Sesquiterpene-Loaded Co-Polymer Hybrid Nanoparticle Effects on Human Mast Cell Surface Receptor Expression, Granule Contents, and Degranulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Plant Material
2.3. Cell Culture
2.4. PLGA/PVA Nanoparticle Formulation, Synthesis and Characterization
2.5. Cytotoxic Analysis of PLGA/PVA Nanoparticle Formulation
2.6. Intracellular Analysis of LAD2 Cells Treated with NP by Electron Microscopy
2.7. Nanoparticle Cellular Internalization and Receptor Expression Analysis by Flow Cytometry
2.8. Nanoparticle Cellular Internalization Analysis by Fluorescence Microscopy
2.9. Real-Time PCR
2.10. Degranulation Assay
2.11. Statistical Analysis
3. Results
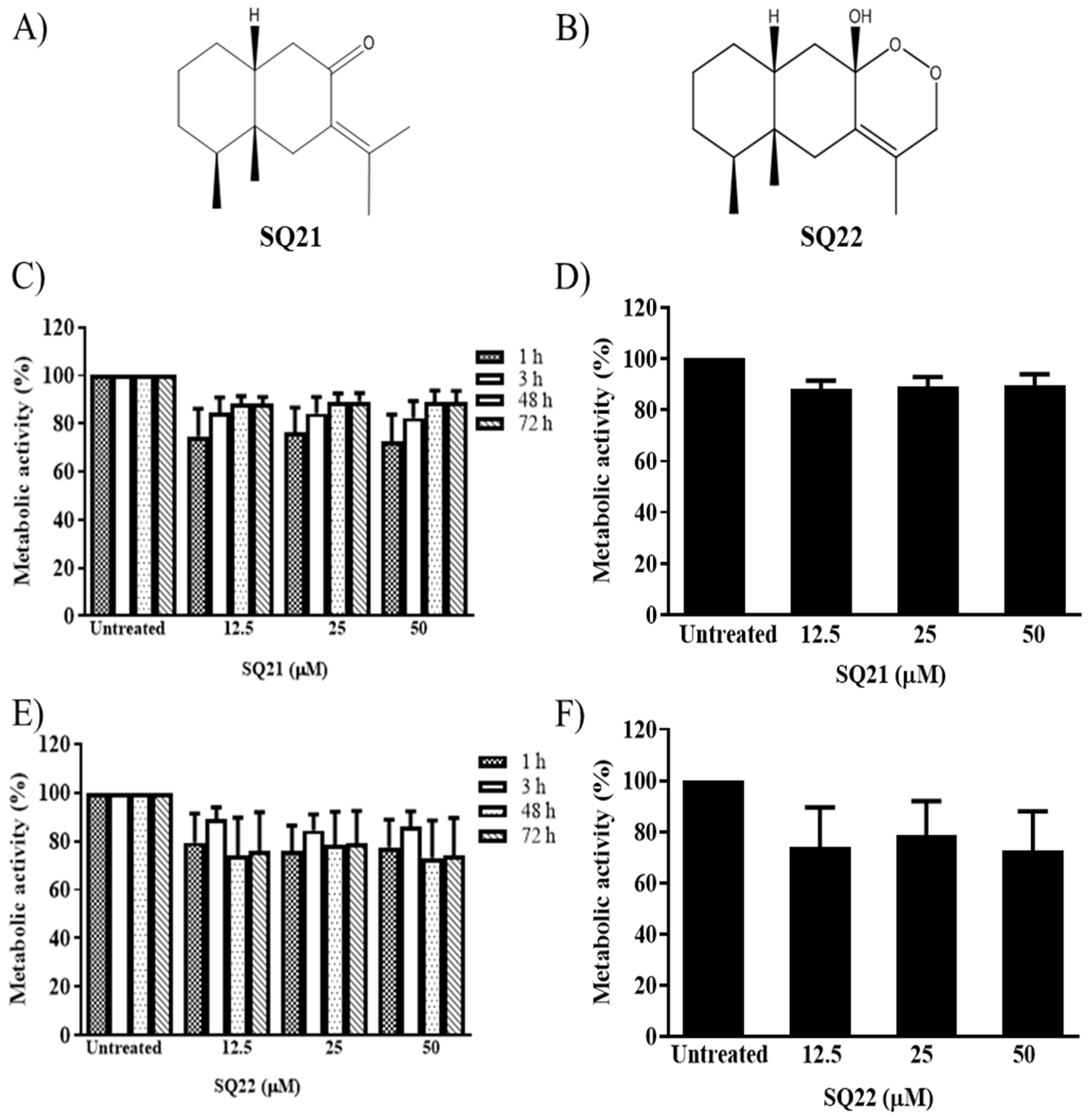
3.1. Effects of Eremophilane-Type SQs on Human Mast Cells
3.2. PLGA/PVA and Eremophilane-Type SQs Generated Uniform Nanoparticles
3.3. Effect of NPs on the Viability of LAD2 Cells
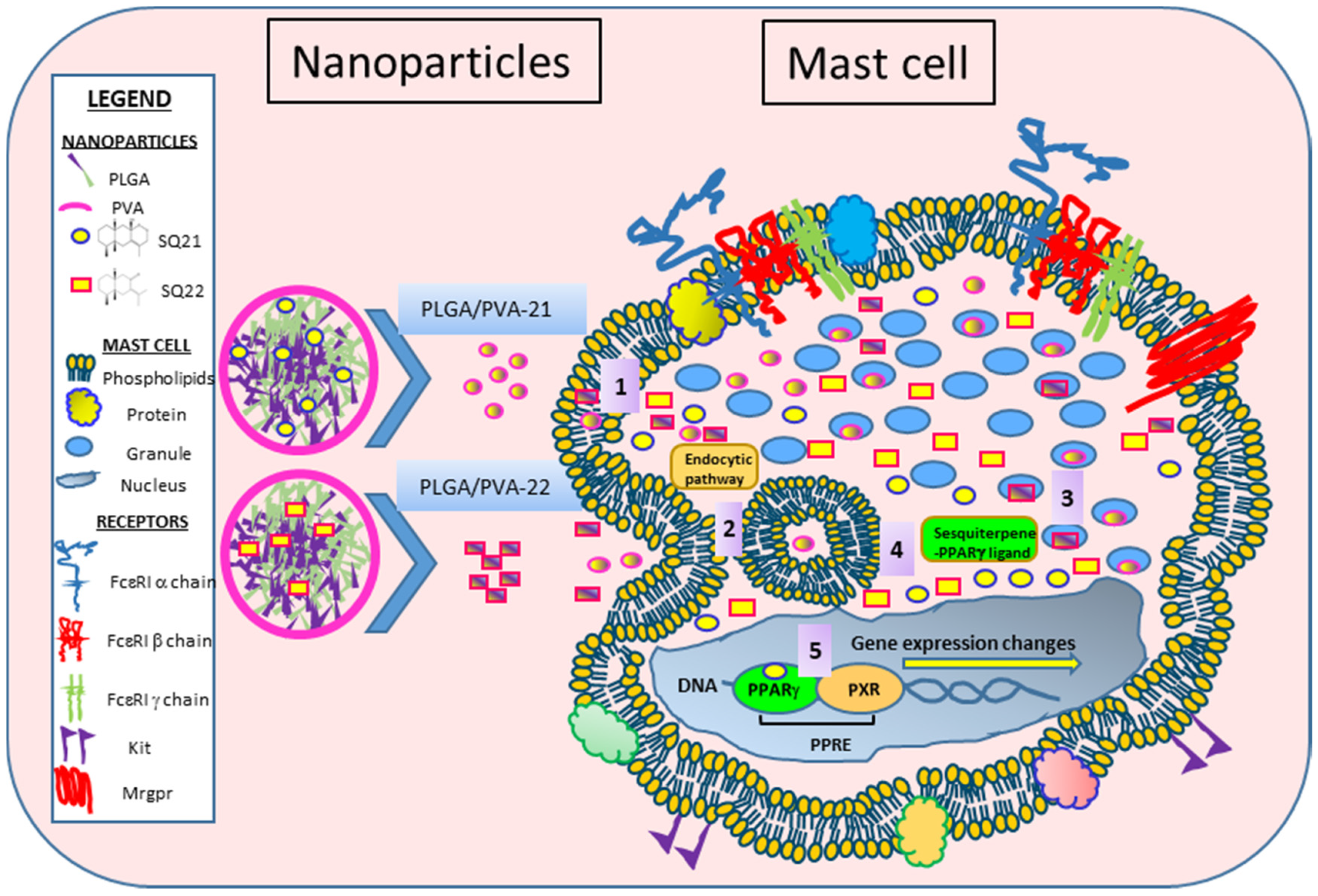
3.4. Internalization of NPs by LAD2 Cells
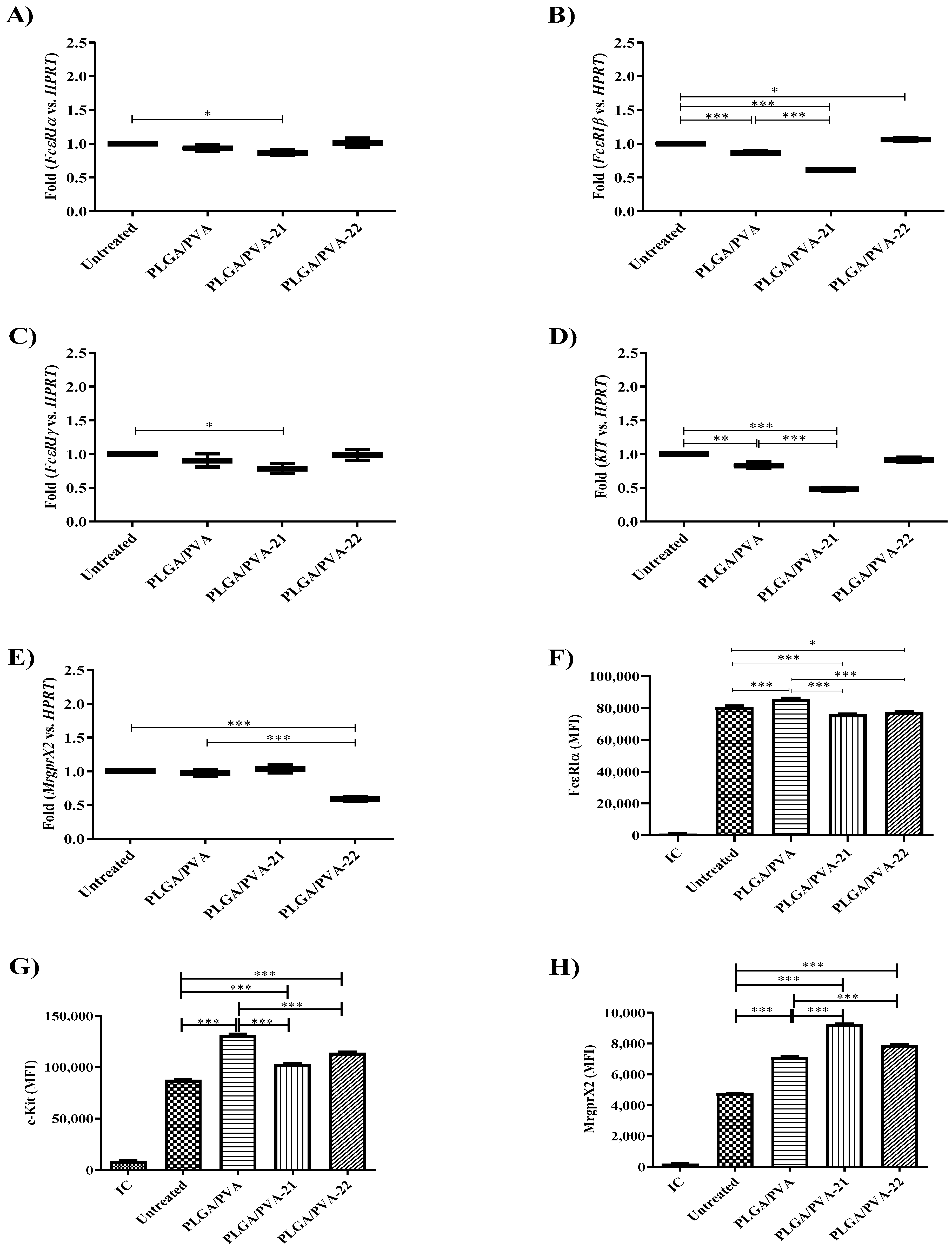
3.5. Hybrid Copolymer NPs Modify Kit and MrgprX2 Gene Expression
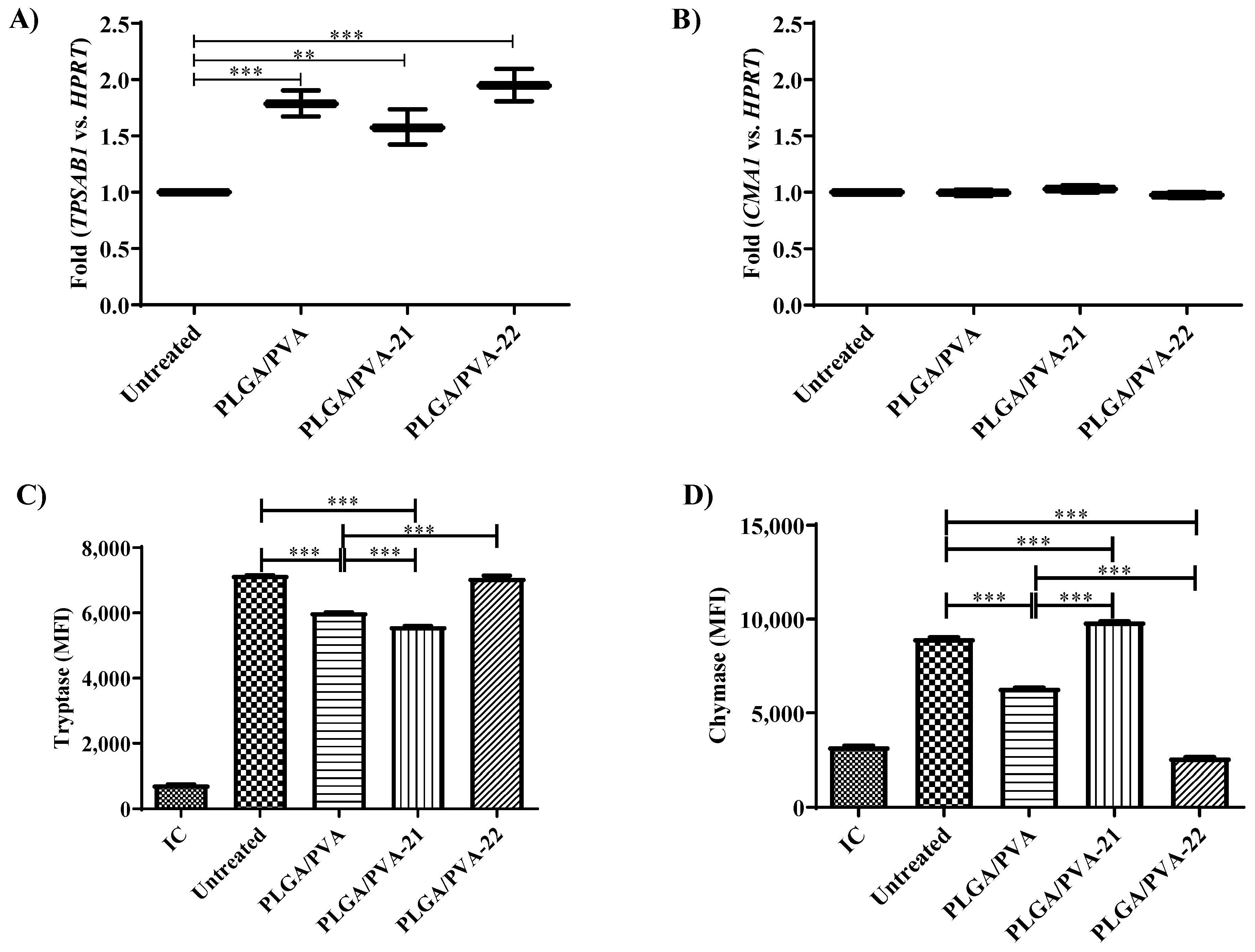
3.6. Effect of Hybrid Copolymer NPs on LAD2 Cell Granule Contents (Tryptase and Chymase Expression)
3.7. Effect of Hybrid Copolymer NPs on Human Mast Cell Degranulation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FDA. Available online: https://www.fda.gov (accessed on April 2020).
- EMA. Available online: https://www.ema.europa.eu (accessed on April 2020).
- Paradossi, G.; Cavalieri, F.; Chiessi, E.; Spagnoli, C.; Cowman, M.K. Poly(vinyl alcohol) as versatile biomaterial for potential biomedical applications. J. Mater. Sci. Mater. Med. 2003, 14, 687–691. [Google Scholar] [CrossRef]
- Alexandre, N.; Ribeiro, J.; Gartner, A.; Pereira, T.; Amorim, I.; Fragoso, J. Biocompatibility and hemocompatibility of polyvinyl alcohol hydrogel used for vascular grafting—In vitro and in vivo studies. J. Biomed. Mater. Res. A 2014, 102, 4262–4275. [Google Scholar] [PubMed]
- Liao, Y.-H.; Brown, M.B.; Jones, S.A.; Nazir, T.; Martin, G.P. The effects of polyvinyl alcohol on the in vitro stability and delivery of spray-dried protein particles from surfactant-free HFA 134a-based pressurised metered dose inhalers. Int. J. Pharm. 2005, 304, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Jampani, V.S.R.; Lagerwall, J.P.F. Realignment of Liquid Crystal Shells Driven by Temperature-Dependent Surfactant Solubility. Langmuir 2019, 35, 11132–11140. [Google Scholar] [CrossRef] [PubMed]
- Bartos, C.; Ambrus, R.; Katona, G.; Sovány, T.; Gáspár, R.; Márki, Á.; Ducza, E.; Ivanov, A.; Tömösi, F.; Janáky, T.; et al. Transformation of Meloxicam Containing Nanosuspension into Surfactant-Free Solid Compositions to Increase the Product Stability and Drug Bioavailability for Rapid Analgesia. Drug Des. Dev. Ther. 2019, 13, 4007–4020. [Google Scholar] [CrossRef] [PubMed]
- Dvorak, A.M. Ultrastructural Studies of Human Basophils and Mast Cells. J. Histochem. Cytochem. 2005, 53, 1043–1070. [Google Scholar] [CrossRef]
- Wernersson, S.; Pejler, G. Mast cell secretory granules: Armed for battle. Nat. Rev. Immunol. 2014, 14, 478–494. [Google Scholar] [CrossRef] [PubMed]
- Pejler, G. The emerging role of mast cell proteases in asthma. Eur. Respir. J. 2019, 54, 1900685. [Google Scholar] [CrossRef]
- Tanaka, S.; Takakuwa, Y. A method for detailed analysis of the structure of mast cell secretory granules by negative contrast imaging. Sci. Rep. 2016, 6, 23369. [Google Scholar] [CrossRef]
- Hou, C.; Kulka, M.; Zhang, J.; Li, Y.; Guo, F. Occurrence and biological activities of eremophilane-type sesquiterpenes. Mini Rev. Med. Chem. 2014, 14, 664–677. [Google Scholar] [CrossRef]
- Arizmendi, N.; Hou, C.; Guo, F.; Li, Y.; Kulka, M. Bicyclic eremophilane-type petasite sesquiterpenes potentiate peroxisome proliferator-activated receptor gamma activator-mediated inhibition of dendritic cells. Int. J. Immunopathol. Pharmacol. 2018, 32, 2058738418787739. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Tada, M.; Takahashi, T.; Horibe, I.; Ishii, H.; Iwata, T.; Kuriyama, K.; Tamura, Y.; Tori, K. NMR and CD spectral studies of conformational isomers of isoligularone and fukinone, naturally occurring cis-decalin derivatives. Chem. Lett. 1977, 6, 1191–1194. [Google Scholar] [CrossRef]
- Saito, Y.; Hattori, M.; Iwamoto, Y.; Takashima, Y.; Mihara, K.; Sasaki, Y.; Fujiwara, M.; Sakaoku, M.; Shimizu, A.; Chao, X.; et al. Overlapping chemical and genetic diversity in Lingularia lamarum and Lingularia subspicata. Isolation of ten new eremophilanes and new seco-bakkane compound. Tetrahedron 2011, 67, 2220–2231. [Google Scholar] [CrossRef]
- Qian, F.; Guo, G.; Li, Y.; Kulka, M. A novel eremophilane lactone inhibits FcepsilonRI-dependent release of pro-inflammatory mediators: Structure-dependent bioactivity. Inflamm. Res. 2016, 65, 303–311. [Google Scholar] [CrossRef] [PubMed]
- Tahara, K.; Tadokoro, S.; Yamamoto, H.; Kawashima, Y.; Hirashima, N. The suppression of IgE-mediated histamine release from mast cells following exocytic exclusion of biodegradable polymeric nanoparticles. Biomaterials 2012, 33, 343–351. [Google Scholar] [CrossRef]
- Compton, S.J.; Cairns, J.A.; Holgate, S.T.; Walls, A.F. Human mast cell tryptase stimulates the release of an IL-8-dependent neutrophil chemotactic activity from human umbilical vein endothelial cells (HUVEC). Clin. Exp. Immunol. 2000, 121, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Monument, M.J.; Hart, D.A.; Salo, P.T.; Befus, A.D.; Hildebrand, K.A. Neuroinflammatory Mechanisms of Connective Tissue Fibrosis: Targeting Neurogenic and Mast Cell Contributions. Adv. Wound Care 2015, 4, 137–151. [Google Scholar] [CrossRef]
- Mullan, C.S.; Riley, M.; Clarke, D.; Tatler, A.; Sutcliffe, A.; Knox, A.J.; Pang, L. Beta-tryptase regulates IL-8 expression in airway smooth muscle cells by a PAR-2-independent mechanism. Am. J. Respir. Cell Mol. Biol. 2008, 38, 600–608. [Google Scholar] [CrossRef]
- Cairns, J.A.; Walls, A.F. Mast cell tryptase is a mitogen for epithelial cells. Stimulation of IL-8 production and intercellular adhesion molecule-1 expression. J. Immunol. 1996, 156, 275–283. [Google Scholar]
- Compton, S.J.; Cairns, J.A.; Holgate, S.T.; Walls, A.F. The role of mast cell tryptase in regulating endothelial cell proliferation, cytokine release, and adhesion molecule expression: Tryptase induces expression of mRNA for IL-1 beta and IL-8 and stimulates the selective release of IL-8 from human umbilical vein endothelial cells. J. Immunol. 1998, 161, 1939–1946. [Google Scholar]
- Burwen, S.J.; Satir, B.H. Plasma membrane folds on the mast cell surface and their relationship to secretory activity. J. Cell Biol. 1977, 74, 690–697. [Google Scholar] [CrossRef]
- Burwen, S.J.; Satir, B.H. A freeze-fracture study of early membrane events during mast cell secretion. J. Cell Biol. 1977, 73, 660–671. [Google Scholar] [CrossRef]
- Chandler, D.E.; Heuser, J.E. Arrest of membrane fusion events in mast cells by quick-freezing. J. Cell Biol. 1980, 86, 666–674. [Google Scholar] [CrossRef]
- Ren, Y.; Yu, J.; Kinghorn, A.D. Development of Anticancer Agents from Plant-Derived Sesquiterpene Lactones. Curr. Med. Chem. 2016, 23, 2397–2420. [Google Scholar] [CrossRef]
- Goto, Y.; Kojima, Y.; Nakayama, T.; Terazawa, M. Allelopathic sesquiterpenoids from rhizomes of Petasites japonicus ssp. giganteus Kitam. Phytochemistry 2001, 57, 109–113. [Google Scholar] [CrossRef]
- Yaoita, Y.N.K.; Suzuki, N.; Kikuchi, M. Structures of Eremophilenolides from the Rhizomes of Petasites japonicus MAXIM. Chem. Pharm. Bull 1992, 40, 3277–3279. [Google Scholar] [CrossRef]
- Tori, M. KMaSM. Eremophilane-type sesquiterpenes from fresh rhizones of Petasites japonicus. Phytochemistry 1998, 47, 401–409. [Google Scholar] [CrossRef]
- Sok, D.; Oh, S.H.; Kim, Y.; Kang, H.; Kim, M.R. Neuroprotection by extract of Petasites japonicus leaves, a traditional vegetable, against oxidative stress in brain of mice challenged with kainic acid. Eur. J. Nutr. 2005, 45, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.S.; Kim, M.R.; Sok, D.-E. Protection by Petaslignolide A, a Major Neuroprotective Compound in the Butanol Extract ofPetasites japonicusLeaves, against Oxidative Damage in the Brains of Mice Challenged with Kainic Acid. J. Agric. Food Chem. 2005, 53, 8526–8532. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.H.; Sok, D.-E.; Kim, M.R. Neuroprotective Effects of Butterbur and Rough Aster Against Kainic Acid-Induced Oxidative Stress in Mice. J. Med. Food 2005, 8, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, H.; Tanaka, J.; Yamada, E.; Morikawa, T.; Kasajima, N.; Yoshikawa, M. Anti Type I Allergic Property of Japanese Butterbur Extract and Its Mast Cell Degranulation Inhibitory Ingredients. J. Agric. Food Chem. 2006, 54, 2915–2920. [Google Scholar] [CrossRef]
- Tobinaga, S.; Takeuchi, N.; Kasama, T.; Yamashita, J.; Aida, Y.; Kaneko, Y. Anti-histaminic and anti-allergic principles of Petasites japonicus Maxim. Chem. Pharm. Bull. 1983, 31, 745–748. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hwang, B.Y.; Lee, J.-H.; Koo, T.H.; Kim, H.S.; Hong, Y.S.; Ro, J.S.; Lee, K.S.; Lee, J.J. Furanoligularenone, an Eremophilane from Ligularia fischeri, Inhibits the LPS-Induced Production of Nitric Oxide and Prostaglandin E2 in Macrophage RAW264.7 Cells. Planta Medica 2002, 68, 101–105. [Google Scholar] [CrossRef]
- Zhao, J.-H.; Shen, T.; Yang, X.; Zhao, H.; Li, X.; Xie, W.-D. Sesquiterpenoids from Farfugium japonicum and their inhibitory activity on NO production in RAW264.7 cells. Arch. Pharmacal Res. 2012, 35, 1153–1158. [Google Scholar] [CrossRef] [PubMed]
- Fadeel, B. Clear and present danger? Engineered nanoparticles and the immune system. Swiss Med. Wkly. 2012, 142. [Google Scholar] [CrossRef] [PubMed]
- Davis, B.K.; Wen, H.; Ting, J.P.-Y. The Inflammasome NLRs in Immunity, Inflammation, and Associated Diseases. Annu. Rev. Immunol. 2011, 29, 707–735. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Du, L.; Zhao, R.; Wang, H.Y.; Zhao, Y.; Nie, G.; Wang, R.-F. Cell-Penetrating Nanoparticles Activate the Inflammasome to Enhance Antibody Production by Targeting Microtubule-Associated Protein 1-Light Chain 3 for Degradation. ACS Nano 2020, 14, 3703–3717. [Google Scholar] [CrossRef] [PubMed]
- Demento, S.L.; Eisenbarth, S.C.; Foellmer, H.G.; Platt, C.; Caplan, M.J.; Saltzman, W.M.; Mellman, I.; Ledizet, M.; Fikrig, E.; Flavell, R.A.; et al. Inflammasome-activating nanoparticles as modular systems for optimizing vaccine efficacy. Vaccine 2009, 27, 3013–3021. [Google Scholar] [CrossRef]
- Deng, Z.J.; Liang, M.; Monteiro, M.; Toth, I.; Minchin, R.F. Nanoparticle-induced unfolding of fibrinogen promotes Mac-1 receptor activation and inflammation. Nat. Nanotechnol. 2010, 6, 39–44. [Google Scholar] [CrossRef]
- Dubreuil, P.; Letard, S.; Ciufolini, M.; Gros, L.; Humbert, M.; Castéran, N.; Borge, L.; Hajem, B.; Lermet, A.; Sippl, W.; et al. Masitinib (AB1010), a Potent and Selective Tyrosine Kinase Inhibitor Targeting KIT. PLoS ONE 2009, 4, e7258. [Google Scholar] [CrossRef]
- Paul, C.; Sans, B.; Suarez, F.; Casassus, P.; Barète, S.; Lanternier, F.; Grandpeix-Guyodo, C.; Dubreuil, P.; Palmérini, F.; Mansfield, C.D.; et al. Masitinib for the treatment of systemic and cutaneous mastocytosis with handicap: A phase 2a study. Am. J. Hematol. 2010, 85, 921–925. [Google Scholar] [CrossRef] [PubMed]
- Savage, D.G.; Antman, K.H. Imatinib mesylate—A new oral targeted therapy. N. Engl. J. Med. 2002, 346, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Avula, M.N.; Rao, A.N.; McGill, L.D.; Grainger, D.W.; Solzbacher, F. Modulation of the foreign body response to implanted sensor models through device-based delivery of the tyrosine kinase inhibitor, masitinib. Biomaterials 2013, 34, 9737–9746. [Google Scholar] [CrossRef] [PubMed]



| Nanoparticle ID | Composition | Dh (nm) | PDI |
|---|---|---|---|
| PLGA/PVA | PLGA + PVA | 72.24 ± 0.7 | 0.14 |
| PLGA/PVA-21 | PLGA + PVA + sesquiterepene21 | 60.15 ± 1.4 | 0.11 |
| PLGA/PVA-22 | PLGA + PVA + sesquiterpene 22 | 60.68 ± 1 | 0.08 |
| PLGA/PVA-C6 | PLGA + PVA + coumarin 6 | 62.96 ± 0.7 | 0.13 |
| ID | Sequence |
|---|---|
| FcεR1α, Homo sapiens | |
| primer | GCA AAC AGA ATC ACC ACC AAC |
| primer | GTT GAA GAC AGT GGA ACC TAC T |
| probe | /5HEX/CTC AGA CTC/ZEN/ATA GTC CAG CTG CCA C/3IABkFQ/ |
| FcεR1β, Homo sapiens | |
| primer | TCT TCA TAA ACA CGA TCC TCT GG |
| primer | GAT GCT GTT TCT CAC CAT TCT G |
| probe | /5HEX/TTG AGT TCT/ZEN/TCC CCA GCT CCA CAG/3IABkFQ/ |
| FcεR1γ, Homo sapiens | |
| primer | CTC ATG CTT CAG AGT CTC GTA |
| primer | GAC TGA AGA TCC AAG TGC GAA |
| probe | /56-FAM/TGG TGC TCA GGC CCG TGT AAA C/36-TAMSp/ |
| Kit, Homo sapiens | |
| primer | TCA GTG CAT AAC AGC CTA ATC TC |
| primer | GTT CTG CTC CTA CTG CTT CG |
| probe | /56-FAM/TTC CCC TGG/ZEN/ACT CAC AGA TGG TTG/3IABkFQ/ |
| MrgprX2, Homo sapiens | |
| primer | GGA CTG AGA AAG TTC AGC AAA TC |
| primer | GAA GCA GTC TGG TGT AGA GAT G |
| probe | /56-FAM/TGT GGC TTT GAG AGG CAA CTT TGC/36-TAMSp/ |
| Tryptase, Homo sapiens | |
| primer | CAG TGG TGT TTT GGA CAG C |
| primer | CGG CCT GGC ATC TAC AC |
| probe | /56-FAM/TGA CTC ACG/ZEN/GCT TTT TGG GGA CAT/3IABkFQ/ |
| Chymase, Homo sapiens | |
| primer | CGT GGT GAA GAG TAG AAG TGT T |
| primer | GTC TAT AAC AGT CAC CCT TGG A |
| probe | /5HEX/AGA GGA AGA/ZEN/AGA CAC ATG GCA GAA GC/3IABkFQ/ |
| HPRT1, Homo sapiens | |
| primer | GCG ATG TCA ATA GGA CTC CAG |
| primer | TTG TTG TAG GAT ATG CCC TTG A |
| probe | /5HEX/AGC CTA AGA/ZEN/TGA GAG TTC AAG TTG AGT TTG G/3IABkFQ/ Or /56-FAM/AGC CTA AGA/ZEN/TGA GAG TTC AAG TTG AGT TTG G/3IABkFQ/ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arizmendi, N.; Qian, H.; Li, Y.; Kulka, M. Sesquiterpene-Loaded Co-Polymer Hybrid Nanoparticle Effects on Human Mast Cell Surface Receptor Expression, Granule Contents, and Degranulation. Nanomaterials 2021, 11, 953. https://doi.org/10.3390/nano11040953
Arizmendi N, Qian H, Li Y, Kulka M. Sesquiterpene-Loaded Co-Polymer Hybrid Nanoparticle Effects on Human Mast Cell Surface Receptor Expression, Granule Contents, and Degranulation. Nanomaterials. 2021; 11(4):953. https://doi.org/10.3390/nano11040953
Chicago/Turabian StyleArizmendi, Narcy, Hui Qian, Yiming Li, and Marianna Kulka. 2021. "Sesquiterpene-Loaded Co-Polymer Hybrid Nanoparticle Effects on Human Mast Cell Surface Receptor Expression, Granule Contents, and Degranulation" Nanomaterials 11, no. 4: 953. https://doi.org/10.3390/nano11040953
APA StyleArizmendi, N., Qian, H., Li, Y., & Kulka, M. (2021). Sesquiterpene-Loaded Co-Polymer Hybrid Nanoparticle Effects on Human Mast Cell Surface Receptor Expression, Granule Contents, and Degranulation. Nanomaterials, 11(4), 953. https://doi.org/10.3390/nano11040953






