Bio-Separated and Gate-Free 2D MoS2 Biosensor Array for Ultrasensitive Detection of BRCA1
Abstract
1. Introduction
2. Materials and Methods
2.1. Biomaterials
- Target detection (BRCA1): 5′-GAACAAAAGGAAGAAAATC-3′,
- Mismatch DNA sequence: 5′-TGCAAGGTGTCAGTATAATCCGACGTTTT-3′.
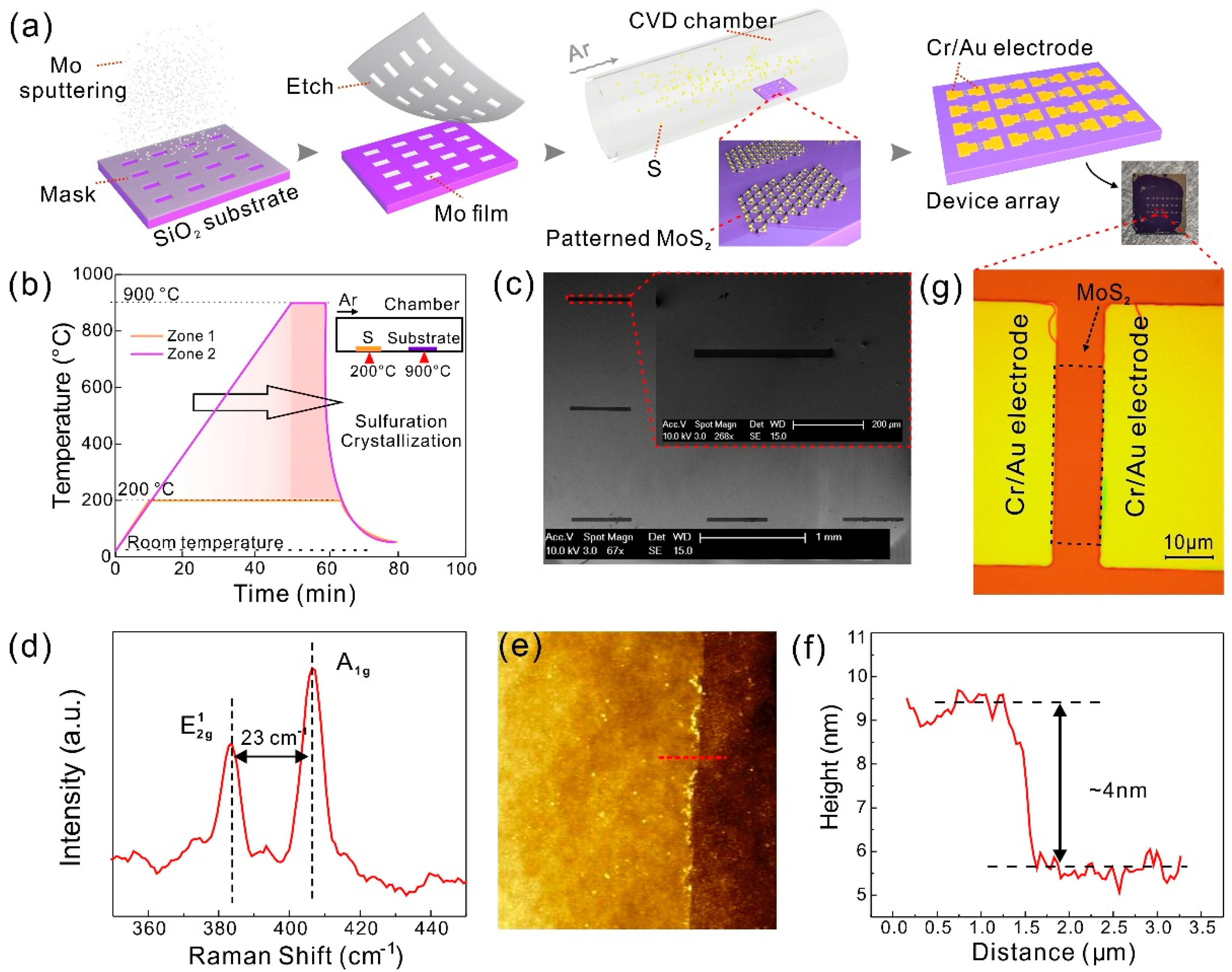
2.2. Synthesis of Patterned MoS2 and Fabrication of MoS2 Device Arrays
2.3. Synthesis of DNA-TPs
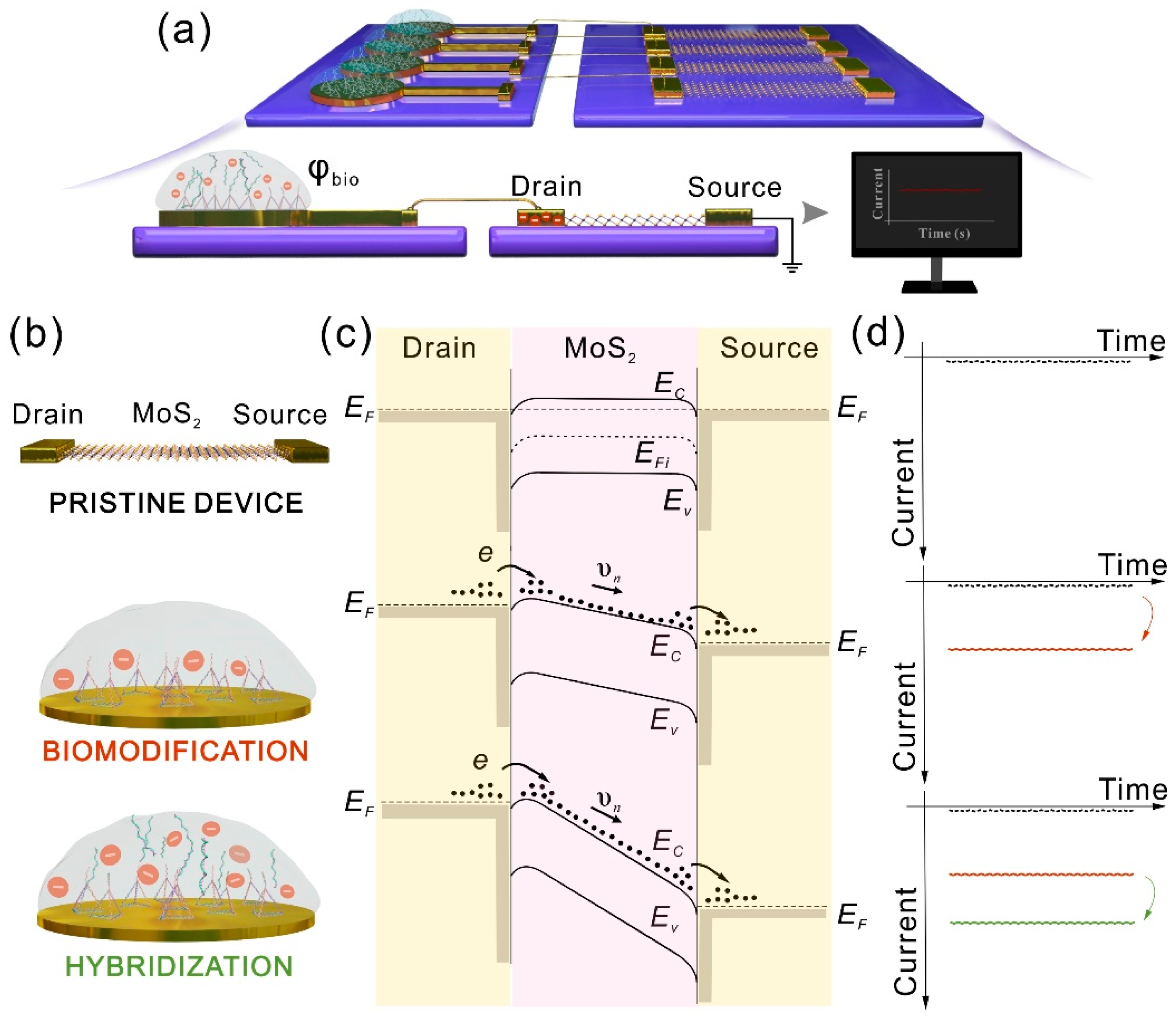
2.4. Development of Separable Biosensor
2.5. Measurement Instruments
3. Results and Discussion
3.1. The Fabrication of MoS2 TFT Arrays
3.2. The Biomodification of the Sensing Part
3.3. Principle of the Separated Gate-Free Sensor
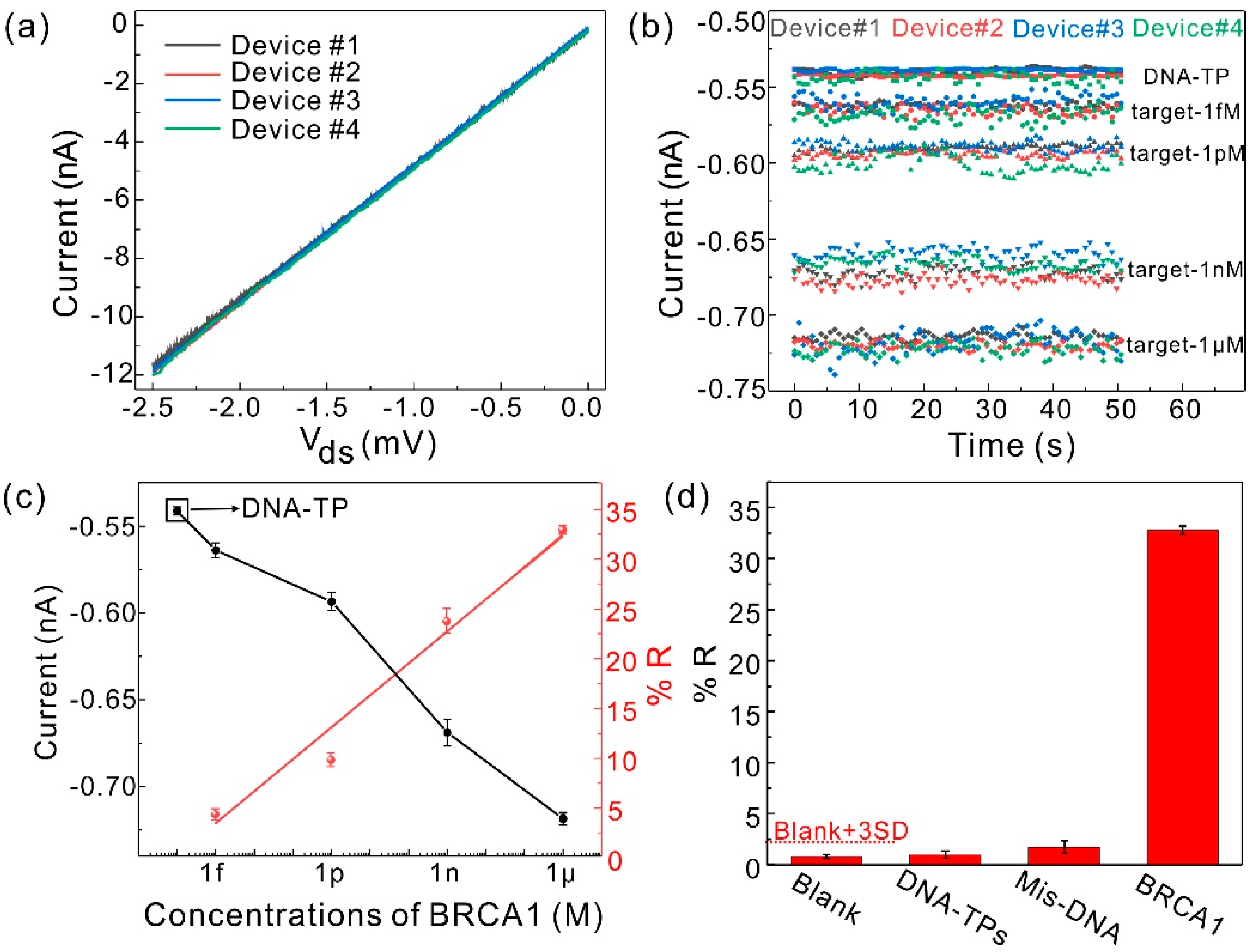
3.4. Detection Performance of the Sensor
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Mathur, P.; Sathishkumar, K.; Chaturvedi, M.; Das, P.; Sudarshan, K.L.; Santhappan, S.; Nallasamy, V.; John, A.; Narasimhan, S.; Roselind, F.S. Cancer Statistics, 2020: Report From National Cancer Registry Programme, India. JCO Glob. Oncol. 2020, 6, 1063–1075. [Google Scholar] [CrossRef]
- Jazayeri, M.H.; Amani, H.; Pourfatollah, A.A.; Pazoki-Toroudi, H.; Sedighimoghaddam, B. Various methods of gold nanoparticles (GNPs) conjugation to antibodies. Sens. Bio-Sens. Res. 2016, 9, 17–22. [Google Scholar] [CrossRef]
- Cai, B.; Wang, S.; Huang, L.; Ning, Y.; Zhang, Z.; Zhang, G.J. Ultrasensitive label-free detection of PNA-DNA hybridization by reduced graphene oxide field-effect transistor biosensor. ACS Nano 2014, 8, 2632–2638. [Google Scholar] [CrossRef] [PubMed]
- Yoo, G.; Park, H.; Kim, M.; Song, W.G.; Jeong, S.; Kim, M.H.; Lee, H.; Lee, S.W.; Hong, Y.K.; Lee, M.G.; et al. Real-time electrical detection of epidermal skin MoS2 biosensor for point-of-care diagnostics. Nano Res. 2016, 10, 767–775. [Google Scholar] [CrossRef]
- Park, H.Y.; Dugasani, S.R.; Kang, D.H.; Jeon, J.; Jang, S.K.; Lee, S.; Roh, Y.; Park, S.H.; Park, J.H. n- and p-Type doping phenomenon by artificial DNA and M-DNA on two-dimensional transition metal dichalcogenides. ACS Nano 2014, 8, 11603–11613. [Google Scholar] [CrossRef]
- Kaisti, M. Detection principles of biological and chemical FET sensors. Biosens. Bioelectron. 2017, 98, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Tian, B. Inorganic semiconductor biointerfaces. Nat. Rev. Mater. 2018, 3, 473–490. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.J.; Sohn, I.Y.; Jung, J.H.; Yoon, O.J.; Lee, N.E.; Park, J.S. Reduced graphene oxide field-effect transistor for label-free femtomolar protein detection. Biosens. Bioelectron. 2013, 41, 621–626. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Wong, J.I.; Palacios, T.; Kong, J.; Yang, H.Y. Functionalized MoS2 nanosheet-based field-effect biosensor for label-free sensitive detection of cancer marker proteins in solution. Small 2014, 10, 1101–1105. [Google Scholar] [CrossRef]
- Mei, J.; Li, Y.T.; Zhang, H.; Xiao, M.M.; Ning, Y.; Zhang, Z.Y.; Zhang, G.J. Molybdenum disulfide field-effect transistor biosensor for ultrasensitive detection of DNA by employing morpholino as probe. Biosens. Bioelectron. 2018, 110, 71–77. [Google Scholar] [CrossRef]
- Xu, S.; Zhan, J.; Man, B.; Jiang, S.; Yue, W.; Gao, S.; Guo, C.; Liu, H.; Li, Z.; Wang, J.; et al. Real-time reliable determination of binding kinetics of DNA hybridization using a multi-channel graphene biosensor. Nat. Commun. 2017, 8, 14902. [Google Scholar] [CrossRef]
- Majd, S.M.; Salimi, A.; Ghasemi, F. An ultrasensitive detection of miRNA-155 in breast cancer via direct hybridization assay using two-dimensional molybdenum disulfide field-effect transistor biosensor. Biosens. Bioelectron. 2018, 105, 6–13. [Google Scholar] [CrossRef]
- Lu, N.; Gao, A.; Dai, P.; Song, S.; Fan, C.; Wang, Y.; Li, T. CMOS-compatible silicon nanowire field-effect transistors for ultrasensitive and label-free microRNAs sensing. Small 2014, 10, 2022–2028. [Google Scholar] [CrossRef] [PubMed]
- Eda, G.; Fujita, T.; Yamaguchi, H.; Voiry, D.; Chen, M.; Chhowalla, M. Coherent atomic and electronic heterostructures of single-layer MoS2. ACS Nano 2012, 6, 7311–7317. [Google Scholar] [CrossRef]
- Wang, X.; Feng, H.; Wu, Y.; Jiao, L. Controlled synthesis of highly crystalline MoS2 flakes by chemical vapor deposition. J. Am. Chem. Soc. 2013, 135, 5304–5307. [Google Scholar] [CrossRef] [PubMed]
- Xiong, K.; Kim, H.; Marstell, R.J.; Göritz, A.; Wipf, C.; Li, L.; Park, J.-H.; Luo, X.; Wietstruck, M.; Madjar, A.; et al. CMOS-compatible batch processing of monolayer MoS2MOSFETs. J. Phys. D Appl. Phys. 2018, 51, 15LT02. [Google Scholar] [CrossRef]
- Mao, S.; Chang, J.; Pu, H.; Lu, G.; He, Q.; Zhang, H.; Chen, J. Two-dimensional nanomaterial-based field-effect transistors for chemical and biological sensing. Chem. Soc. Rev. 2017, 46, 6872–6904. [Google Scholar] [CrossRef] [PubMed]
- Gan, X.; Zhao, H.; Quan, X. Two-dimensional MoS2: A promising building block for biosensors. Biosens. Bioelectron. 2017, 89, 56–71. [Google Scholar] [CrossRef] [PubMed]
- Spanu, A.; Lai, S.; Cosseddu, P.; Tedesco, M.; Martinoia, S.; Bonfiglio, A. An organic transistor-based system for reference-less electrophysiological monitoring of excitable cells. Sci Rep. 2015, 5, 8807. [Google Scholar] [CrossRef]
- Thakur, B.; Zhou, G.; Chang, J.; Pu, H.; Jin, B.; Sui, X.; Yuan, X.; Yang, C.H.; Magruder, M.; Chen, J. Rapid detection of single E. coli bacteria using a graphene-based field-effect transistor device. Biosens. Bioelectron. 2018, 110, 16–22. [Google Scholar] [CrossRef]
- Huang, Y.-W.; Wu, C.-S.; Chuang, C.-K.; Pang, S.-T.; Pan, T.-M.; Yang, Y.-S.; Ko, F.-H. Real-Time and Label-Free Detection of the Prostate-Specific Antigen in Human Serum by a Polycrystalline Silicon Nanowire Field-Effect Transistor Biosensor. Anal. Chem. 2013, 85, 7912–7918. [Google Scholar] [CrossRef]
- Yu, J.; Xu, M.; Liang, L.; Guan, M.; Zhang, Y.; Yan, F.; Cao, H. Separative extended-gate AlGaAs/GaAs HEMT biosensors based on capacitance change strategy. Appl. Phys. Lett. 2020, 116, 123704. [Google Scholar] [CrossRef]
- Luo, J.; Li, S.; Xu, M.; Guan, M.; Yang, M.; Ren, J.; Zhang, Y.; Zeng, Y. Real-time detection of cardiac troponin I and mechanism analysis of AlGaAs/GaAs high electron mobility transistor biosensor. AIP Adv. 2020, 10, 115205. [Google Scholar] [CrossRef]
- Zheng, C.; Huang, L.; Zhang, H.; Sun, Z.; Zhang, Z.; Zhang, G.-J. Fabrication of Ultrasensitive Field-Effect Transistor DNA Biosensors by a Directional Transfer Technique Based on CVD-Grown Graphene. ACS Appl. Mater. Interfaces 2016, 8, 27546–27552. [Google Scholar] [CrossRef]
- Kaisti, M.; Kerko, A.; Aarikka, E.; Saviranta, P.; Boeva, Z.; Soukka, T.; Lehmusvuori, A. Real-time wash-free detection of unlabeled PNA-DNA hybridization using discrete FET sensor. Sci Rep. 2017, 7, 15734. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Wang, Z.; Meng, Z.; Wang, P.; San, L.; Wang, W.; Aldalbahi, A.; Li, L.; Shen, J.; Mi, X. DNA Tetrahedral Nanostructure-Based Electrochemical miRNA Biosensor for Simultaneous Detection of Multiple miRNAs in Pancreatic Carcinoma. ACS Appl Mater. Interfaces 2017, 9, 24118–24125. [Google Scholar] [CrossRef] [PubMed]
- Napoli, C.; Lai, S.; Giannetti, A.; Tombelli, S.; Baldini, F.; Barbaro, M.; Bonfiglio, A. Electronic Detection of DNA Hybridization by Coupling Organic Field-Effect Transistor-Based Sensors and Hairpin-Shaped Probes. Sensors 2018, 18, 990. [Google Scholar] [CrossRef]
- Manzano, M.; Viezzi, S.; Mazerat, S.; Marks, R.S.; Vidic, J. Rapid and label-free electrochemical DNA biosensor for detecting hepatitis A virus. Biosens. Bioelectron. 2018, 100, 89–95. [Google Scholar] [CrossRef]
- Liu, J.; Chen, X.; Wang, Q.; Xiao, M.; Zhong, D.; Sun, W.; Zhang, G.; Zhang, Z. Ultrasensitive Monolayer MoS2 Field-Effect Transistor Based DNA Sensors for Screening of Down Syndrome. Nano Lett. 2019, 19, 1437–1444. [Google Scholar] [CrossRef]
- Feng, D.; Su, J.; He, G.; Xu, Y.; Wang, C.; Zheng, M.; Qian, Q.; Mi, X. Electrochemical DNA Sensor for Sensitive BRCA1 Detection Based on DNA Tetrahedral-Structured Probe and Poly-Adenine Mediated Gold Nanoparticles. Biosensors 2020, 10, 78. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Zhang, H.; Zhu, D.; Li, J.; San, L.; Wang, Z.; Wang, C.; Wang, Y.; Wang, L.; Zuo, X.; et al. A novel ultrasensitive electrochemical DNA sensor based on double tetrahedral nanostructures. Biosens. Bioelectron. 2015, 71, 434–438. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.; Zhang, C.; Xu, Y.; Su, J.; Lu, X.; Shi, J.; Wang, L.; Landry, M.P.; Zhu, Y.; et al. A DNA tetrahedral structure-mediated ultrasensitive fluorescent microarray platform for nucleic acid test. Sens. Actuators B 2020, 321, 128538. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, D.; Xu, Y.; Yin, Z.; Dou, W.; Habiba, U.E.; Pan, C.; Zhang, Z.; Mou, H.; Deng, H.; et al. DNA-Based Functionalization of Two-Dimensional MoS2 FET Biosensor for Ultrasensitive Detection of PSA. Appl. Surf. Sci. 2021, 149169. [Google Scholar] [CrossRef]
- Splendiani, A.; Sun, L.; Zhang, Y.; Li, T.; Kim, J.; Chim, C.Y.; Galli, G.; Wang, F. Emerging photoluminescence in monolayer MoS2. Nano Lett 2010, 10, 1271–1275. [Google Scholar] [CrossRef]
- Naylor, C.H.; Kybert, N.J.; Schneier, C.; Xi, J.; Romero, G.; Saven, J.G.; Liu, R.; Johnson, A.T. Scalable Production of Molybdenum Disulfide Based Biosensors. ACS Nano 2016, 10, 6173–6179. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Xu, P.; Zhou, D.; Sun, Y.; Li, Y.C.; Nguyen, M.A.T.; Terrones, M.; Mallouk, T.E. Fast and efficient preparation of exfoliated 2H MoS2 nanosheets by sonication-assisted lithium intercalation and infrared laser-induced 1T to 2H phase reversion. Nano Lett. 2015, 15, 5956–5960. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Yan, H.; Brus, L.E.; Heinz, T.F.; Hone, J.; Ryu, S. Anomalous lattice vibrations of single- and few-layer MoS2. ACS Nano 2010, 4, 2695–2700. [Google Scholar] [CrossRef]
- Pei, H.; Li, F.; Wan, Y.; Wei, M.; Liu, H.; Su, Y.; Chen, N.; Huang, Q.; Fan, C. Designed diblock oligonucleotide for the synthesis of spatially isolated and highly hybridizable functionalization of DNA-gold nanoparticle nanoconjugates. J. Am. Chem. Soc. 2012, 134, 11876–11879. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, Z.; Lu, Y. Functional nucleic acid sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, B.; Wang, D.; Wen, Y.; Liu, G.; Dong, H.; Song, S.; Fan, C. DNA nanostructure-based universal microarray platform for high-efficiency multiplex bioanalysis in biofluids. ACS Appl Mater. Interfaces 2014, 6, 17944–17953. [Google Scholar] [CrossRef] [PubMed]
- Mikhlin, Y.; Likhatski, M.; Tomashevich, Y.; Romanchenko, A.; Erenburg, S.; Trubina, S. XAS and XPS examination of the Au–S nanostructures produced via the reduction of aqueous gold(III) by sulfide ions. J. Electron. Spectrosc. Relat. Phenom. 2010, 177, 24–29. [Google Scholar] [CrossRef]
- Neamen, D.A. Semiconductor Physics and Devices: Basic Principles; McGraw-Hill: New York, NY, USA, 2012. [Google Scholar]
- Jiang, W.; Wang, X.; Chen, Y.; Wu, G.; Ba, K.; Xuan, N.; Sun, Y.; Gong, P.; Bao, J.; Shen, H.; et al. Large-area high quality PtSe2 thin film with versatile polarity. InfoMat 2019. [Google Scholar] [CrossRef]

| Detection Technique | Probe | Sensing Range | Detection Limit | REF |
|---|---|---|---|---|
| FET–biosensor | PNA | 1 fM~100 pM | 1 fM | [25] |
| Electrochemical | PNA | 10 nM~1 μM | 10 nM | [26] |
| Electrochemical | DNA-TPs | 10 fM~10 nM | 10 fM | [27] |
| FET–biosensor | ssDNA | 10 pM~10 nM | 10 pM | [28] |
| Electrochemical | ssDNA | 10 fg/μL~100 pg/μL | 10 fg/μL | [29] |
| FET–biosensor | ssDNA | 0.1 fM~10 fM | 0.1 fM | [30] |
| Electrochemical | DNA-TPs | 1 fM~1 nM | 0.1 fM | [31] |
| TFT–biosensor | DNA-TPs | 1 fM~1 μM | 1 fM | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Jiang, W.; Feng, D.; Wang, C.; Xu, Y.; Shan, Y.; Wang, J.; Yin, Z.; Deng, H.; Mi, X.; et al. Bio-Separated and Gate-Free 2D MoS2 Biosensor Array for Ultrasensitive Detection of BRCA1. Nanomaterials 2021, 11, 545. https://doi.org/10.3390/nano11020545
Zhang Y, Jiang W, Feng D, Wang C, Xu Y, Shan Y, Wang J, Yin Z, Deng H, Mi X, et al. Bio-Separated and Gate-Free 2D MoS2 Biosensor Array for Ultrasensitive Detection of BRCA1. Nanomaterials. 2021; 11(2):545. https://doi.org/10.3390/nano11020545
Chicago/Turabian StyleZhang, Yi, Wei Jiang, Dezhi Feng, Chenguang Wang, Yi Xu, Yufeng Shan, Jianlu Wang, Ziwei Yin, Huiyong Deng, Xianqiang Mi, and et al. 2021. "Bio-Separated and Gate-Free 2D MoS2 Biosensor Array for Ultrasensitive Detection of BRCA1" Nanomaterials 11, no. 2: 545. https://doi.org/10.3390/nano11020545
APA StyleZhang, Y., Jiang, W., Feng, D., Wang, C., Xu, Y., Shan, Y., Wang, J., Yin, Z., Deng, H., Mi, X., & Dai, N. (2021). Bio-Separated and Gate-Free 2D MoS2 Biosensor Array for Ultrasensitive Detection of BRCA1. Nanomaterials, 11(2), 545. https://doi.org/10.3390/nano11020545






