Radiation Crosslinked Smart Peptide Nanoparticles: A New Platform for Tumor Imaging
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Solid-Phase Synthesis of the Peptides
2.3. Preparation and Irradiation of the Synthetic Peptide Aqueous Solution
2.4. Kinetics Study
2.5. HPLC and Ion Chromatography Analysis
2.6. Dynamic Light Scattering (DLS) and ELS Measurement
2.7. Biodegradability
2.8. Cellular Uptake Test
3. Results and Discussion
3.1. Peptide Synthesis
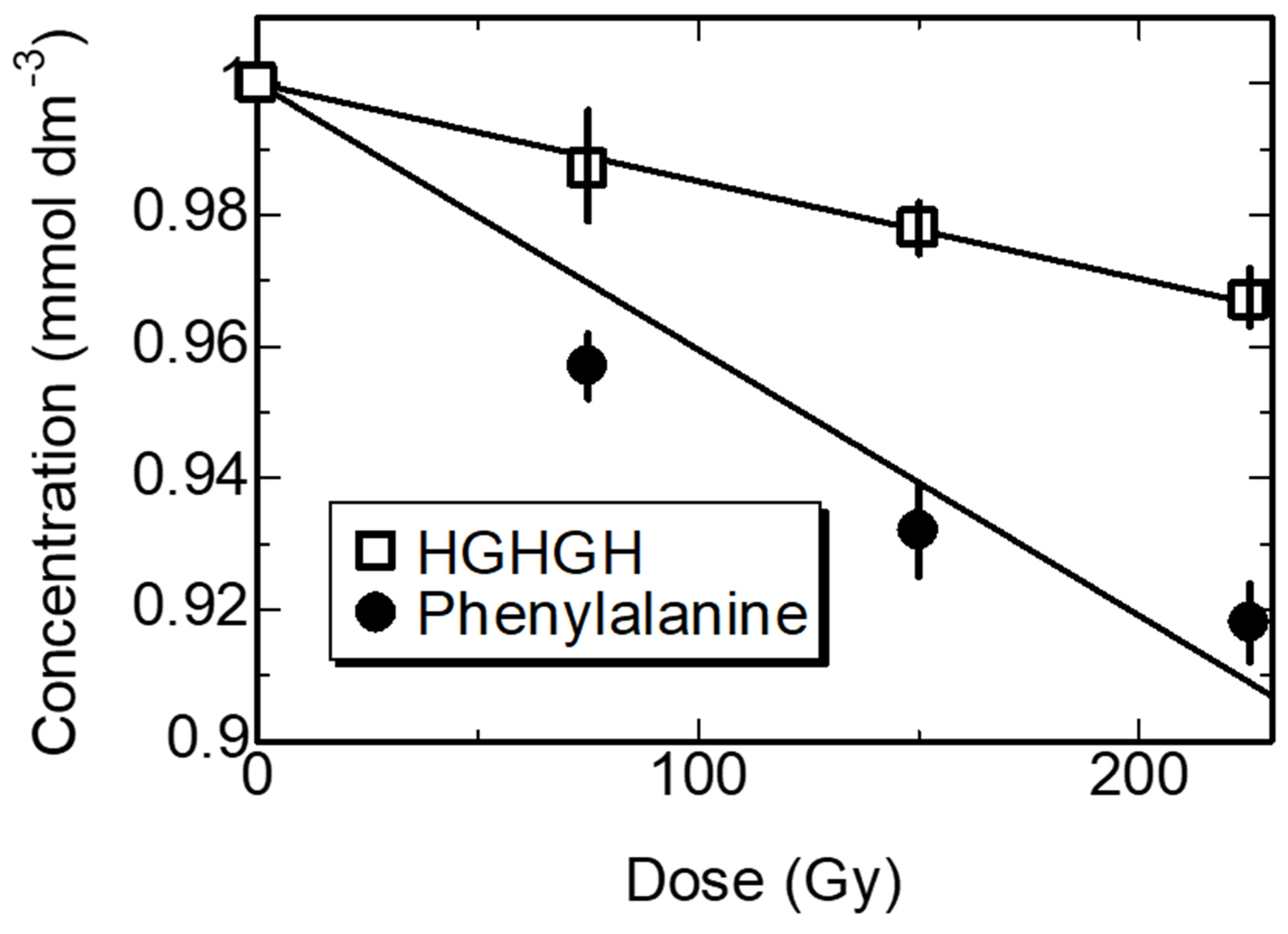
3.2. Kinetics of the Synthesized Peptides in Water by γ-ray Irradiation

HOOCCH(NH2)CH2-C6H4-C6H4-CH2CH(NH2)COOH
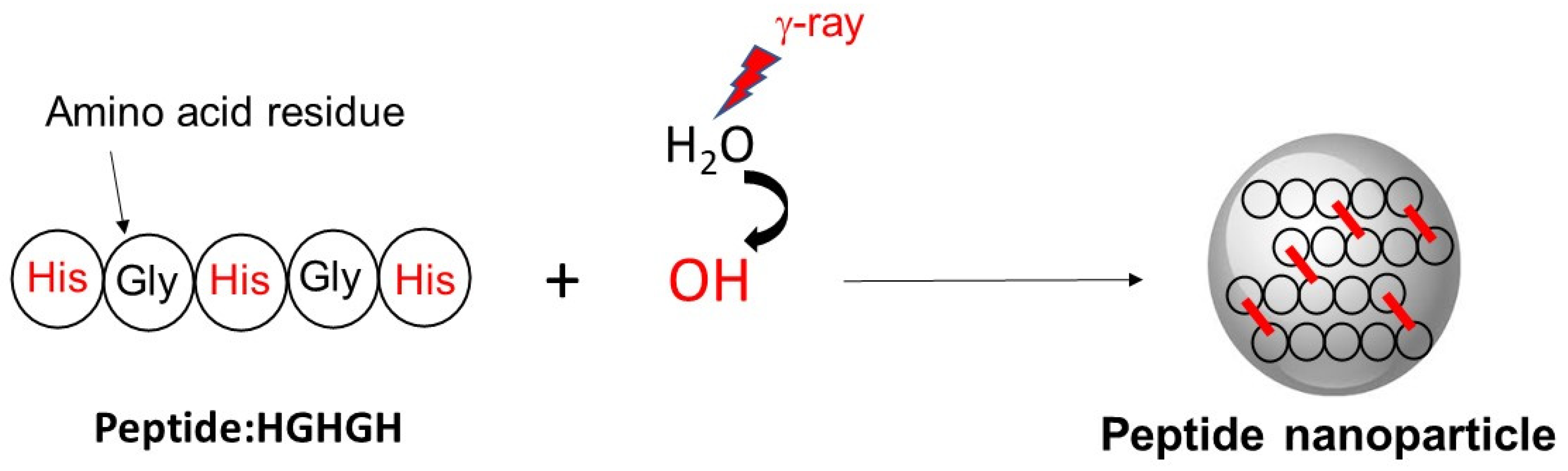
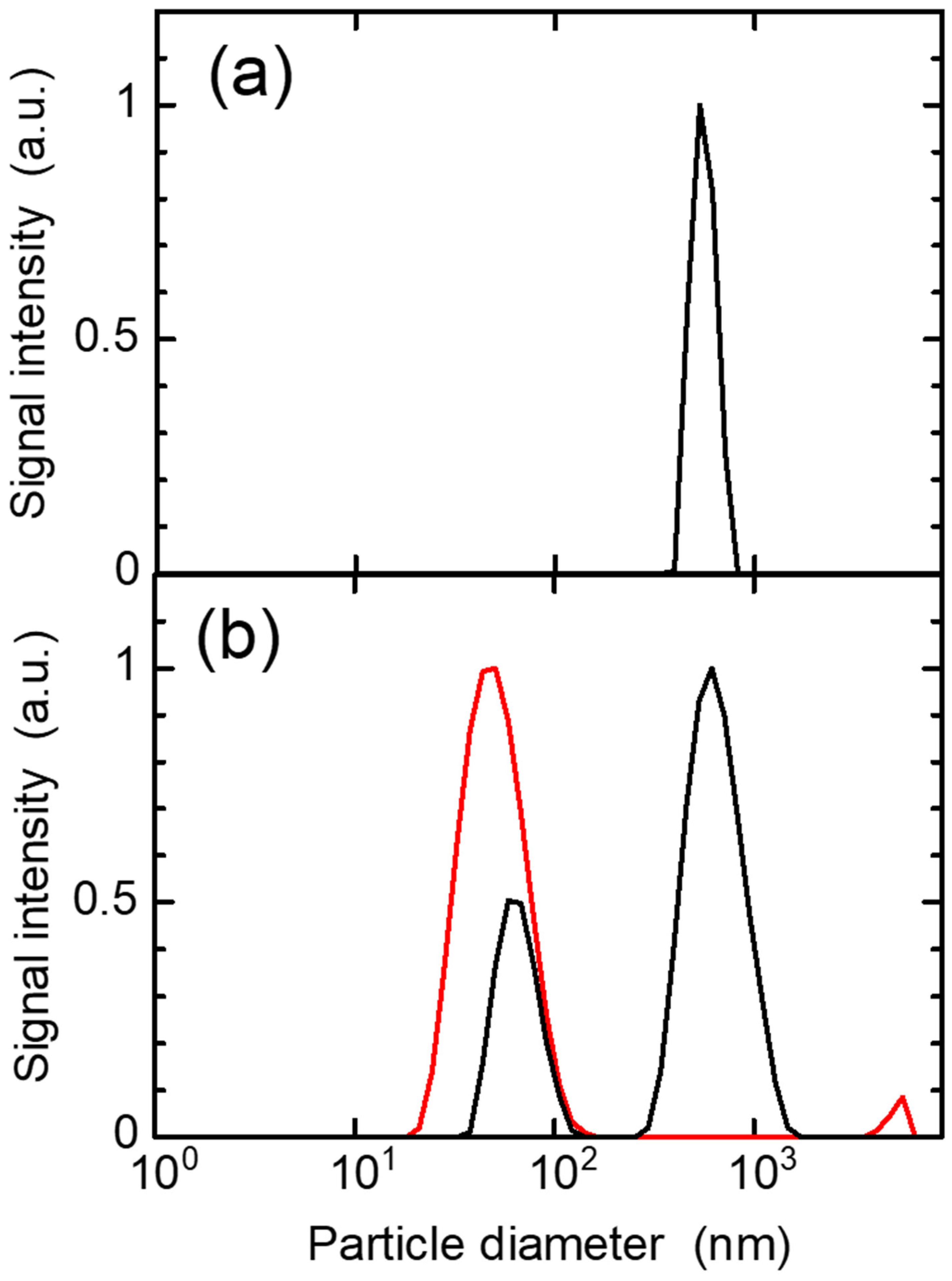
3.3. Production of Peptide Nanoparticles by Ionizing Radiation
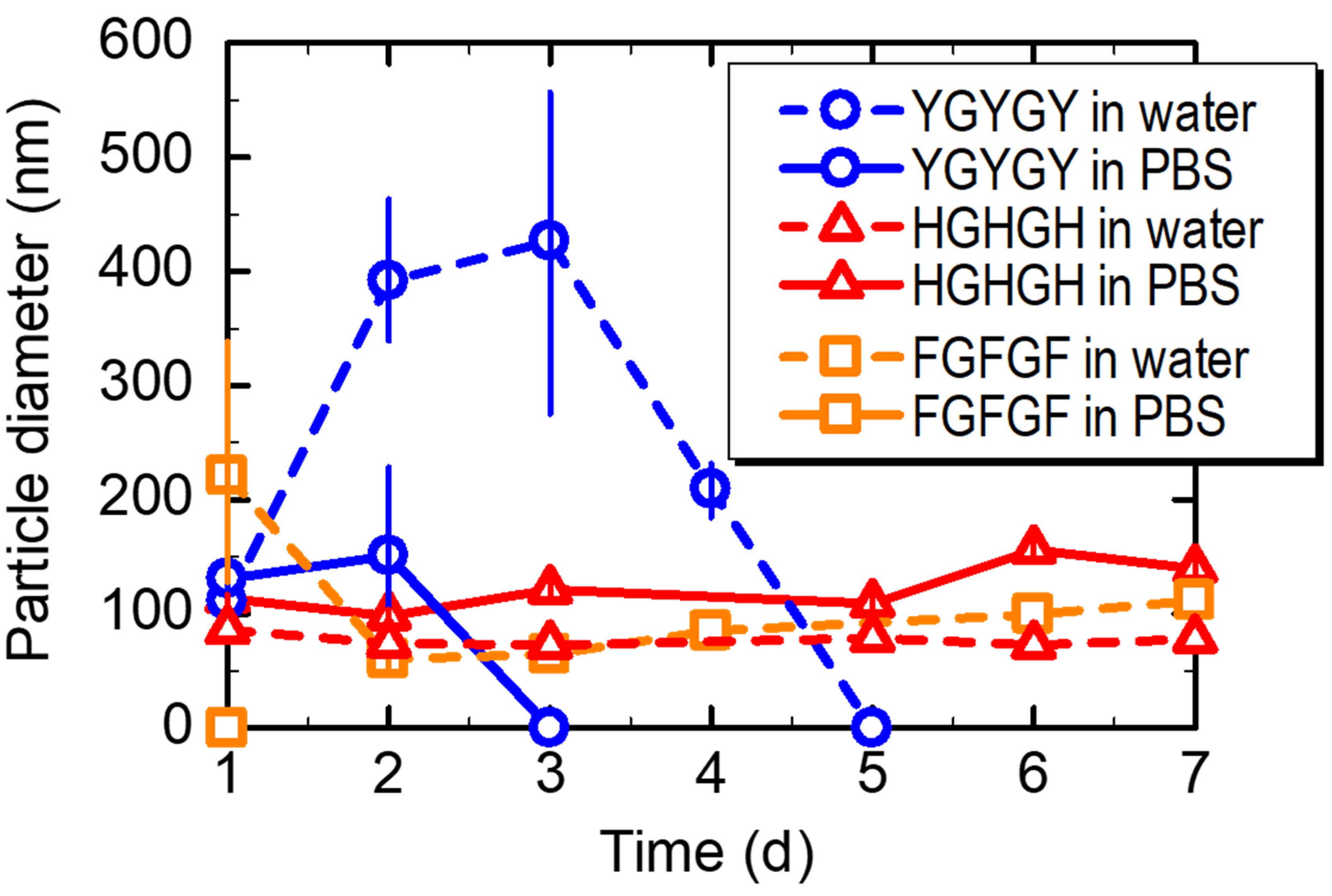
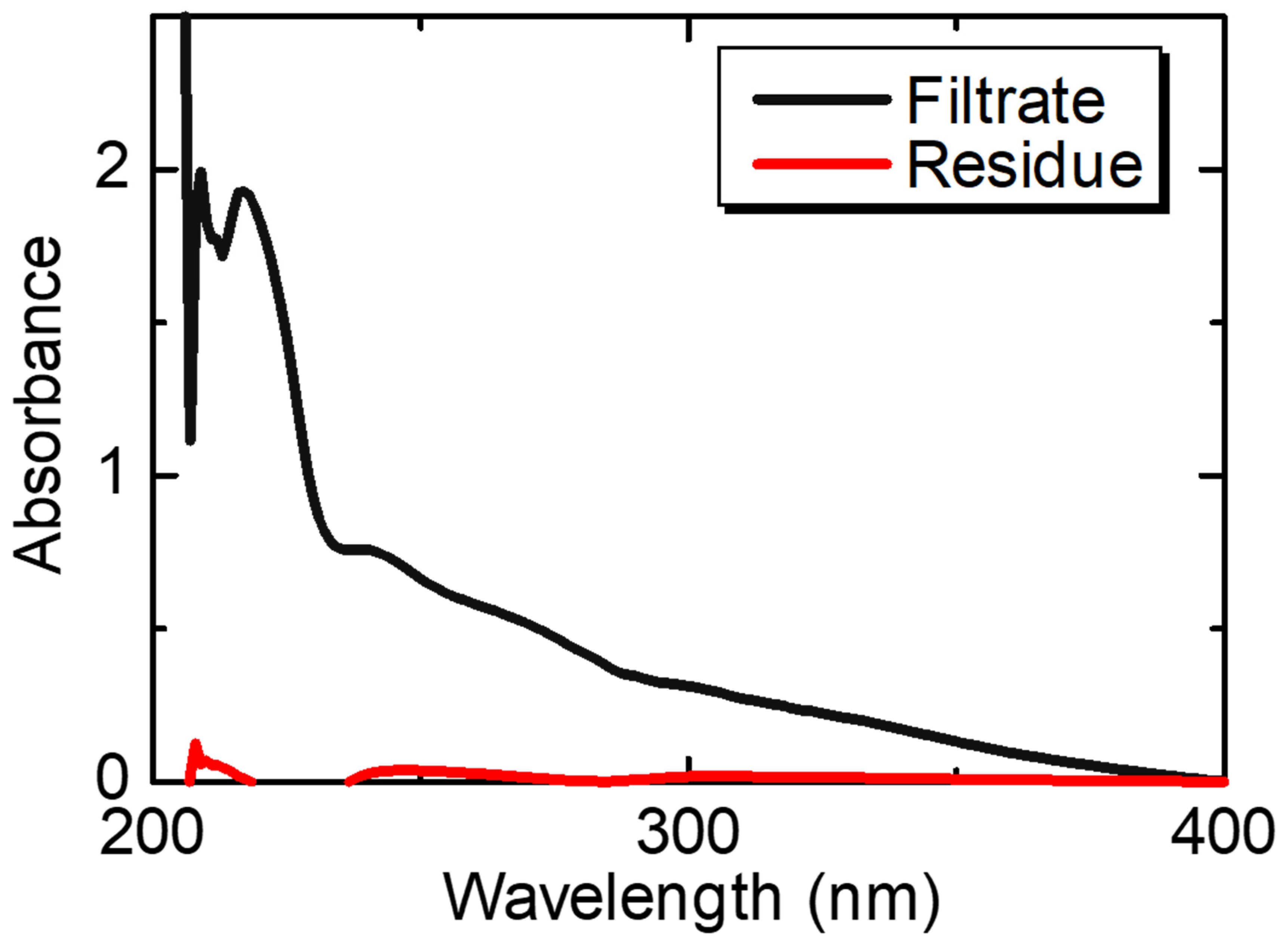
3.4. Stability and Biodegradability of the Peptide Nanoparticles
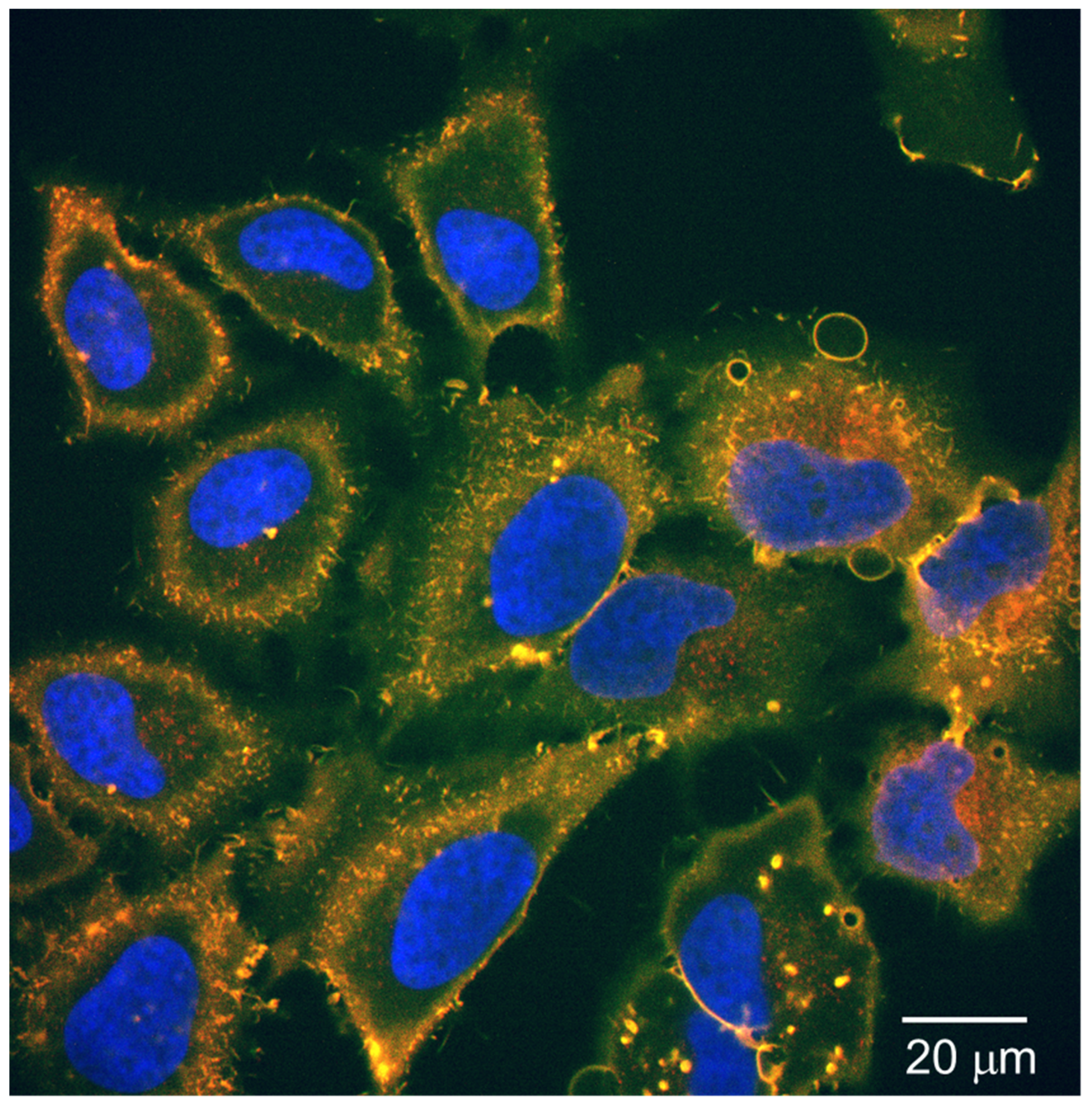
3.5. Cellular Uptake Test for the Fluorescent-Labeled Peptide Nanoparticles
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shi, Y.; van der Meel, R.; Chen, X.; Lammers, T. The EPR effect and beyond: Strategies to improve tumor targeting and cancer nanomedicine treatment efficacy. Theranostics 2020, 10, 7921–7924. [Google Scholar] [CrossRef]
- Kang, H.; Rho, S.; Stiles, W.R.; Hu, S.; Baek, Y.; Hwang, D.W.; Kashiwagi, S.; Kim, M.S.; Choi, H.S. Size-dependent EPR effect of polymeric nanoparticles on tumor targeting. Adv. Healthcare Mater. 2020, 9, 1901223. [Google Scholar] [CrossRef] [PubMed]
- Nasery, M.M.; Abadi, B.; Poormoghadam, D.; Zarrabi, A.; Keyhanvar, P.; Khanbabaei, H.; Ashrafizadeh, M.; Mohammadinejad, R.; Tavakol, S.; Sethi, G. Curcumin delivery mediated by bio-based nanoparticles: A review. Molecules 2020, 25, 689. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Lee, J.; Chen, Y.; Hoshino, K. Studies of nanoparticle delivery with in vitro bio-engineered microtissues. Bioactive Mater. 2020, 5, 924–937. [Google Scholar] [CrossRef]
- Fröhlich, E. The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed. 2012, 7, 5577–5591. [Google Scholar] [CrossRef]
- Han, X.; Xu, K.; Taratula, O.; Farsad, K. Applications of nanoparticles in biomedical imaging. Nanoscale 2019, 11, 799–819. [Google Scholar] [CrossRef]
- Wang, H.; Yi, J.; Mukherjee, S.; Banerjee, P.; Zhou, S. Magnetic/NIR-thermally responsive hybrid nanogels for optical temperature sensing, tumor cell imaging and triggered drug release. Nanoscale 2014, 6, 13001–13011. [Google Scholar] [CrossRef]
- Veloso, S.R.S.; Andrade, R.G.D.; Ribeiro, B.C.; Fernandes, A.V.F.; Rodrigues, A.R.O.; Martins, J.A.; Ferreira, P.M.T.; Coutinho, P.J.G.; Castanheira, E.M.S. Magnetoliposomes incorporated in peptide-based hydrogels: Towards development of magnetolipogels. Nanomaterials 2020, 10, 1702. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.-Q.; Wang, C.-C. Biodegradable smart nanogels: A new platform for targeting drug delivery and biomedical diagnostics. Langmuir 2016, 32, 6211–6225. [Google Scholar] [CrossRef] [PubMed]
- Saadat, M.; Zahednezhad, F.; Zakeri-Milani, P.; Heidari, H.R.; Shahbazi-Mojarrad, J.; Valizadeh, H. Drug targeting strategies based on charge dependent uptake of nanoparticles into cancer cells. J. Pharm. Pharm. Sci. 2019, 22, 131–364. [Google Scholar] [CrossRef] [PubMed]
- Anegelova, A.; Drechsler, M.; Garamus, V.M.; Angelov, B. Pep-lipid cubosomes and vesicles compartmentalized by micelles from self-assembly of multiple neuroprotective building blocks including a large peptide hormone PACAP-DHA. ChemNanoMat 2019, 5, 1381–1389. [Google Scholar] [CrossRef]
- Mohd-Zahid, M.H.; Mohamud, R.; Abdullah, C.A.C.; Lim, J.; Alem, H.; Hanaffi, W.N.W.; Iskandar, Z.A. Colorectal cancer stem cells: A review of targeted drug delivery by gold nanoparticles. RSC Adv. 2020, 10, 973–985. [Google Scholar] [CrossRef]
- Harush-Frenkel, O.; Debotton, N.; Benita, S.; Altschuler, Y. Targeting of nanoparticles to the clathrin-mediated endocytic pathway. Biochem. Biophys. Res. Commun. 2007, 353, 26–32. [Google Scholar] [CrossRef]
- Cabral, H.; Matsumoto, Y.; Mizuno, K.; Chen, Q.; Murakami, M.; Kimura, M.; Terada, Y.; Kano, M.R.; Miyazono, K.; Uesaka, M.; et al. Accumulation of sub-100 nm polymeric micelles in poorly permeable tumors depends on size. Nat. Nanotechnol. 2011, 6, 815–823. [Google Scholar] [CrossRef]
- Zein, R.; Sharrouf, W.; Selting, K. Physical properties of nanoparticles that result in improved cancer targeting. J. Oncol. 2020, 5194780. [Google Scholar] [CrossRef]
- Yue, Z.-G.; Wei, W.; Lv, P.-P.; Yue, H.; Wang, L.-Y.; Su, Z.-G.; Ma, G.-H. Surface charge affects cellular uptake and intracellular trafficking of chitosan-based nanoparticles. Biomacromolecules 2011, 12, 2440–2446. [Google Scholar] [CrossRef] [PubMed]
- Asati, A.; Santra, S.; Kaittanis, C.; Perez, J.M. Surface-charge-dependent cell localization and cytotoxicity of cerium oxide nanoparticles. ACS Nano 2010, 4, 5321. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Hu, Y.; Yin, L.; Tang, C.; Yin, C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials 2010, 31, 3657–3666. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Jothar, L.; Barendrecht, A.D.; de Graaff, A.M.; Oliveira, S.; van Nostrum, C.F.; Schiffelers, R.M.; Hennink, W.E.; Fens, M.H.A.M. Endothelial cell targeting by cRGD-functionalized polymeric nanoparticles under static and flow conditions. Nanomaterials 2020, 10, 1353. [Google Scholar] [CrossRef]
- Poon, W.; Zhang, Y.-N.; Ouyang, B.; Kingston, B.R.; Wu, J.L.Y.; Wilhelm, S.; Chan, W.C.W. Elimination pathways of nanoparticles. ACS Nano 2019, 13, 5785–5798. [Google Scholar] [CrossRef]
- Pentlavalli, S.; Coulter, S.; Laverty, G. Peptide nanomaterials for drug delivery applications. Curr. Protein Pept. Sci. 2020, 21, 401–412. [Google Scholar] [CrossRef]
- Oyama, T.G.; Kimura, A.; Nagasawa, N.; Oyama, K.; Taguchi, M. Development of advanced biodevices using quantum beam microfabrication technology. Quantum Beam Sci. 2020, 4, 14. [Google Scholar] [CrossRef]
- Oyama, T.G.; Oyama, K.; Taguchi, M. A simple method for production of hydrophilic, rigid, and sterilized multi-layer 3D integrated polydimethylsiloxane microfluidic chips. Lab Chip 2020, 20, 2354–2363. [Google Scholar] [CrossRef] [PubMed]
- Oyama, T.G.; Barba, B.J.D.; Hosaka, Y.; Taguchi, M. Single-step fabrication of polydimethylsiloxane microwell arrays with long-lasting hydrophilic inner surfaces. Appl. Phys. Lett. 2018, 112, 213704. [Google Scholar] [CrossRef]
- Sasaki, S.; Omata, S.; Murakami, T.; Nagasawa, N.; Taguchi, M.; Suzuki, A. Effect of gamma ray irradiation on friction property of poly(vinyl alcohol) cast-drying on freeze-thawed hybrid gel. Gels 2018, 4, 30. [Google Scholar] [CrossRef]
- Haema, K.; Oyama, T.G.; Kimura, A.; Taguchi, M. Radiation stability and modification of gelatin for biological and medical applications. Radiat. Phys. Chem. 2014, 103, 126–130. [Google Scholar] [CrossRef]
- Kimura, A.; Nagasawa, N.; Taguchi, M. Synthesis of polysaccharide hybrid gel in ionic liquids via radiation-induced crosslinking. Polym. Degrad. Stab. 2019, 159, 133–138. [Google Scholar] [CrossRef]
- Nagasawa, N.; Kimura, A.; Idesaki, A.; Yamada, N.; Koka, M.; Satoh, T.; Ishii, Y.; Taguchi, M. Microfabrication of biocompatible hydrogels by proton beam writing. Nucl. Instrum. Methods Phys. Res. Sect. B 2017, 409, 102–106. [Google Scholar] [CrossRef]
- Bessho, M.; Kojima, T.; Okuda, S.; Hara, M. Radiation-induced cross-linking of gelatin by using γ-rays: Insoluble gelatin hydrogel formation. Chem. Soc. Jap. 2007, 80, 979–985. [Google Scholar] [CrossRef]
- Terao, K.; Nagasawa, N.; Nishida, H.; Furusawa, K.; Mori, Y.; Yoshii, F.; Dobashi, T. Reagent-free crosslinking of aqueous gelatin: Manufacture and characteristics of gelatin gels irradiated with gamma-ray and electron beam. J. Biomater. Sci. Polym. Ed. 2003, 14, 1197–1208. [Google Scholar] [CrossRef]
- Inoue, N.; Bessho, M.; Furuta, M.; Kojima, T.; Okuda, S.; Hara, M. A novel collagen hydrogel cross-linked by gamma-ray irradiation in acidic pH conditions. J. Biomater. Sci. Polym. Ed. 2006, 17, 837–858. [Google Scholar] [CrossRef]
- Kimura, A.; Yoshida, F.; Ueno, M.; Taguchi, M. Application of radiation crosslinking technique to development of gelatin scaffold for tissue engineering. Radiat. Phys. Chem. 2021, 180, 109287. [Google Scholar] [CrossRef]
- Furusawa, K.; Terao, K.; Nagasawa, N.; Yoshii, F.; Kubota, K.; Dobashi, T. Nanometer-sized gelatin particles prepared by means of gamma-ray irradiation. Colloid Polym. Sci. 2004, 283, 229–233. [Google Scholar]
- Akiyama, Y.; Fujiwara, T.; Takeda, S.-I.; Izumi, Y.; Nishijima, S. Preparation of stimuli-responsive protein nanogel by quantum-ray irradiation. Colloid Polym. Sci. 2007, 285, 801–807. [Google Scholar]
- Kimura, A.; Jo, J.; Yoshida, F.; Hong, Z.; Tabata, Y.; Sumiyoshi, A.; Taguchi, M.; Aoki, I. Ultra-small size gelatin nanogel as a blood brain barrier impermeable contrast agent for magnetic resonance imaging. Acta Biomater. 2021, in press. [Google Scholar] [CrossRef]
- Kaga, C.; Okochi, M.; Tomita, Y.; Kato, R.; Honda, H. Computationally assisted screening and design of cell-interactive peptides by a cell-based assay using peptide arrays and a fuzzy neural network algorithm. Biotechniques 2008, 44, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Garrison, W.M. Reaction mechanisms in the radiolysis of peptides, polypeptides, and proteins. Chem. Rev. 1987, 87, 381–398. [Google Scholar] [CrossRef]
- Hrnčiř, S.; Kopoldová, J. Gamma-radiolysis of aqueous solution of histidine. Z. Naturforsch. 1977, 32c, 482–487. [Google Scholar] [CrossRef]
- Kimura, A.; Osawa, M.; Taguchi, M. Decomposition of persistent pharmaceuticals in wastewater by ionizing radiation. Radiat. Phys. Chem. 2012, 81, 1508–1512. [Google Scholar] [CrossRef]
- Masuda, T.; Nakano, S.; Kondo, M. Rate constants for the reactions of OH radicals with the enzyme proteins as determined by the p-nitrosodimethylaniline method. J. Radiat. Res. 1973, 14, 339–345. [Google Scholar] [CrossRef]
- Ye, M.; Shuler, R.H. Second-order combination reactions of phenoxyl radicals. J. Phys. Chem. 1989, 93, 1898–1902. [Google Scholar] [CrossRef]
- Phung, P.V.; Burton, M. Radiolysis of aqueous solutions of hydrocarbons benzene, benzene-d6, cyclohexane. J. Radiat. Res. 1957, 7, 199–216. [Google Scholar] [CrossRef]


| Abbreviation | Peptide Sequence |
|---|---|
| HGHGH | His-Gly-His-Gly-His |
| YGYGY | Tyr-Gly-Tyr-Gly-Tyr |
| FGFGF | Phe-Gly-Phe-Gly-Phe |
| FG4 | Phe-Gly-Gly-Gly-Gly |
| FG3F | Phe-Gly-Gly-Gly-Phe |
| G5 | Gly-Gly-Gly-Gly-Gly |
| Amino Acids and Peptides | Abbreviation | Rate Constant with ·OH Radicals (mol−1 dm3 s−1) |
|---|---|---|
| Phenylalanine | F | 7.2 × 109 |
| Histidine | H | 4.3 × 109 |
| Tyrosine | Y | 10.5 × 109 |
| Glycine | G | 1.7 × 107 |
| His-Gly-His-Gly-His | HGHGH | 2.6 × 109 |
| Tyr-Gly-Tyr-Gly-Tyr | YGYGY | 4.1 × 109 |
| Phe-Gly-Phe-Gly-Phe | FGFGF | 6.8 × 109 |
| Peptides | Before Fluorescent Staining /mV | After Fluorescent Staining /mV |
|---|---|---|
| HGHGH | +23.4 | −13.9 |
| HGHGH (double amount) | +23.4 | −14.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kimura, A.; Ueno, M.; Arai, T.; Oyama, K.; Taguchi, M. Radiation Crosslinked Smart Peptide Nanoparticles: A New Platform for Tumor Imaging. Nanomaterials 2021, 11, 714. https://doi.org/10.3390/nano11030714
Kimura A, Ueno M, Arai T, Oyama K, Taguchi M. Radiation Crosslinked Smart Peptide Nanoparticles: A New Platform for Tumor Imaging. Nanomaterials. 2021; 11(3):714. https://doi.org/10.3390/nano11030714
Chicago/Turabian StyleKimura, Atsushi, Miho Ueno, Tadashi Arai, Kotaro Oyama, and Mitsumasa Taguchi. 2021. "Radiation Crosslinked Smart Peptide Nanoparticles: A New Platform for Tumor Imaging" Nanomaterials 11, no. 3: 714. https://doi.org/10.3390/nano11030714
APA StyleKimura, A., Ueno, M., Arai, T., Oyama, K., & Taguchi, M. (2021). Radiation Crosslinked Smart Peptide Nanoparticles: A New Platform for Tumor Imaging. Nanomaterials, 11(3), 714. https://doi.org/10.3390/nano11030714







