin, C.C.; Krejsa, C.M.; Pierce, R.H.; White, C.C.; Fausto, N.; Kavanagh, T.J. Caspase-3-Dependent Cleavage of the Glutamate-l-Cysteine Ligase Catalytic Subunit during Apoptotic Cell Death. Am. J. Pathol. 2002, 160, 1887–1894. [Google Scholar] [CrossRef] [Green Version]
- Traverso, N.; Ricciarelli, R.; Nitti, M.; Marengo, B.; Furfaro, A.L.; Pronzato, M.A.; Marinari, U.M.; Domenicotti, C. Role of Glutathione in Cancer Progression and Chemoresistance. Oxidative Med. Cell. Longev. 2013, 2013, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.-G.; Xu, H.; Suo, S.-S.; Xu, X.-L.; Ni, M.-W.; Gu, L.-H.; Chen, W.; Wang, L.-Y.; Zhao, Y.; Tian, B.; et al. The Essential Role of H19 Contributing to Cisplatin Resistance by Regulating Glutathione Metabolism in High-Grade Serous Ovarian Cancer. Sci. Rep. 2016, 6, 26093. [Google Scholar] [CrossRef] [Green Version]
- Brouard, S.; Otterbein, L.E.; Anrather, J.; Tobiasch, E.; Bach, F.H.; Choi, A.M.K.; Soares, M.P. Carbon Monoxide Generated by Heme Oxygenase 1 Suppresses Endothelial Cell Apoptosis. J. Exp. Med. 2000, 192, 1015–1026. [Google Scholar] [CrossRef]
- Lin, C.-W.; Shen, S.-C.; Hou, W.-C.; Yang, L.-Y.; Chen, Y.-C. Heme Oxygenase-1 Inhibits Breast Cancer Invasion via Suppressing the Expression of Matrix Metalloproteinase-9. Mol. Cancer Ther. 2008, 7, 1195–1206. [Google Scholar] [CrossRef] [Green Version]
- Yee, K.S.; Vousden, K.H. Complicating the Complexity of P53. Carcinogenesis 2005, 26, 1317–1322. [Google Scholar] [CrossRef]
- Riley, T.; Sontag, E.; Chen, P.; Levine, A. Transcriptional Control of Human P53-Regulated Genes. Nat. Rev. Mol. Cell Biol. 2008, 9, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Vousden, K.H. PUMA, a Novel Proapoptotic Gene, Is Induced by P53. Mol. Cell 2001, 7, 683–694. [Google Scholar] [CrossRef]
- Han, J.-w.; Flemington, C.; Houghton, A.B.; Gu, Z.; Zambetti, G.P.; Lutz, R.J.; Zhu, L.; Chittenden, T. Expression of Bbc3, a pro-Apoptotic BH3-Only Gene, Is Regulated by Diverse Cell Death and Survival Signals. Proc. Natl. Acad. Sci. USA 2001, 98, 11318–11323. [Google Scholar] [CrossRef] [Green Version]
- Oda, E. Noxa, a BH3-Only Member of the Bcl-2 Family and Candidate Mediator of P53-Induced Apoptosis. Science 2000, 288, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.-W. Mediation of Poly(ADP-Ribose) Polymerase-1-Dependent Cell Death by Apoptosis-Inducing Factor. Science 2002, 297, 259–263. [Google Scholar] [CrossRef] [PubMed]
- Jeffers, J.R.; Parganas, E.; Lee, Y.; Yang, C.; Wang, J.; Brennan, J.; MacLean, K.H.; Han, J.; Chittenden, T.; Ihle, J.N.; et al. Puma Is an Essential Mediator of P53-Dependent and -Independent Apoptotic Pathways. Cancer Cell 2003, 4, 321–328. [Google Scholar] [CrossRef] [Green Version]
- Yu, J.; Zhang, L. No PUMA, No Death. Cancer Cell 2003, 4, 248–249. [Google Scholar] [CrossRef] [Green Version]
- Papaianni, E.; El Maadidi, S.; Schejtman, A.; Neumann, S.; Maurer, U.; Marino-Merlo, F.; Mastino, A.; Borner, C. Phylogenetically Distant Viruses Use the Same BH3-Only Protein Puma to Trigger Bax/Bak-Dependent Apoptosis of Infected Mouse and Human Cells. PLoS ONE 2015, 10, e0126645. [Google Scholar] [CrossRef] [Green Version]
- Iles, N.; Rulten, S.; El-Khamisy, S.F.; Caldecott, K.W. APLF (C2orf13) Is a Novel Human Protein Involved in the Cellular Response to Chromosomal DNA Strand Breaks. MCB 2007, 27, 3793–3803. [Google Scholar] [CrossRef] [Green Version]
- Masson, M.; Niedergang, C.; Schreiber, V.; Muller, S.; Menissier-de Murcia, J.; de Murcia, G. XRCC1 Is Specifically Associated with Poly(ADP-Ribose) Polymerase and Negatively Regulates Its Activity Following DNA Damage. Mol. Cell. Biol. 1998, 18, 3563–3571. [Google Scholar] [CrossRef] [Green Version]
- Ahel, D.; Horejsi, Z.; Wiechens, N.; Polo, S.E.; Garcia-Wilson, E.; Ahel, I.; Flynn, H.; Skehel, M.; West, S.C.; Jackson, S.P.; et al. Poly(ADP-Ribose)-Dependent Regulation of DNA Repair by the Chromatin Remodeling Enzyme ALC1. Science 2009, 325, 1240–1243. [Google Scholar] [CrossRef] [Green Version]
- Beik, J.; Khateri, M.; Khosravi, Z.; Kamrava, S.K.; Kooranifar, S.; Ghaznavi, H.; Shakeri-Zadeh, A. Gold Nanoparticles in Combinatorial Cancer Therapy Strategies. Coord. Chem. Rev. 2019, 387, 299–324. [Google Scholar] [CrossRef]
- Meier, J.D.; Oliver, D.A.; Varvares, M.A. Surgical Margin Determination in Head and Neck Oncology: Current Clinical Practice. The Results of an International American Head and Neck Society Member Survey. Head Neck 2005, 27, 952–958. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug Resistance in Cancer: Role of ATP–Dependent Transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef] [Green Version]
- Peer, D.; Margalit, R. Fluoxetine and Reversal of Multidrug Resistance. Cancer Lett. 2006, 237, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.M.; Wilson, W.R. Exploiting Tumour Hypoxia in Cancer Treatment. Nat. Rev. Cancer 2004, 4, 437–447. [Google Scholar] [CrossRef]
- Moeller, B.J.; Richardson, R.A.; Dewhirst, M.W. Hypoxia and Radiotherapy: Opportunities for Improved Outcomes in Cancer Treatment. Cancer Metastasis Rev. 2007, 26, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Tian, G.; Zhang, X.; Gu, Z.; Zhao, Y. Recent Advances in Upconversion Nanoparticles-Based Multifunctional Nanocomposites for Combined Cancer Therapy. Adv. Mater. 2015, 27, 7692–7712. [Google Scholar] [CrossRef]
- Lukianova-Hleb, E.Y.; Ren, X.; Sawant, R.R.; Wu, X.; Torchilin, V.P.; Lapotko, D.O. On-Demand Intracellular Amplification of Chemoradiation with Cancer-Specific Plasmonic Nanobubbles. Nat. Med. 2014, 20, 778–784. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Lu, J.; Lin, W. Hybrid Nanoparticles for Combination Therapy of Cancer. J. Control. Release 2015, 219, 224–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manchado, E.; Weissmueller, S.; Morris, J.P.; Chen, C.-C.; Wullenkord, R.; Lujambio, A.; de Stanchina, E.; Poirier, J.T.; Gainor, J.F.; Corcoran, R.B.; et al. A Combinatorial Strategy for Treating KRAS-Mutant Lung Cancer. Nature 2016, 534, 647–651. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Sun, Z.; Wang, X.-J.; Jiang, T.; Huang, Z.; Fang, D.; Zhang, D.D. Direct Interaction between Nrf2 and P21Cip1/WAF1 Upregulates the Nrf2-Mediated Antioxidant Response. Mol. Cell 2009, 34, 663–673. [Google Scholar] [CrossRef] [Green Version]
- Asher, G. A Mechanism of Ubiquitin-Independent Proteasomal Degradation of the Tumor Suppressors P53 and P73. Genes Dev. 2005, 19, 316–321. [Google Scholar] [CrossRef] [Green Version]
- Kalo, E.; Kogan-Sakin, I.; Solomon, H.; Bar-Nathan, E.; Shay, M.; Shetzer, Y.; Dekel, E.; Goldfinger, N.; Buganim, Y.; Stambolsky, P.; et al. Mutant P53 R273H Attenuates the Expression of Phase 2 Detoxifying Enzymes and Promotes the Survival of Cells with High Levels of Reactive Oxygen Species. J. Cell Sci. 2012, 125, 5578–5586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, T.; Chen, N.; Zhao, F.; Wang, X.-J.; Kong, B.; Zheng, W.; Zhang, D.D. High Levels of Nrf2 Determine Chemoresistance in Type II Endometrial Cancer. Cancer Res. 2010, 70, 5486–5496. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Test | Cell Culture Supports | N° Cells | Experimental Treatments |
|---|---|---|---|
| Viability Assay (MTT) | 96 MW | 10 × 103/well | 1.5–9 µg/mL CCDP Combos: 1.5–9 µg/mL CCDP+ 1.5–9 mg/mL NK600 |
| Proliferation Assay (DNA Assay) | 12 MW | 80 × 103/well | 1.5–9 µg/mL CCDP Combos: 1.5–9 µg/mL CCDP+ 1.5–9 mg/mL NK600 |
| qPCR, WB, GSH | Flask 75 cm2 | 150 × 103/well | 6 mg/mL NK600 6 µg/mL CCDP Combo: 6 µg/mL CCDP+ 6 mg/mL NK600 |
| Gene | Assay Type | Amplicon |
|---|---|---|
| BCL2L11 | SYBR Green | AGACCAAATGGCAAAGCAACCTTCTGATGTAAGTTCTGAGTGTGACCGAGAAGGTA GACAATTGCAGCCTGCGGAGAGGCCTCCCCAGCTCAGACCTGGGGCCCCTACCTCC CTACAGACAGAGCCACAAGGTAATCC |
| CASP3 | SYBR Green | TTCTGAATGTTTCCCTGAGGTTTGCTGCATCGACATCTGTACCAGACCGAGATGTCA TTCCAGTGCTTTTATGAAAATTCTTATTATTAATTATTATACATAAACCCATCTCAGGAT AATCCATTTTATAACTGTTGTCCAGGGATATTCCAGAGTCCATTGATTCGCTTCCAT |
| GCLC | SYBR Green | GAAGTTATTGTGCAAAGAGCCTGATTTTCTTCTAATATAGAAGTAGCCTCCTTCCGGC GTTTTCGCATGTTGGCCTCAACTGTATTGAACTCGGACATTGTTCCTCCGTAGGGCT GTCCTGGTGTCCCTTCAATCATGTAACTCCCATACTCTGGTCTCCAAAGGGTAGGAT GGTTTGGGTTTGTCCTTTCCCCCTTCTCTTGCAGAGTTTCAAGAACT |
| GSTP1 | SYBR Green | CTCACCCTGTACCAGTCCAATACCATCCTGCGTCACCTGGGCCGCACCCTTGGGCTC TATGGGAAGGACCAGCAGGAGGCAGCCCTGGTGGACATGGTGAATGACGGCGTGG AGGACCTCC |
| HMOX1 | SYBR Green | CATTGCCAGTGCCACCAAGTTCAAGCAGCTCTACCGCTCCCGCATGAACTCCCTGG AGATGACTCCCGCAGTCAGGCAGAGGGTGATAGAAGAGGCCAAGACTGCGTTCCTG CTCAACATCCAGCTCTTTGAGGAGTTGCAGGAGCTGCTGACCCATGACACCAAGGA C |
| LDOC1 | SYBR Green | TGGCATTTTCCAGGTTGGTCCTGATCTCGAAAGGTGGTAGTCTGTAGGTGGGATGTG GTGAGTGGATGTGAAGTGGCAGCATAGTTCTCTGGGAAG |
| GAPDH | SYBR Green | GTATGACAACGAATTTGGCTACAGCAACAGGGTGGTGGACCTCATGGCCCACATGG CCTCCAAGGAGTAAGACCCCTGGACCACCAGCCCCAGCAAGAGCACAAGAGGAAG AGAGAGACCCTCACTGCTGGGGAGTCCCTGCCACAC |
| Protein | MW (kDa) | Company |
|---|---|---|
| NRF2 (A-10): sc-365949 | 61 | Santa Cruz Biotechnology, Santa Cruz, CA, USA |
| NF-kB (D14E12) | 65 | Cell Signaling Technology, Danvers, MA, USA |
| NF-kB-P (Ser536-93H1) | 65 | Cell Signaling Technology, Danvers, MA, USA |
| PARP (46D11) | 116/89 | Cell Signaling Technology, Danvers, MA, USA |
| PUMA(D30C10) | 23 | Cell Signaling Technology, Danvers, MA, USA |
| P53 | 53 | Cell Signaling Technology, Danvers, MA, USA |
| HO-1 (TA327035) | 32 | Origene, Rockville, MD, USA |
| HDAC1 (H-11): sc-8410 | 60 | Santa Cruz Biotechnology, Santa Cruz, CA, USA |
| GAPDH (0411): sc-47724 | 37 | Santa Cruz Biotechnology, Santa Cruz, CA, USA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vernazza, S.; Dellacasa, E.; Tirendi, S.; Pastorino, L.; Bassi, A.M. Lipoperoxide Nanoemulsion as Adjuvant in Cisplatin Cancer Therapy: In Vitro Study on Human Colon Adenocarcinoma DLD-1 Cells. Nanomaterials 2021, 11, 1365. https://doi.org/10.3390/nano11061365
Vernazza S, Dellacasa E, Tirendi S, Pastorino L, Bassi AM. Lipoperoxide Nanoemulsion as Adjuvant in Cisplatin Cancer Therapy: In Vitro Study on Human Colon Adenocarcinoma DLD-1 Cells. Nanomaterials. 2021; 11(6):1365. https://doi.org/10.3390/nano11061365
Chicago/Turabian StyleVernazza, Stefania, Elena Dellacasa, Sara Tirendi, Laura Pastorino, and Anna Maria Bassi. 2021. "Lipoperoxide Nanoemulsion as Adjuvant in Cisplatin Cancer Therapy: In Vitro Study on Human Colon Adenocarcinoma DLD-1 Cells" Nanomaterials 11, no. 6: 1365. https://doi.org/10.3390/nano11061365
APA StyleVernazza, S., Dellacasa, E., Tirendi, S., Pastorino, L., & Bassi, A. M. (2021). Lipoperoxide Nanoemulsion as Adjuvant in Cisplatin Cancer Therapy: In Vitro Study on Human Colon Adenocarcinoma DLD-1 Cells. Nanomaterials, 11(6), 1365. https://doi.org/10.3390/nano11061365







