Recent Advances in Nanomaterial-Based Aptasensors in Medical Diagnosis and Therapy
Abstract
1. Introduction
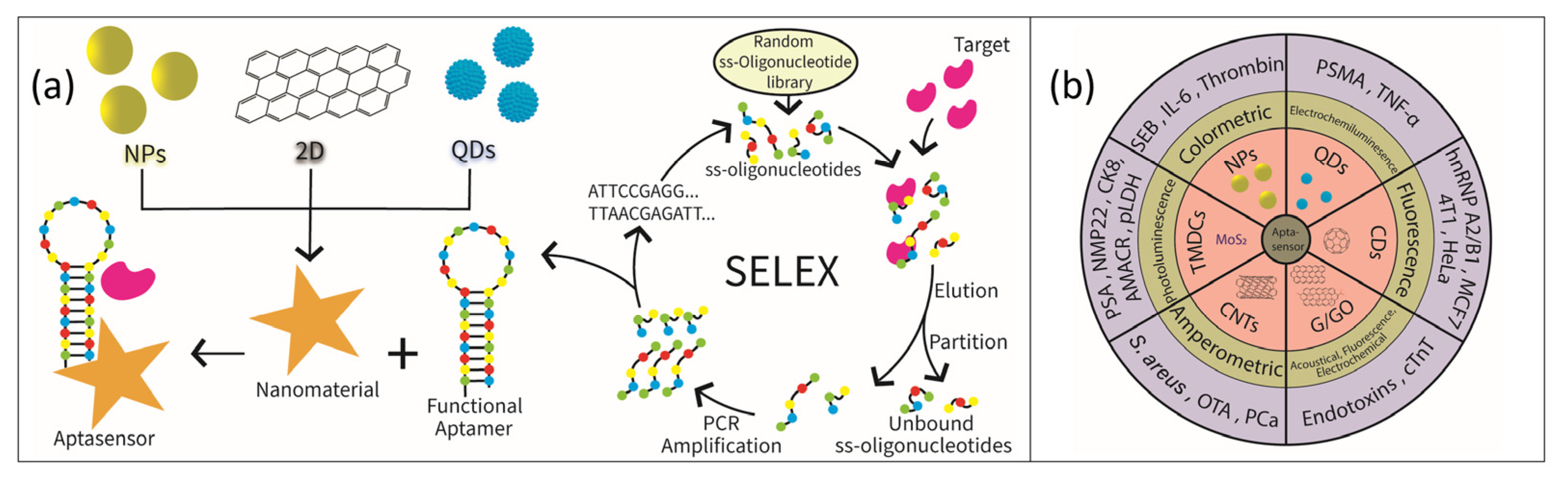
2. Selection Protocol
3. Nanomaterials Based Aptamer Sensors as Diagnostic Tool
3.1. Aptamer–Gold Nanoparticle Aptasensors
3.2. Quantum Dot-Based Aptasensor
3.3. Carbon Quantum Dot-Based Aptasensor
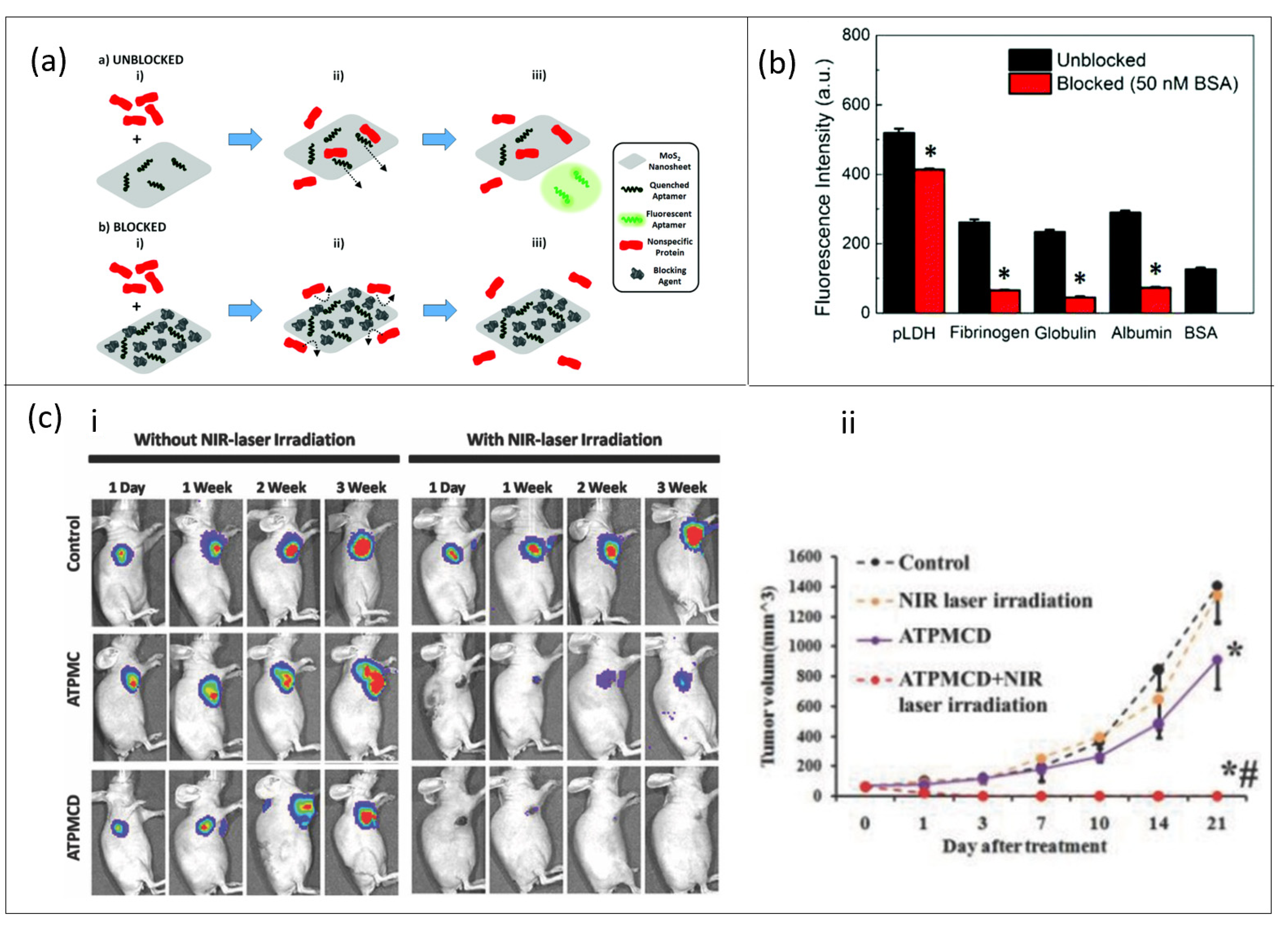
3.4. MoS2-Based Aptasensor
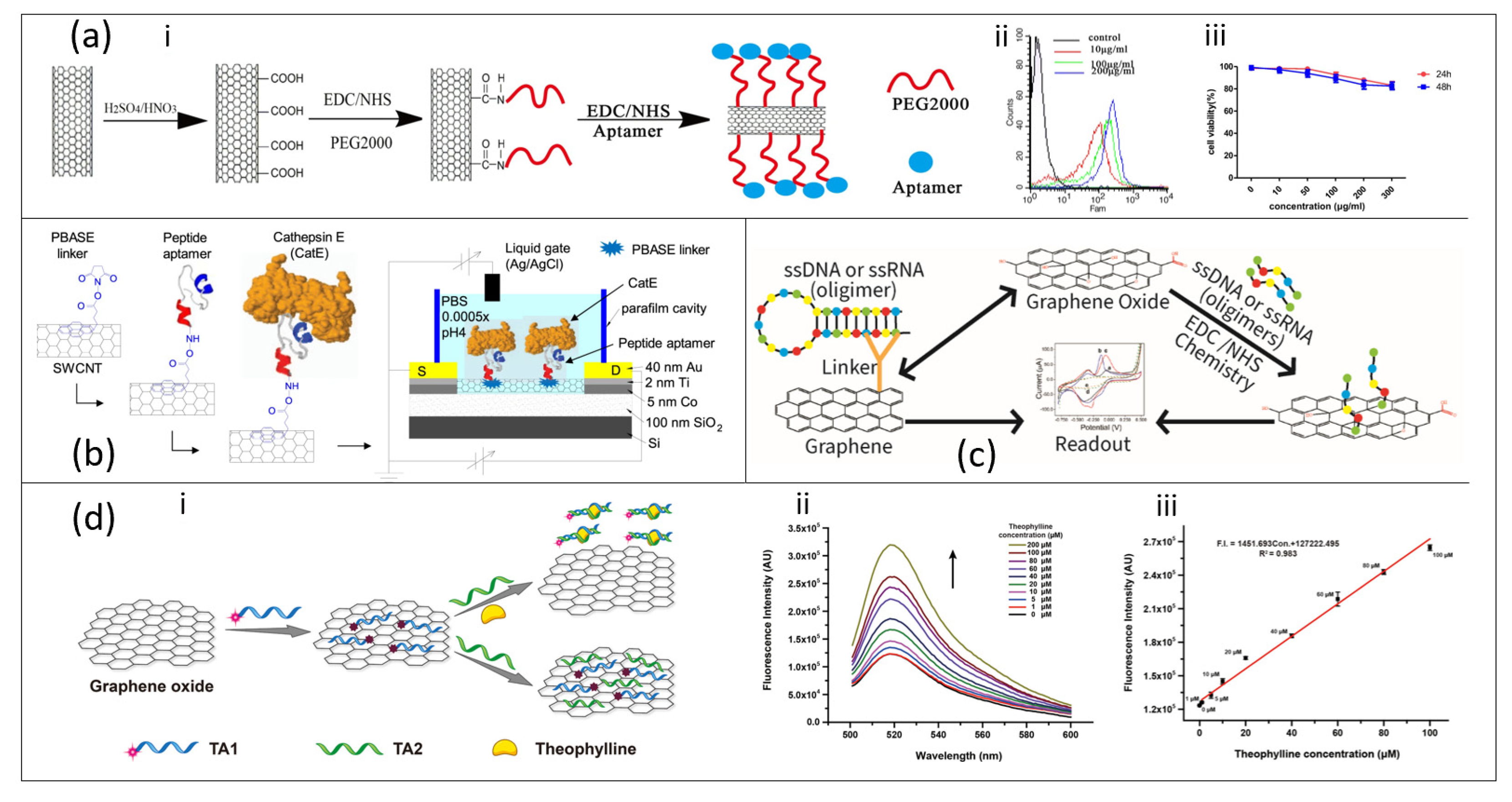
3.5. Carbon Nanotube-Based Aptasensor
3.6. Graphene/Graphene Oxide-Based Aptasensor
3.7. Other Nanomaterial-Based Aptasensor
4. Future Perspectives and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, Y.; Zhang, T.; Lin, X.; Feng, J.; Luo, F.; Gao, H.; Wu, Y.; Deng, R.; He, Q. Graphene/Aptamer Probes for Small Molecule Detection: From in Vitro Test to in Situ Imaging. Microchim. Acta 2020, 187, 179. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, X.; Wang, Z.; Jiang, H.; Lv, Z.; Shen, J.; Xia, G.; Wen, K. Universal Simultaneous Multiplex ELISA of Small Molecules in Milk Based on Dual Luciferases. Anal. Chim. Acta 2018, 1001, 125–133. [Google Scholar] [CrossRef]
- Zhang, Z.; Oni, O.; Liu, J. New Insights into a Classic Aptamer: Binding Sites, Cooperativity and More Sensitive Adenosine Detection. Nucleic Acids Res. 2017, 45, 7593–7601. [Google Scholar] [CrossRef]
- Deng, R.; Dong, Y.; Xia, X.; Dai, Y.; Zhang, K.; He, Q.; Zeng, W.; Ren, X.; Li, J. Recognition-Enhanced Metastably Shielded Aptamer for Digital Quantification of Small Molecules. Anal. Chem. 2018, 90, 14347–14354. [Google Scholar] [CrossRef]
- Masson, J.-F. Surface Plasmon Resonance Clinical Biosensors for Medical Diagnostics. ACS Sens. 2017, 2, 16–30. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Long, R.; Prezhdo, O.V. Why Chemical Vapor Deposition Grown MoS2 Samples Outperform Physical Vapor Deposition Samples: Time-Domain Ab Initio Analysis. Nano Lett. 2018, 18, 4008–4014. [Google Scholar] [CrossRef]
- Roisin, S.; Huang, T.-D.; de Mendonça, R.; Nonhoff, C.; Bogaerts, P.; Hites, M.; Delaere, B.; Hamels, S.; de Longueville, F.; Glupczynski, Y.; et al. Prospective Evaluation of a High Multiplexing Real-Time Polymerase Chain Reaction Array for the Rapid Identification and Characterization of Bacteria Causative of Nosocomial Pneumonia from Clinical Specimens: A Proof-of-Concept Study. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2018, 37, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Florea, A.; Melinte, G.; Simon, I.; Cristea, C. Electrochemical Biosensors as Potential Diagnostic Devices for Autoimmune Diseases. Biosensors 2019, 9, 38. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Xu, H.; Zhao, Y.; Han, Y.; Zhang, Y.; Zhang, J.; Xu, C.; Wang, W.; Guo, Q.; Ge, J. Aptamer Based High Throughput Colorimetric Biosensor for Detection of Staphylococcus Aureus. Sci. Rep. 2020, 10, 9190. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Farid, S.; Meshik, X.; Xu, K.; Choi, M.; Ranginwala, S.; Wang, Y.Y.; Burke, P.; Dutta, M.; Stroscio, M.A. Detection of Immunoglobulin E with a Graphene-Based Field-Effect Transistor Aptasensor. Available online: https://www.hindawi.com/journals/js/2018/3019259/ (accessed on 22 February 2021).
- Iliuk, A.B.; Hu, L.; Tao, W.A. Aptamer in Bioanalytical Applications. Anal. Chem. 2011, 83, 4440–4452. [Google Scholar] [CrossRef] [PubMed]
- Pehlivan, Z.S.; Torabfam, M.; Kurt, H.; Ow-Yang, C.; Hildebrandt, N.; Yüce, M. Aptamer and Nanomaterial Based FRET Biosensors: A Review on Recent Advances (2014-2019). Mikrochim. Acta 2019, 186, 563. [Google Scholar] [CrossRef]
- Saad, M.; Chinerman, D.; Tabrizian, M.; Faucher, S.P. Identification of Two Aptamers Binding to Legionella Pneumophila with High Affinity and Specificity. Sci. Rep. 2020, 10, 9145. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Liang, Z.; Zhou, N. Design Strategies for Aptamer-Based Biosensors. Sensors 2010, 10, 4541–4557. [Google Scholar] [CrossRef]
- Rozenblum, G.T.; Lopez, V.G.; Vitullo, A.D.; Radrizzani, M. Aptamers: Current Challenges and Future Prospects. Expert Opin. Drug Discov. 2016, 11, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Lipi, F.; Chen, S.; Chakravarthy, M.; Rakesh, S.; Veedu, R.N. In Vitro Evolution of Chemically-Modified Nucleic Acid Aptamers: Pros and Cons, and Comprehensive Selection Strategies. RNA Biol. 2016, 13, 1232–1245. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.; Ban, C. Aptamer–Nanoparticle Complexes as Powerful Diagnostic and Therapeutic Tools. Exp. Mol. Med. 2016, 48, e230. [Google Scholar] [CrossRef]
- Zhou, W.; Huang, P.-J.J.; Ding, J.; Liu, J. Aptamer-Based Biosensors for Biomedical Diagnostics. Analyst 2014, 139, 2627–2640. [Google Scholar] [CrossRef]
- Liu, M.; Yin, Q.; Chang, Y.; Zhang, Q.; Brennan, J.D.; Li, Y. In Vitro Selection of Circular DNA Aptamers for Biosensing Applications. Angew. Chem. Int. Ed. 2019, 58, 8013–8017. [Google Scholar] [CrossRef]
- Zhuo, Z.; Yu, Y.; Wang, M.; Li, J.; Zhang, Z.; Liu, J.; Wu, X.; Lu, A.; Zhang, G.; Zhang, B. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017, 18, 2142. [Google Scholar] [CrossRef]
- Park, J.-W.; Lee, S.J.; Ren, S.; Lee, S.; Kim, S.; Laurell, T. Acousto-Microfluidics for Screening of SsDNA Aptamer. Sci. Rep. 2016, 6, 27121. [Google Scholar] [CrossRef]
- Lee, S.; Kim, B.W.; Shin, H.-S.; Go, A.; Lee, M.-H.; Lee, D.-K.; Kim, S.; Jeong, O.C. Aptamer Affinity-Bead Mediated Capture and Displacement of Gram-Negative Bacteria Using Acoustophoresis. Micromachines 2019, 10, 770. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent Advances in Aptamer Discovery and Applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef]
- Yang, J.; Bowser, M.T. Capillary Electrophoresis–SELEX Selection of Catalytic DNA Aptamers for a Small-Molecule Porphyrin Target. Anal. Chem. 2013, 85, 1525–1530. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Zhou, H.; Jiang, H.; Ou, H.; Li, X.; Zhang, L. Development of a Fraction Collection Approach in Capillary Electrophoresis SELEX for Aptamer Selection. Analyst 2015, 140, 2664–2670. [Google Scholar] [CrossRef] [PubMed]
- Quang, N.N.; Miodek, A.; Cibiel, A.; Ducongé, F. Selection of Aptamers Against Whole Living Cells: From Cell-SELEX to Identification of Biomarkers. In Synthetic Antibodies: Methods and Protocols; Tiller, T., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 253–272. ISBN 978-1-4939-6857-2. [Google Scholar]
- Ray, P.; White, R.R. Cell-SELEX Identifies a “Sticky” RNA Aptamer Sequence. J. Nucleic Acids 2017, 2017, e4943072. [Google Scholar] [CrossRef]
- Ouellet, E.; Foley, J.H.; Conway, E.M.; Haynes, C. Hi-Fi SELEX: A High-Fidelity Digital-PCR Based Therapeutic Aptamer Discovery Platform. Biotechnol. Bioeng. 2015, 112, 1506–1522. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Wu, X.; Ho, M.; Chomchan, P.; Rossi, J.J.; Burnett, J.C.; Zhou, J. High Throughput Sequencing Analysis of RNA Libraries Reveals the Influences of Initial Library and PCR Methods on SELEX Efficiency. Sci. Rep. 2016, 6, 33697. [Google Scholar] [CrossRef]
- Pfeiffer, F.; Tolle, F.; Rosenthal, M.; Brändle, G.M.; Ewers, J.; Mayer, G. Identification and Characterization of Nucleobase-Modified Aptamers by Click-SELEX. Nat. Protoc. 2018, 13, 1153–1180. [Google Scholar] [CrossRef]
- Pleiko, K.; Saulite, L.; Parfejevs, V.; Miculis, K.; Vjaters, E.; Riekstina, U. Differential Binding Cell-SELEX Method to Identify Cell-Specific Aptamers Using High-Throughput Sequencing. Sci. Rep. 2019, 9, 8142. [Google Scholar] [CrossRef]
- Kaur, H.; Shorie, M. Nanomaterial Based Aptasensors for Clinical and Environmental Diagnostic Applications. Nanoscale Adv. 2019, 1, 2123–2138. [Google Scholar] [CrossRef]
- Şahin, S.; Caglayan, M.O.; Üstündağ, Z. Recent Advances in Aptamer-Based Sensors for Breast Cancer Diagnosis: Special Cases for Nanomaterial-Based VEGF, HER2, and MUC1 Aptasensors. Microchim. Acta 2020, 187, 549. [Google Scholar] [CrossRef] [PubMed]
- Alves, R.S.; Sigoli, F.A.; Mazali, I.O. Aptasensor based on a flower-shaped silver magnetic nanocomposite enables the sensitive and label-free detection of troponin I (cTnI) by SERS. Nanotechnology 2020, 31, 505505. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Zhang, H.; Wang, Z.; Cao, H.; Zhang, K.; Li, X.; Yang, Z. Nanomaterial-Based Aptamer Sensors for Arsenic Detection. Biosens. Bioelectron. 2020, 148, 111785. [Google Scholar] [CrossRef]
- Mondal, B.; Ramlal, S.; Lavu, P.S.; N, B.; Kingston, J. Highly Sensitive Colorimetric Biosensor for Staphylococcal Enterotoxin B by a Label-Free Aptamer and Gold Nanoparticles. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Giorgi-Coll, S.; Marín, M.J.; Sule, O.; Hutchinson, P.J.; Carpenter, K.L.H. Aptamer-Modified Gold Nanoparticles for Rapid Aggregation-Based Detection of Inflammation: An Optical Assay for Interleukin-6. Mikrochim. Acta 2020, 187. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Li, L.; Mu, X.; Guo, L. Aptamer-Gold Nanoparticle-Based Colorimetric Assay for the Sensitive Detection of Thrombin. Sens. Actuators B Chem. 2013, 177, 818–825. [Google Scholar] [CrossRef]
- Chen, Y.; Xiang, J.; Liu, B.; Chen, Z.; Zuo, X. Gold Nanoparticle-Engineered Electrochemical Aptamer Biosensor for Ultrasensitive Detection of Thrombin. Anal. Methods 2020, 12, 3729–3733. [Google Scholar] [CrossRef]
- Kitte, S.A.; Tafese, T.; Xu, C.; Saqib, M.; Li, H.; Jin, Y. Plasmon-Enhanced Quantum Dots Electrochemiluminescence Aptasensor for Selective and Sensitive Detection of Cardiac Troponin I. Talanta 2021, 221, 121674. [Google Scholar] [CrossRef]
- Isildak, I.; Navaeipour, F.; Afsharan, H.; Kanberoglu, G.S.; Agir, I.; Ozer, T.; Annabi, N.; Totu, E.E.; Khalilzadeh, B. Electrochemiluminescence Methods Using CdS Quantum Dots in Aptamer-Based Thrombin Biosensors: A Comparative Study. Microchim. Acta 2019, 187, 25. [Google Scholar] [CrossRef]
- Li, H.; Guo, L.; Huang, A.; Xu, H.; Liu, X.; Ding, H.; Dong, J.; Li, J.; Wang, C.; Su, X.; et al. Nanoparticle-Conjugated Aptamer Targeting HnRNP A2/B1 Can Recognize Multiple Tumor Cells and Inhibit Their Proliferation. Biomaterials 2015, 63, 168–176. [Google Scholar] [CrossRef]
- Kong, R.-M.; Ding, L.; Wang, Z.; You, J.; Qu, F. A Novel Aptamer-Functionalized MoS2 Nanosheet Fluorescent Biosensor for Sensitive Detection of Prostate Specific Antigen. Anal. Bioanal. Chem. 2015, 407, 369–377. [Google Scholar] [CrossRef]
- Liu, X.; Tang, Y.; Liu, P.; Yang, L.; Li, L.; Zhang, Q.; Zhou, Y.; Khan, M.Z.H. A Highly Sensitive Electrochemical Aptasensor for Detection of Microcystin-LR Based on a Dual Signal Amplification Strategy. Analyst 2019, 144, 1671–1678. [Google Scholar] [CrossRef] [PubMed]
- Tung, N.T.; Tue, P.T.; Thi Ngoc Lien, T.; Ohno, Y.; Maehashi, K.; Matsumoto, K.; Nishigaki, K.; Biyani, M.; Takamura, Y. Peptide Aptamer-Modified Single-Walled Carbon Nanotube-Based Transistors for High-Performance Biosensors. Sci. Rep. 2017, 7, 17881. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.; Pang, Y.; Li, D.; Huang, Z.; Zhang, Z.; Xue, N.; Xu, Y.; Mu, X. An Aptamer-Based Shear Horizontal Surface Acoustic Wave Biosensor with a CVD-Grown Single-Layered Graphene Film for High-Sensitivity Detection of a Label-Free Endotoxin. Microsyst. Nanoeng. 2020, 6, 1–11. [Google Scholar] [CrossRef]
- Ling, K.; Jiang, H.; Li, Y.; Tao, X.; Qiu, C.; Li, F.-R. A Self-Assembling RNA Aptamer-Based Graphene Oxide Sensor for the Turn-on Detection of Theophylline in Serum. Biosens. Bioelectron. 2016, 86, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Jang, J. Flexible Electrical Aptasensor Using Dielectrophoretic Assembly of Graphene Oxide and Its Subsequent Reduction for Cardiac Biomarker Detection. Sci. Rep. 2019, 9, 5970. [Google Scholar] [CrossRef] [PubMed]
- Alsager, O.A.; Alotaibi, K.M.; Alswieleh, A.M.; Alyamani, B.J. Colorimetric Aptasensor of Vitamin D3: A Novel Approach to Eliminate Residual Adhesion between Aptamers and Gold Nanoparticles. Sci. Rep. 2018, 8, 12947. [Google Scholar] [CrossRef]
- Jiang, Y.; Shi, M.; Liu, Y.; Wan, S.; Cui, C.; Zhang, L.; Tan, W. Aptamer/AuNP Biosensor for Colorimetric Profiling of Exosomal Proteins. Angew. Chem. Int. Ed. 2017, 56, 11916–11920. [Google Scholar] [CrossRef]
- Rosi, N.L.; Mirkin, C.A. Nanostructures in Biodiagnostics. Chem. Rev. 2005, 105, 1547–1562. [Google Scholar] [CrossRef]
- Truong, P.L.; Ma, X.; Sim, S.J. Resonant Rayleigh Light Scattering of Single Au Nanoparticles with Different Sizes and Shapes. Nanoscale 2014, 6, 2307–2315. [Google Scholar] [CrossRef]
- Shafiqa, A.R.; Aziz, A.A.; Mehrdel, B. Nanoparticle Optical Properties: Size Dependence of a Single Gold Spherical Nanoparticle. J. Phys. Conf. Ser. 2018, 1083, 012040. [Google Scholar] [CrossRef]
- Gopinath, S.C.B.; Lakshmipriya, T.; Awazu, K. Colorimetric Detection of Controlled Assembly and Disassembly of Aptamers on Unmodified Gold Nanoparticles. Biosens. Bioelectron. 2014, 51, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Lee, S.J.; Moskovits, M. Aptamer-Mediated Surface-Enhanced Raman Spectroscopy Intensity Amplification. Nano Lett. 2010, 10, 4181–4185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Ma, X.; Liu, Y.; Duan, N.; Wu, S.; Wang, Z.; Xu, B. Gold Nanoparticles Enhanced SERS Aptasensor for the Simultaneous Detection of Salmonella Typhimurium and Staphylococcus Aureus. Biosens. Bioelectron. 2015, 74, 872–877. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, T.-H. Quantum Dot Enabled Molecular Sensing and Diagnostics. Theranostics 2012, 2, 631–654. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; He, L.; Zhang, G.; Lv, A.; Wang, R.; Zhang, X.; Tan, W. Aptamer-Assembled Nanomaterials for Fluorescent Sensing and Imaging. Nanophotonics 2017, 6, 109–121. [Google Scholar] [CrossRef]
- Bagalkot, V.; Zhang, L.; Levy-Nissenbaum, E.; Jon, S.; Kantoff, P.W.; Langer, R.; Farokhzad, O.C. Quantum Dot-Aptamer Conjugates for Synchronous Cancer Imaging, Therapy, and Sensing of Drug Delivery Based on Bi-Fluorescence Resonance Energy Transfer. Nano Lett. 2007, 7, 3065–3070. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Liao, Z.; Zhang, Y.; Fu, F.; Hao, M.; Song, Y.; Song, E. Aptamer-Quantum Dots and Teicoplanin-Gold Nanoparticles Constructed FRET Sensor for Sensitive Detection of Staphylococcus Aureus. Chin. Chem. Lett. 2020. [Google Scholar] [CrossRef]
- Ghosh, S.; Chen, Y.; Sebastian, J.; George, A.; Dutta, M.; Stroscio, M.A. A Study on the Response of FRET Based DNA Aptasensors in Intracellular Environment. Sci. Rep. 2020, 10, 13250. [Google Scholar] [CrossRef]
- Wen, L.; Qiu, L.; Wu, Y.; Hu, X.; Zhang, X. Aptamer-Modified Semiconductor Quantum Dots for Biosensing Applications. Sensors 2017, 17, 1736. [Google Scholar] [CrossRef]
- Duan, Q.; Che, M.; Hu, S.; Zhao, H.; Li, Y.; Ma, X.; Zhang, W.; Zhang, Y.; Sang, S. Rapid Cancer Diagnosis by Highly Fluorescent Carbon Nanodots-Based Imaging. Anal. Bioanal. Chem. 2019, 411, 967–972. [Google Scholar] [CrossRef]
- Shen, X.; Xu, L.; Zhu, W.; Li, B.; Hong, J.; Zhou, X. A Turn-on Fluorescence Aptasensor Based on Carbon Dots for Sensitive Detection of Adenosine. New J. Chem. 2017, 41, 9230–9235. [Google Scholar] [CrossRef]
- Yousefi, S.; Saraji, M. Developing a Fluorometric Aptasensor Based on Carbon Quantum Dots and Silver Nanoparticles for the Detection of Adenosine. Microchem. J. 2019, 148, 169–176. [Google Scholar] [CrossRef]
- Gao, X.; Du, C.; Zhuang, Z.; Chen, W. Carbon Quantum Dot-Based Nanoprobes for Metal Ion Detection. J. Mater. Chem. C 2016, 4, 6927–6945. [Google Scholar] [CrossRef]
- Li, M.; Chen, T.; Gooding, J.J.; Liu, J. Review of Carbon and Graphene Quantum Dots for Sensing. ACS Sens. 2019, 4, 1732–1748. [Google Scholar] [CrossRef]
- Molaei, M.J. Principles, Mechanisms, and Application of Carbon Quantum Dots in Sensors: A Review. Anal. Methods 2020, 12, 1266–1287. [Google Scholar] [CrossRef]
- Yoo, D.; Park, Y.; Cheon, B.; Park, M.-H. Carbon Dots as an Effective Fluorescent Sensing Platform for Metal Ion Detection. Nanoscale Res. Lett. 2019, 14, 272. [Google Scholar] [CrossRef]
- Gao, Y.; Gao, F.; Zhang, G.; Chen, L.; Wu, Q.; Liu, X. Sensor Array Based on Single Carbon Quantum Dot for Fluorometric Differentiation of All Natural Amino Acids. Microchim. Acta 2019, 186, 858. [Google Scholar] [CrossRef]
- Wu, W.; Ding, L.; Lin, H.; Yu, S.; Huang, J.; Xia, Z. A Highly Sensitive Fluorescence Sensor for Adrenaline Detection Based on Modified Carbon Quantum Dots. In Proceedings of the Tenth International Conference on Information Optics and Photonics, Beijing, China, 8–11 July 2018; Volume 10964, p. 109645F. [Google Scholar]
- Xu, B.; Zhao, C.; Wei, W.; Ren, J.; Miyoshi, D.; Sugimoto, N.; Qu, X. Aptamer Carbon Nanodot Sandwich Used for Fluorescent Detection of Protein. Analyst 2012, 137, 5483–5486. [Google Scholar] [CrossRef]
- Lee, C.H.; Rajendran, R.; Jeong, M.-S.; Ko, H.Y.; Joo, J.Y.; Cho, S.; Chang, Y.W.; Kim, S. Bioimaging of Targeting Cancers Using Aptamer-Conjugated Carbon Nanodots. Chem. Commun. 2013, 49, 6543–6545. [Google Scholar] [CrossRef]
- Li, H.; Ding, H.-M.; Li, J.; Xu, A.-X.; Su, X.-T.; Liang, C.; Zhang, L.-Q.; Zhao, Q.; Li, S.-H.; Shao, N.-H. Selection and Identification of SsDNA Aptamers Specific to Rat ROS1728 Cells. Lett. Biotechnol. 2015. Available online: http://en.cnki.com.cn/Article_en/CJFDTotal-SWTX201501017.htm (accessed on 22 February 2021).
- Motaghi, H.; Mehrgardi, M.A.; Bouvet, P. Carbon Dots-AS1411 Aptamer Nanoconjugate for Ultrasensitive Spectrofluorometric Detection of Cancer Cells. Sci. Rep. 2017, 7, 10513. [Google Scholar] [CrossRef]
- Kong, T.; Zhou, R.; Zhang, Y.; Hao, L.; Cai, X.; Zhu, B. AS1411 Aptamer Modified Carbon Dots via Polyethylenimine-Assisted Strategy for Efficient Targeted Cancer Cell Imaging. Cell Prolif. 2020, 53, e12713. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhu, H. Two-Dimensional MoS2: Properties, Preparation, and Applications. J. Materiomics 2015, 1, 33–44. [Google Scholar] [CrossRef]
- An, J.H.; Jang, J. A Highly Sensitive FET-Type Aptasensor Using Flower-like MoS2 Nanospheres for Real-Time Detection of Arsenic(III). Nanoscale 2017, 9, 7483–7492. [Google Scholar] [CrossRef] [PubMed]
- Kalantar-zadeh, K.; Ou, J.Z. Biosensors Based on Two-Dimensional MoS2. ACS Sens. 2016, 1, 5–16. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Wong, J.I.; Palacios, T.; Kong, J.; Yang, H.Y. Functionalized MoS2 Nanosheet-Based Field-Effect Biosensor for Label-Free Sensitive Detection of Cancer Marker Proteins in Solution. Small 2014, 10, 1101–1105. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zeng, B.; Li, Y.; Liang, H.; Yang, Y.; Yuan, Q. Construction of MoS2 Field Effect Transistor Sensor Array for the Detection of Bladder Cancer Biomarkers. Sci. China Chem. 2020, 63, 997–1003. [Google Scholar] [CrossRef]
- Ying, Z.; Feng, L.; Ji, D.; Zhang, Y.; Chen, W.; Dai, Y.; Janyasupab, M.; Li, X.; Wen, W.; Liu, C.-C. Phase-Regulated Sensing Mechanism of MoS2 Based Nanohybrids toward Point-of-Care Prostate Cancer Diagnosis. Small 2020, 16, 2000307. [Google Scholar] [CrossRef]
- Park, H.; Han, G.; Lee, S.W.; Lee, H.; Jeong, S.H.; Naqi, M.; AlMutairi, A.; Kim, Y.J.; Lee, J.; Kim, W.; et al. Label-Free and Recalibrated Multilayer MoS2 Biosensor for Point-of-Care Diagnostics. ACS Appl. Mater. Interfaces 2017, 9, 43490–43497. [Google Scholar] [CrossRef]
- Ge, J.; Ou, E.-C.; Yu, R.-Q.; Chu, X. A Novel Aptameric Nanobiosensor Based on the Self-Assembled DNA–MoS2 Nanosheet Architecture for Biomolecule Detection. J. Mater. Chem. B 2014, 2, 625–628. [Google Scholar] [CrossRef]
- Zhu, C.; Zeng, Z.; Li, H.; Li, F.; Fan, C.; Zhang, H. Single-Layer MoS2-Based Nanoprobes for Homogeneous Detection of Biomolecules. J. Am. Chem. Soc. 2013, 135, 5998–6001. [Google Scholar] [CrossRef] [PubMed]
- Geldert, A.; Kenry; Zhang, X.; Zhang, H.; Lim, C.T. Enhancing the Sensing Specificity of a MoS2 Nanosheet-Based FRET Aptasensor Using a Surface Blocking Strategy. Analyst 2017, 142, 2570–2577. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Liu, Z.; Cao, Y.; Dai, W.; Zhang, K.; Dong, H.; Feng, X.; Zhang, X. Fabricating Aptamer-Conjugated PEGylated-MoS2/Cu1.8S Theranostic Nanoplatform for Multiplexed Imaging Diagnosis and Chemo-Photothermal Therapy of Cancer. Adv. Funct. Mater. 2017, 27, 1605592. [Google Scholar] [CrossRef]
- Zhao, L.; Kong, D.; Wu, Z.; Liu, G.; Gao, Y.; Yan, X.; Liu, F.; Liu, X.; Wang, C.; Cui, J.; et al. Interface Interaction of MoS2 Nanosheets with DNA Based Aptameric Biosensor for Carbohydrate Antigen 15–3 Detection. Microchem. J. 2020, 155, 104675. [Google Scholar] [CrossRef]
- Chen, X.; Hao, S.; Zong, B.; Liu, C.; Mao, S. Ultraselective Antibiotic Sensing with Complementary Strand DNA Assisted Aptamer/MoS2 Field-Effect Transistors. Biosens. Bioelectron. 2019, 145, 111711. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.; Flahaut, E.; Golzio, M. Overview of Carbon Nanotubes for Biomedical Applications. Materials 2019, 12, 624. [Google Scholar] [CrossRef]
- Antman-Passig, M.; Ignatova, T.; Heller, D.A. Carbon Nanotube Optical Probes and Sensors. Electrochem. Soc. Interface 2019, 28, 61. [Google Scholar] [CrossRef]
- Choi, H.-K.; Lee, J.; Park, M.-K.; Oh, J.-H. Development of Single-Walled Carbon Nanotube-Based Biosensor for the Detection of Staphylococcus Aureus. Available online: https://www.hindawi.com/journals/jfq/2017/5239487/ (accessed on 22 February 2021).
- Khan, F.; He, M.; Taussig, M.J. Double-Hexahistidine Tag with High-Affinity Binding for Protein Immobilization, Purification, and Detection on Ni−Nitrilotriacetic Acid Surfaces. Anal. Chem. 2006, 78, 3072–3079. [Google Scholar] [CrossRef]
- Gutierrez, F.A.; Rubianes, M.D.; Rivas, G.A. Electrochemical Sensor for Amino Acids and Glucose Based on Glassy Carbon Electrodes Modified with Multi-Walled Carbon Nanotubes and Copper Microparticles Dispersed in Polyethylenimine. J. Electroanal. Chem. 2016, 765, 16–21. [Google Scholar] [CrossRef]
- Abnous, K.; Danesh, N.M.; Alibolandi, M.; Ramezani, M.; Taghdisi, S.M. Amperometric Aptasensor for Ochratoxin A Based on the Use of a Gold Electrode Modified with Aptamer, Complementary DNA, SWCNTs and the Redox Marker Methylene Blue. Microchim. Acta 2017, 184, 1151–1159. [Google Scholar] [CrossRef]
- Gu, F.; Hu, C.; Xia, Q.; Gong, C.; Gao, S.; Chen, Z. Aptamer-Conjugated Multi-Walled Carbon Nanotubes as a New Targeted Ultrasound Contrast Agent for the Diagnosis of Prostate Cancer. J. Nanoparticle Res. Interdiscip. Forum Nanoscale Sci. Technol. 2018, 20, 303. [Google Scholar] [CrossRef]
- Kim, S.-W.; Lee, Y.K.; Lee, J.Y.; Hong, J.H.; Khang, D. PEGylated Anticancer-Carbon Nanotubes Complex Targeting Mitochondria of Lung Cancer Cells. Nanotechnology 2017, 28, 465102. [Google Scholar] [CrossRef] [PubMed]
- Geyik, C.; Evran, S.; Timur, S.; Telefoncu, A. The Covalent Bioconjugate of Multiwalled Carbon Nanotube and Amino-Modified Linearized Plasmid DNA for Gene Delivery. Biotechnol. Prog. 2014, 30, 224–232. [Google Scholar] [CrossRef] [PubMed]
- Kavosi, A.; Hosseini Ghale Noei, S.; Madani, S.; Khalighfard, S.; Khodayari, S.; Khodayari, H.; Mirzaei, M.; Kalhori, M.R.; Yavarian, M.; Alizadeh, A.M.; et al. The Toxicity and Therapeutic Effects of Single-and Multi-Wall Carbon Nanotubes on Mice Breast Cancer. Sci. Rep. 2018, 8, 8375. [Google Scholar] [CrossRef] [PubMed]
- Peña-Bahamonde, J.; Nguyen, H.N.; Fanourakis, S.K.; Rodrigues, D.F. Recent Advances in Graphene-Based Biosensor Technology with Applications in Life Sciences. J. Nanobiotechnology 2018, 16, 75. [Google Scholar] [CrossRef] [PubMed]
- Torrente-Rodríguez, R.M.; Lukas, H.; Tu, J.; Min, J.; Yang, Y.; Xu, C.; Rossiter, H.B.; Gao, W. SARS-CoV-2 RapidPlex: A Graphene-Based Multiplexed Telemedicine Platform for Rapid and Low-Cost COVID-19 Diagnosis and Monitoring. Matter 2020, 3, 1981–1998. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Su, S.; Wu, N.; Wan, H.; Wan, S.; Bi, H.; Sun, L. Graphene-Based Sensors for Human Health Monitoring. Front. Chem. 2019, 7. [Google Scholar] [CrossRef]
- Pu, Z.; Tu, J.; Han, R.; Zhang, X.; Wu, J.; Fang, C.; Wu, H.; Zhang, X.; Yu, H.; Li, D. A Flexible Enzyme-Electrode Sensor with Cylindrical Working Electrode Modified with a 3D Nanostructure for Implantable Continuous Glucose Monitoring. Lab. Chip 2018, 18, 3570–3577. [Google Scholar] [CrossRef]
- Andoy, N.M.; Filipiak, M.S.; Vetter, D.; Gutiérrez-Sanz, Ó.; Tarasov, A. Graphene-Based Electronic Immunosensor with Femtomolar Detection Limit in Whole Serum. Adv. Mater. Technol. 2018, 3, 1800186. [Google Scholar] [CrossRef]
- Suvarnaphaet, P.; Pechprasarn, S. Graphene-Based Materials for Biosensors: A Review. Sensors 2017, 17, 2161. [Google Scholar] [CrossRef]
- Tang, L.; Wang, Y.; Li, J. The Graphene/Nucleic Acid Nanobiointerface. Chem. Soc. Rev. 2015, 44, 6954–6980. [Google Scholar] [CrossRef]
- Furukawa, K.; Ueno, Y.; Takamura, M.; Hibino, H. Graphene FRET Aptasensor. ACS Sens. 2016, 1, 710–716. [Google Scholar] [CrossRef]
- Vishnubhotla, R.; Ping, J.; Gao, Z.; Lee, A.; Saouaf, O.; Vrudhula, A.; Johnson, A.T.C. Scalable Graphene Aptasensors for Drug Quantification. AIP Adv. 2017, 7, 115111. [Google Scholar] [CrossRef]
- You, H.; Mu, Z.; Zhao, M.; Zhou, J.; Chen, Y.; Bai, L. Voltammetric Aptasensor for Sulfadimethoxine Using a Nanohybrid Composed of Multifunctional Fullerene, Reduced Graphene Oxide and Pt@Au Nanoparticles, and Based on Direct Electron Transfer to the Active Site of Glucose Oxidase. Microchim. Acta 2018, 186, 1. [Google Scholar] [CrossRef] [PubMed]
- Divsar, F.; Habibzadeh, K.; Shariati, S.; Shahriarinour, M. Aptamer Conjugated Silver Nanoparticles for the Colorimetric Detection of Arsenic Ions Using Response Surface Methodology. Anal. Methods 2015, 7, 4568–4576. [Google Scholar] [CrossRef]
- Huang, K.-J.; Liu, Y.-J.; Zhang, J.-Z.; Liu, Y.-M. A Novel Aptamer Sensor Based on Layered Tungsten Disulfide Nanosheets and Au Nanoparticles Amplification for 17β-Estradiol Detection. Anal. Methods 2014, 6, 8011–8017. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Xia, H.; Huang, K.-J.; Wu, X.; Ma, Y.-Y.; Deng, R.; Lu, Y.-F.; Han, Z.-W. Ultrasensitive Determination of Thrombin by Using an Electrode Modified with WSe2 and Gold Nanoparticles, Aptamer-Thrombin-Aptamer Sandwiching, Redox Cycling, and Signal Enhancement by Alkaline Phosphatase. Mikrochim. Acta 2018, 185, 502. [Google Scholar] [CrossRef] [PubMed]
- Yousefi, S.; Saraji, M. Optical Aptasensor Based on Silver Nanoparticles for the Colorimetric Detection of Adenosine. Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 2019, 213, 1–5. [Google Scholar] [CrossRef]
- Li, A.; Zhang, J.; Qiu, J.; Zhao, Z.; Wang, C.; Zhao, C.; Liu, H. A Novel Aptameric Biosensor Based on the Self-Assembled DNA–WS2 Nanosheet Architecture. Talanta 2017, 163, 78–84. [Google Scholar] [CrossRef]
- Hu, Z.; Tan, J.; Lai, Z.; Zheng, R.; Zhong, J.; Wang, Y.; Li, X.; Yang, N.; Li, J.; Yang, W.; et al. Aptamer Combined with Fluorescent Silica Nanoparticles for Detection of Hepatoma Cells. Nanoscale Res. Lett. 2017, 12, 96. [Google Scholar] [CrossRef]
- Grechkin, Y.A.; Grechkina, S.L.; Zaripov, E.A.; Fedorenko, S.V.; Mustafina, A.R.; Berezovski, M.V. Aptamer-Conjugated Tb(III)-Doped Silica Nanoparticles for Luminescent Detection of Leukemia Cells. Biomedicines 2020, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.-J.; Shuai, H.-L.; Chen, Y.-X. Layered Molybdenum Selenide Stacking Flower-like Nanostructure Coupled with Guanine-Rich DNA Sequence for Ultrasensitive Ochratoxin A Aptasensor Application. Sens. Actuators B Chem. 2016, 225, 391–397. [Google Scholar] [CrossRef]
- Huang, K.-J.; Liu, Y.-J.; Zhang, J.-Z.; Cao, J.-T.; Liu, Y.-M. Aptamer/Au Nanoparticles/Cobalt Sulfide Nanosheets Biosensor for 17β-Estradiol Detection Using a Guanine-Rich Complementary DNA Sequence for Signal Amplification. Biosens. Bioelectron. 2015, 67, 184–191. [Google Scholar] [CrossRef]
- Yin, X.; Cai, J.; Feng, H.; Wu, Z.; Zou, J.; Cai, Q. A Novel VS2 Nanosheet-Based Biosensor for Rapid Fluorescence Detection of Cytochrome c. New J. Chem. 2015, 39, 1892–1898. [Google Scholar] [CrossRef]
- Huang, K.-J.; Liu, Y.-J.; Zhang, J.-Z.; Liu, Y.-M. A Sequence-Specific DNA Electrochemical Sensor Based on Acetylene Black Incorporated Two-Dimensional CuS Nanosheets and Gold Nanoparticles. Sens. Actuators B Chem. 2015, 209, 570–578. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, T.; Gu, Y.; Yan, X.; Lu, N.; Liu, H.; Xu, Z.; Xing, Y.; Song, Y.; Zhang, Z.; et al. An Electrochemical Thrombin Aptasensor Based on the Use of Graphite-like C3N4 Modified with Silver Nanoparticles. Microchim. Acta 2020, 187, 163. [Google Scholar] [CrossRef]
- Chen, X.; Chen, S.; Hu, T.; Ma, Q. Fluorescent Aptasensor for Adenosine Based on the Use of Quaternary CuInZnS Quantum Dots and Gold Nanoparticles. Microchim. Acta 2017, 184, 1361–1367. [Google Scholar] [CrossRef]
- Ardekani, L.S.; Moghadam, T.T.; Thulstrup, P.W.; Ranjbar, B. Design and Fabrication of a Silver Nanocluster-Based Aptasensor for Lysozyme Detection. Plasmonics 2019, 14, 1765–1774. [Google Scholar] [CrossRef]
- Roushani, M.; Ghanbari, K. An Electrochemical Aptasensor for Streptomycin Based on Covalent Attachment of the Aptamer onto a Mesoporous Silica Thin Film-Coated Gold Electrode. Microchim. Acta 2019, 186, 115. [Google Scholar] [CrossRef]

| Modified SELEX | Description of Modification | Selection Rounds | Mean Kd (nM) | Advantages | Drawbacks | Reference |
|---|---|---|---|---|---|---|
| Hi-Fidelity SELEX | Hi-Fi SELEX utilized a fixed-region blocking elements to safeguard functional diversity of the SELEX library. The chemistry of aptamers is engineered such that non-specific retention of aptamers is strongly inhibited by modification of the target-display surface and composition of the equilibration solvent. Integration of novel qPCR into the Hi-Fi SELEX workflow allowed for rapid sequencing during selection rounds. | 3 selection rounds. 107–108 | ~2 and 20 | Partition efficiencies approaching 106 are realized. High potential value in screening a small amount of retained aptamers for putative therapeutics. | High reagent volume is required to sufficiently amplify library members between each selection round. | [28] |
| HT-SELEX | High throughput sequencing technology and bioinformatics analysis coupled with SELEX (HT-SELEX) assisted in understanding the effect of initial library and PCR methods in the RNA aptamer identification. The analysis revealed that a distinct sequence and nucleotide existed in the initial, unselected libraries and the fate of “biased sequences” was target-dependent during selection. Amplification by either PCR-driven SELEX or droplet digital PCR (ddPCR)-driven SELEX did result in molecular evolution, during which highly enriched aptamers were produced after the 5th round of selection. | 5–7 rounds | PCR-driven SELEX = 65.2 ddPCR-driven SELEX = 111.2 | ddPCR-driven selection allowed preservation of molecular diversity and chances of obtaining highly structural sequences are increased. | ddPCR requires extra steps: (1) droplet generation, (2) extraction of the amplicon by organic solvent. | [29] |
| Click SELEX | A chemical modification of nucleic acid libraries carried out using copper-catalyzed alkyne-azide cycloaddition (CuAAC) or click chemistry allowed for the introduction of a wide range of possible functionalities. The interaction properties of the resultant DNA aptamers are not accessible with the cononical set of nucleotides. The modified DNA is incubated with the target molecule and the best binding sequences are recovered after subsequent selection sequence. The chemical modification is removed during the amplification process. | 15 cycles, ~1 day for each selection cycle | _ | Relies only on well-established and commercially available building blocks. This feature makes click-SELEX accessible to many laboratories, even if in-house synthesis is not available. | The azide of choice must be stable under the conditions used for CuAAC and during the selection process; in addition, it must quantitatively react with the alkyne-modified DNA strand to avoid non-functionalized nucleobases during the selection process. | [30] |
| Cell-SELEX | A differential binding Cell-SELEX workflow that adapts the FASTAptamer toolbox and bioinformatics edgeR is employed to achieve more informative metrics about the selection process. The high-throughput (HT) aptamer identification method is coupled with the Cell-SELEX technique to increase the aptamer selection rate against live cells. | 11 selection cycles | _ | Shorter time for aptamer identification. Selection of aptamer sequences that can selectively bind to the target and control cells. | High round of selection cycles, at the 11th round, aptamer’ binding was non-specific. | [31] |
| Aptasensor | Signal Type | Target Molecule | Linear Range | Detection Limit | Reference |
|---|---|---|---|---|---|
| AuNPs-SEB aptamer | Colorimetry | Staphylococcal enterotoxin B | 50 µg/mL–0.5 ng/mL | 50 ng/mL | [36] |
| AuNPs-IL-6 aptamer | Colorimetry | Interleukin-6 | 3.3–125 µg/mL | 1.95 µg/mL | [37] |
| AuNPs-thio/27-mer aptamer | Colorimetry | Thrombin | 5 pM–2 nM | 5 pM | [38] |
| AuNPs-[Ru(NH3)6]3+-TBA2 aptamer | Electrochemical | Thrombin | 1 fM–6 pM | 0.1429 fM | [39] |
| CDS-QDs/AuNPs/Tro6 aptamer | Electrochemiluminescence | Cardiac troponin 1 | 1 fg/mL–10 ng/mL | 0.75 fg/mL | [40] |
| CdS-NCs/AuNPs/luminol aptamer | Ratiometric ECL | Thrombin | - | 500 fg/mL | [41] |
| CDs/AS1411 aptamer | Spectrofluorometry | Cancer cells | - | ~100 cells/mL | [42] |
| MoS2-NS aptamer | Fluorescence | PSA | 0.2 ng/mL | [43] | |
| MoS2-AuNPs/TiONBs/MC-LR aptamer | Electrochemical | Microcystin-LR | 0.005–30 nM | 0.002 nM6 | [44] |
| SWCNTs-PBASE aptamer | FET | Capthepsin K | 2.3 pM–0.23 nM | - | [45] |
| Graphene/SH-SAW aptamer | Surface Acoustic Wave | Endotoxins | 0–100 ng/mL | 3.53 ng/mL | [46] |
| GO/33-mer aptamer | Fluorescence | Theophylline | 1–100 µM | 0.155 µM | [47] |
| rGO-PET/cTnT aptamer | Electrical | Cardiac troponin T | 0.001–10 ng/mL | 1.2–1.7 pg/mL | [48] |
| Aptasensor | Signal Type | Target Molecule | Linear Range | Detection Limit | Reference |
|---|---|---|---|---|---|
| Tungsten diselenide/AuNPs based- thrombin aptamer (WSe2/AuNPs/TBA1 apt) | Electrochemical | Thrombin | 0–1 ngmL−1 | 190 fgmL−1 | [112] |
| Streptavidin-conjugated fluorescent silica nanoparticles-based biotin aptamer (SA-FSiNPs/Bio-TLS11a apt) | Fluorescence | HepG2 cell | - | - | [115] |
| Amino- and carboxyl-modified silica-coated terbium (III) thiacalix[4]arenesulfonate-based Sgc8 aptamer ([Tb(TCAS)]-SiNPs/Sgc8 apt) | Luminescence | Leukemia cell | - | - | [116] |
| Molybdenum diselenide modified AuNPs-based ochratoxin A aptamer (MoSe2/AuNPs/OTA apt) | Electrochemical | ochratoxin A | 0.0001–1 nM | 0.08 pM | [117] |
| Tungsten disulfide nanosheets/Au nanoparticles-modified glassy carbon electrode -based estradiol aptamer (GC-WS2/AuNPs/estrad apt) | Electrochemical | 17b-estradiol | 1.0 × 10−11–5.0 × 10−9 M | 2.0 × 10−12 | [118] |
| Vanadium disulfide-based cytochrome c aptamer (VS2/Cyt c apt) | Fluorescence | Cytochrome c | 0.75 nM–50 µM | 0.5 nM | [119] |
| Cobalt sulfide/Au nanoparticles modified electrode-based 17β-estradiol aptamer (CoS/AuNPs/17β-estrad apt) | Electrochemical | 17β-estradiol | 1.0 × 10−9 −1.0 × 10−12 M | 7.0 × 10−13 M | [111] |
| Acetylene black-copper sulfide nanosheets/Au modified electrode-based DNA aptamer (CuS-AB/Au/DNA apt) | Electrochemical | DNA | 0.1 pM–1 nM | 20 fM | [120] |
| Silver nanoparticles modified graphite-like carbon nitride-based thrombin aptamer (AgNPs-gr/C3N4 apt) | Electrochemical | Thrombin | 100 fM–20 nM | 38 fM | [121] |
| Quaternary CuInZnS quantum dots modified Au nanoparticles-based adenosine aptamer (CulnZnS-QDs/AuNPs apt) | Fluorescence | Adenosine | 50–400 µM | 1.1 µM | [122] |
| Silver nanoclusters based complementary DNA aptamer (AgNCs-cDNA apt) | Fluorescence | Lysozyme | 2–25 nM | 5.6 nM | [123] |
| Au electrode coated mesoporous silica film/silver nanoparticles based-streptomycin aptamer (MSF/Au/AgNPs strept apt) | Electrochemical | Streptomycin | 1 fg/mL–6.2 ng/mL | 0.33 fg/mL | [124] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayodele, O.O.; Adesina, A.O.; Pourianejad, S.; Averitt, J.; Ignatova, T. Recent Advances in Nanomaterial-Based Aptasensors in Medical Diagnosis and Therapy. Nanomaterials 2021, 11, 932. https://doi.org/10.3390/nano11040932
Ayodele OO, Adesina AO, Pourianejad S, Averitt J, Ignatova T. Recent Advances in Nanomaterial-Based Aptasensors in Medical Diagnosis and Therapy. Nanomaterials. 2021; 11(4):932. https://doi.org/10.3390/nano11040932
Chicago/Turabian StyleAyodele, Olubunmi O., Adeyinka O. Adesina, Sajedeh Pourianejad, Jared Averitt, and Tetyana Ignatova. 2021. "Recent Advances in Nanomaterial-Based Aptasensors in Medical Diagnosis and Therapy" Nanomaterials 11, no. 4: 932. https://doi.org/10.3390/nano11040932
APA StyleAyodele, O. O., Adesina, A. O., Pourianejad, S., Averitt, J., & Ignatova, T. (2021). Recent Advances in Nanomaterial-Based Aptasensors in Medical Diagnosis and Therapy. Nanomaterials, 11(4), 932. https://doi.org/10.3390/nano11040932






