Abstract
Zinc oxide nanoparticles (ZnO NPs) are applied in various applications in catalysis, biosensing, imaging, and as antibacterial agents. Here we prepared ZnO nanomaterials decorated by γ-aminobutyric acid (GABA), curcumin derivatives (CurBF2) and silver nanoparticles (CurBF2-AgNPs). The structures of all ZnO nanostructures were characterized using Fourier transform infrared (FT-IR) spectroscopy, X-ray diffraction (XRD), UV–VIS spectrophotometry, fluorescence spectrophotometry, scanning electron microscopy (SEM), transmission electron microscopy (TEM), and high-resolution transmission electron microscopy (HR-TEM). Further, their antibacterial activities against Gram-negative (Escherichia coli) and Gram-positive (Staphylococcus aureus) bacteria were investigated through analysis of minimum inhibitory concentration (MIC) method. Among the prepared nanostructures, the ZnO NPs-GABA/CurBF2-AgNPs showed excellent antibacterial activity against both Gram-positive and Gram-negative bacteria. ZnO NPs fabricated here may have potential use in future anti-bacterial compositions and coatings technologies.
1. Introduction
Zinc oxide nanoparticles (ZnO NPs) are an important example of inorganic metal oxide NPs that exhibit significant size- and shape-dependent antibacterial activity for a broad range of bacterial species [1,2,3]. ZnO shows better antimicrobial activity when particle size is decreased to the nanometer range (1–100 nm) [4] because nanoscale ZnO can interact with either the surface or the core of bacteria with consequent bactericidal effects [5]. Some studies suggest that the antibacterial function of ZnO may derive from disturbances of the cell membrane [6]. Others suggest that ZnO can, under certain conditions, stimulate the production of reactive oxygen species (ROS), which can destroy bacteria [7]. It has been shown that ZnO can be activated by UV and visible light, leading to ROS, including hydroxide ions (OH−), hydrogen peroxide (H2O2), and oxygen superoxide (O22−). The hydroxyl radicals and superoxide can bind to the cell surface, whilst hydrogen peroxide can penetrate bacterial cells and do damage from within [2].
ZnO NPs also have the high surface area, low toxicity, appropriate biocompatibility, and high chemical stability to make them of considerable interest in biomedical studies [8]. ZnO NPs have also been employed for applications such as biosensing [9], catalysis [10,11], photocatalysis [12,13], and imaging [14]. The specific properties required for each application can be obtained by various synthetic methods that control ZnO NP size and morphology. ZnO nanorods, nanospheres, nanotubes, nanowires, and nanoneedles have been obtained by tuning the chemical and physical parameters (solvent, precursors, pH, temperature, etc.) during synthesis [15,16,17]. Different reagents have been employed as capping agents, altering growth kinetics at certain facets and driving morphological diversity. These agents include mercaptoethanol (resulting in spherical particles) [18], 3-aminopropyltriethoxysilane (spherical) [19], monoethanolamine (nanosheet) [20], fatty acids (nanorods) [21], curcumin (semi-spherical core-shell) [8], mercaptoacetic acid (spherical) [22], and enzymes (spherical) [23]. In addition, the incorporation of ZnO NPs and silver nanoparticles was reported previously. Dhara, S. et al. investigated the decoration of AgNPs on ZnO nanoflowers impregnated on eggshell membranes and Eshraghi et al. reported the effect of the incorporation of ZnO and AgNPs on bacterial biofilms. These studies showed that AgNPs decorated on ZnO enhanced the latter’s antimicrobial activity [24,25].
Gamma-amino butyric acid (GABA), is a small organic molecule composed of amino and carboxylic functional groups. It is a non-protein amino acid that is distributed broadly in biological systems [26] and a major inhibitory neurotransmitter in the human central nervous system [27,28]. Due to the non-toxicity of GABA, its simplicity, and its bi-functionality, we utilize it here as a modifying surface agent in the synthesis of ZnO NPs for controlling the size and morphology of the ZnO NPs.
Curcumin can be obtained by the extraction of the rhizomes of Curcuma longa and is a yellow-orange dye with low toxicity [29]. It is an active ingredient of turmeric and has been widely evaluated for the treatment of wounds [30], Alzheimer’s disease [31], diabetes [32], inflammatory bowel syndrome [33], and rheumatoid arthritis [34]. Curcumin has carbonyl group functionality and undergoes keto-enol tautomerization. The addition of boron difluoride (BF2) to the carbonyl groups of curcumin can inhibit the keto-enol tautomerization, giving CurBF2. The latter has vastly different photophysical properties to curcumin, being highly fluorescent, and having markedly higher stability relative to curcumin [35]. Patra et al. [36], reported the functionalization of ZnO nanostructures with curcumin that changed the photophysical properties of the ZnO NPs. The NPs were then applied to sense arsenic ion contamination in water. In this work, we investigate for the first time of the functionalization of ZnO NPs with CurBF2 and its resultant properties.
The present work focuses on the preparation of ZnO nanostructures using γ-aminobutyric acid (GABA) and a curcumin derivative (CurBF2). Moreover, silver nanoparticles stabilized by CurBF2 (CurBF2-AgNPs) are synthesized and their decoration on ZnO NPs is achieved. Several characterization techniques are used to analyze the structures of these novel nanomaterials as well as their electronic properties. Further, we investigate the antibacterial efficiency of all the as-prepared ZnO nanostructures toward both Gram positive (S. aureus) and Gram negative (E. coli) bacteria by minimum inhibitory concentration (MIC) procedures. This work is a part of a continuing drive in our research group to synthesize and study new nanomaterials as well as to investigate their antibacterial abilities by natural chemical and nanoscale modifications.
2. Materials and Methods
2.1. Materials
Zinc chloride anhydrous (ZnCl2, 98%) was purchased from Loba Chemie (Mumbai, India). Sodium hydroxide (NaOH, 99%) and methanol (MeOH, 99.9%) were purchased from RCL Labscan (Bangkok, Thailand). Boron trifluoride diethyl etherate (BF3O(C2H5)2, 48%) and 1,1′-Carbonyldiimidazole (CDI, C7H6N4O, 97%) were purchased from Acros Organic (Fair Lawn, NJ, USA). γ-Amino butyric acid (GABA, C4H9NO2, >99%) was obtained from Sigma-Aldrich (Missouri, MO, USA). Synthetic grade curcumin (C21H20O6, pure > 97%) was purchased from TCI (Tokyo, Japan). Dimethyl sulfoxide (DMSO, C2H6OS, >95%) and deuterated dimethyl sulfoxide (DMSO-d6, C2D6OS, 99.8%) were purchased from Fisher Scientific (Leicestershire, UK) and Eurisotop (Derbyshire, UK), respectively. Ethanol (C2H5OH, 99.9%) and potassium carbonate (K2CO3, ≥99.0%) were obtained from Merck (Darmstadt, Germany). Silver nitrate (AgNO3, 99.9%) was purchased from POCH™ (Avantor Performance Materials, Sowińskiego, Poland). Deionized (DI) water with specific resistivity of 18.2 MΩ.cm was obtained from a RiOsTM Type I Simplicity 185 (Millipore water purification system), ELGA Labwater (Lane End, UK). Bacterial strains (Staphylococcus aureus (S. aureus) ATCC25923 and Escherichia coli (E. coli) O157:H7 were purchased from Department of Medical Sciences, Ministry of Public Health (Bangkok, Thailand).
2.2. Methods
2.2.1. Synthesis of CurBF2
CurBF2 was synthesized by the addition of boron trifluoride diethyl etheate to the enolate unit of curcumin [35]. Briefly, a solution of curcumin (1.5336 g, 4.16 mmol) in methanol 50 mL was added to a 250 mL two-necked round bottom flask equipped with a magnetic bar and a reflux condenser. Next, the solution was heated to 60 °C and then boron trifluoride diethyl etheate (0.77 mL, 6.24 mmol) was added. The color of the solution changed immediately from yellow to dark red. The reaction was stirred and refluxed overnight until a red precipitate formed. The desired product (CurBF2) was obtained as a red solid (38.4% yield) after washing with cold methanol and drying under vacuum. 1H NMR (Figure S1b) (400 MHz, d6-DMSO) δ (in ppm): 10.12 (s, 2H), 7.94 (d, J = 15.2 Hz, 2H), 7.49 (s, 2H), 7.36 (d, J = 8.4 Hz, 2H), 7.04 (d, J = 15.2 Hz, 2H), 6.90 (d, J = 8.4 Hz, 2H), 6.47 (s, 1H), 3.87 (s, 6H).
2.2.2. Synthesis of γ-Aminobutyric Acid-Capped ZnO NPs (ZnO NPs-GABA)
ZnO NPs were synthesized by a co-precipitation method [8]. Briefly, 2.0584 g of zinc chloride was dissolved in 60 mL of water: ethanol (1:1) solution in a round bottom flask equipped with magnetic stirrer. Then, a solution of GABA (0.4043 g in 1 mL in DI water) was added dropwise to the zinc chloride solution while stirring. The pH of the mixture was adjusted to 11 using 2 M NaOH aqueous solution. Subsequently, this mixture was stirred at room temperature overnight. The solution was then centrifuged at 6000 rpm for 20 min. The resulting precipitate pellet was re-dispersed and washed with a mixture of DI water and ethanol (ratio 1:1) before centrifuging and washing one more time. Finally, the precipitate was dried in an oven at 60 °C for 24 h to obtain 1.1345 g of the desired product (ZnO NPs-GABA). ZnO NPs without GABA surface modification were also synthesized using the same method but without adding GABA.
2.2.3. Functionalization of CurBF2 on ZnO NPs-GABA
ZnO NPs-GABA (0.5 g) were dispersed in 15 mL dimethyl sulfoxide in a two-neck round bottom flask. Then, 0.5 g of 1,1′-carbonyldiimidazole (CDI) was added to the DMSO/ZnO NPs-GABA suspension to activate the carboxyl group on the surface of ZnO nanoparticles. Then, after refluxing for 2 h, 0.2 g of CurBF2 was added to the solution, changing its color from colorless to blue-purple. The mixture was shaken for 24 h at room temperature. The desired product was purified by centrifugation at 20,000 rpm for 5 min and washed with DI water. The product was dried by evaporating under vacuum and kept in a desiccator for three days to obtain the product (1.0422 g).
2.2.4. The synthesis of Silver Nanoparticles (CurBF2-AgNPs) and Their Decoration on ZnO NPs-GABA
The silver nanoparticles were prepared via a modification of a previous report [37]. Briefly, a solution of 20 mM CurBF2 in DMSO (750 μL) was added to 68 mL of DI water in a 250 mL round bottom flask. The solution pH was then adjusted to 9 by K2CO3. Then, the solution was heated to 100 °C and 7.5 mL of 10 mM AgNO3 was quickly added to the solution mixture. The mixture was stirred vigorously at 100 °C for 1 h and filtered by micro filter to obtain the CurBF2-AgNPs. The concentration of CurBF2-AgNPs was estimated to be 59.13 nM based on an extinction coefficient of 1.84 × 108 M−1·cm−1 at 392 nm for 8 nm diameter citrate-silver nanoparticles [38]. The CurBF2-AgNPs decorated on ZnO NPs-GABA were prepared by distribution of ZnO NPs-GABA (0.5 g) in 4 mL of CurBF2-AgNPs solution. The solution was sonicated for 10 min to obtain the final product.
2.2.5. Bacteria Strains Preparation
Escherichia coli O157:H7 and Staphylococcus aureus ATCC25923 were purchased from the Department of Medical Sciences, Ministry of Public Health (Bangkok, Thailand). The bacteria strains were cultured in Mueller Hinton broth (MHB) at 37 °C overnight and then subcultured in 5 mL of the same medium at 37 °C in a 180-rpm shaker-incubator for 3 h to yield a mid-logarithmic growth phase culture.
2.2.6. Antibacterial Screening by Well Diffusion
Antimicrobial tests were performed by the well diffusion method. First, a single colony was grown in Mueller Hinton Broth (MHB) at 37 °C for 24 h. After inoculation for 3 h, the bacteria were diluted in the same media at an inoculum of 1 × 106 CFU/mL. The, bacteria were then swabbed onto three-dimensional MH agar plate. The agar was cut to produce wells of 6 mm diameter. ZnO NPs, Curcumin, CurBF2, ZnO NPs-GABA and ZnO NPs GABA-CurBF2, ZnO-GABA/CurBF2-AgNPs, CurBF2-AgNPs and standard drug were dropped into the wells at 30 µL and incubated at 37 °C for 24 h. Gentamicin was used as the positive control and DI water as the negative control. After 24 h of incubation, the inhibition zone was measured on the millimeter (mm) scale.
2.2.7. Minimum Inhibitory Concentrations (MICs) by Broth Microdilution Method
The MICs were determined by the broth microdilution method. Briefly, a range of concentrations of ZnO NPs, CurBF2, ZnO NPs-GABA and ZnO NPs GABA-CurBF2, ZnO-GABA/CurBF2-AgNPs, CurBF2-AgNPs and standard drug were prepared by serial dilution of two-fold. The solutions were then added to an equal volume of bacterial suspension (50 μL) in each well of a 96-well plate with the final cell concentration ranging from 106–107 CFU/mL. The plates were incubated at 37 °C for 24 h. The MICs were taken at the lowest concentration of antibacterial agents that inhibits ≥ 99% of the growth of bacteria [39,40].
2.2.8. Characterization Methods
NMR spectra were recorded using a Bruker Avance 400 MHz spectrometer (Bruker, Fällanden, Switzerland) using d6-DMSO as a solvent. The phase characterization of pristine and functionalized ZnO powder was studied by an X-ray diffractometer (Rigaku, model SmartLab, Tokyo, Japan) with the X-ray source of CuKα radiation (λ = 1.5418 nm). The analysis was carried out by a conventional 2theta scan over the range 10–80 degrees. The morphology of ZnO NPs was determined by transmission electron microscope (TEM) and high-resolution transmission electron microscope (HRTEM) using a Thermo Scientific TALOS F200X system (Eindhoven, Netherlands). Fourier transform infrared (FTIR) spectra were recorded with a Perkin Elmer system (spectrum one model, Waltham, MA, USA) using KBr pellets, over the region 400–4000 cm−1. The solution mixtures were shaken using a Vision Scientific shaker (VS-201D, Daejeon, Korea). UV-VIS diffuse reflectance spectra were record using UV–VIS diffuse reflectance spectroscopy (Shimadzu UV-3101PC, Tokyo, Japan) over the range of 200–800 nm. UV-VIS spectrum of CurBF2-AgNPs was monitored using an Agilent Technologies Cary 60 UV–VIS spectrophotometer (Penang, Malaysia) with 1.0 cm pathlength quartz cells over the range 200–800 nm. Fluorescence spectra were examined by a fluorescence spectrophotometer (Shimazu RF-5301PC, Tokyo, Japan) with slit widths at 5 nm/5 nm using 1.0 cm quartz cells. Particle sizing was determined by dynamic light scattering (DLS), and particle zeta potential by electrophoretic light scattering, using a Malvern Zetasizer Nano series (Nano ZS, Worcestershire, UK) for all samples. Determinations were repeated in triplicate and the data presented as the mean ± standard deviation (SD). All samples for DLS and zeta potential measurements were prepared by dispersing 0.1 g of ZnO NPs in 10 mL DI water and sonicating at room temperature for 10 min before measurement. The morphology and the elemental constitution were investigated by focused ion beam scanning electron microscopes (FIB-SEM, FEI Helios NanoLab G3 CX, Prague, Czech Republic). The elemental composition of AgNPs decorated on ZnO NPs-GABA was investigated by energy dispersive X-ray analysis (EDX) using transmission electron microscopy (Tecnai G2 20 S-Twin, Prague, Czech Republic).
3. Results and Discussion
3.1. ZnO Nanoparticles Functionalization by γ-Aminobutyric Acid (GABA) and CurBF2 and the Decoration of CurBF2 on ZnO NPs
Firstly, we prepared the fluoroboronated curcumin derivative CurBF2 as reported in the literature [35]. It was synthesized by the addition of curcumin to boron trifluoride diethyl etherate ((C2H5)2OBF3) in refluxing methanol at 60 °C as shown in Scheme 1. The product was obtained as a red solid and characterized by 1H-NMR spectroscopy, showing slight downfield shifts of all peaks in the 1H-NMR spectrum compared with 1H-NMR peaks of pure curcumin (Figure S1a,b).
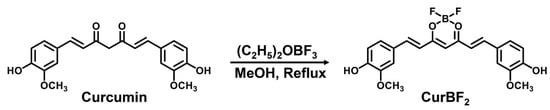
Scheme 1.
Synthetic scheme of the fluoroboronated curcumin derivative CurBF2.
Next, zinc oxide nanostructures were prepared by a co-precipitation method using ZnCl2 and NaOH as the zinc source and the precipitation agent. A solution of ZnCl2 was prepared and γ-aminobutyric acid (GABA) solution was added dropwise to it. Aqueous NaOH solution was then added to adjust the pH to 11. A proposed reaction mechanism is depicted in Equations (1)–(3). We expected that the surface modification of ZnO NPs with γ-aminobutyric acid (GABA) occurred during the ZnO NPs formation to acquire ZnO NPs-GABA.
ZnCl2 + 2NaOH → Zn(OH)2 + 2NaCl
Zn(OH)2 → ZnO + H2O
ZnO + GABA → ZnO NPs-GABA
In the second part of this study, we aimed to functionalize the surface of the ZnO NPs-GABA, on which we expected carboxylic groups (R-COOH) of GABA to be available for binding with CurBF2. To do this, the activating agent 1,1′-carbonyldiimidazole (CDI) was used to generate reactive carbonyl groups as imidazole carbamate moieties on the surface of ZnO NPs-GABA as depicted in Scheme 2. The hydroxyl groups on the aromatic part of CurBF2 can then react with reactive carbonyl, forming an ester linkage. In addition, chains of CurBF2 might form at the ZnO NPs-GABA via hydrogen bonding.
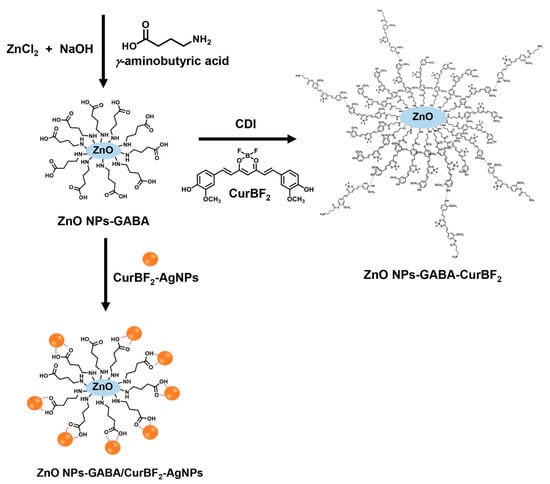
Scheme 2.
Schematic representation of synthesis of ZnO NPs-GABA, ZnO NPs-GABA-CurBF2 and CurBF2-AgNPs decorated on ZnO NPs-GABA. GABA binds to the ZnO NP surface via an amine group during synthesis, driving NP growth towards rod-like shapes and leaving carboxylic groups available at the particle surface. Following activation of those carboxylic groups, a fluoroboronated curcumin could be coupled via an ester linkage.
Lastly, we aim to improve the dispersion of ZnO NPs in water by the decoration of AgNPs on the surface of ZnO NPs (Scheme 2) as well as to improve their antibacterial activity (see antibacterial study section). CurBF2-AgNPs was firstly prepared and characterized by UV–VIS spectroscopy as shown in Figure 1a. It illustrates the surface plasmon resonance band at 419 nm. Moreover, the morphology of CurBF2-AgNPs was investigated by TEM (Figure 1b) revealing good monodispersity with a size distribution of 7.75 ± 2.26 nm (n = 70). Next, CurBF2-AgNPs decorated on ZnO NPs-GABA were prepared by the dispersion of ZnO NPs-GABA in CurBF2-AgNPs solution. The solution was then homogenized using ultrasonication for 10 min. The morphology of the ZnO NPs-GABA/CurBF2-AgNPs from SEM (Figure 2d) indicates the formation of rod-shaped nanomaterials. The TEM images in Figure 3f show that spheroidal particles of CurBF2-AgNPs settle on the surface of the ZnO NPs-GABA. The particle size of CurBF2-AgNPs decorated on the surface of ZnO NPs-GABA is 22.48 ± 4.15 (n = 32), larger than CurBF2-AgNPs alone. This could be the aggregation of CurBF2-AgNPs upon surface binding. The interaction between ZnO NPs-GABA and CurBF2-AgNPs is driven at least in part by electrostatic interactions, with the zeta potentials of the AgNPs and ZnO NPs-GABA being negative and positive, respectively. The interaction drives association of the particles at room temperature and is complete within 10 min of mixing and sonication. The presence of elemental silver in ZnO NPs-GABA/CurBF2-AgNPs was confirmed by energy-dispersive X-ray spectroscopy (EDX) as shown in Figure S2. The results indicated a major peak due to elemental silver near 3 keV [41] being 7.1% of total content. In addition, elemental mapping of ZnO NPs-GABA/CurBF2-AgNPs (Figure S3) also confirms the presence of zinc, oxygen, nitrogen, boron, fluorine, and silver.
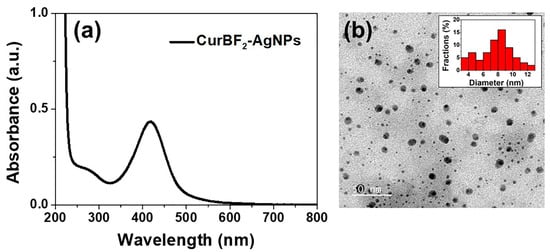
Figure 1.
(a) UV–VIS spectrum of CurBF2-AgNPs, (b) TEM image of CurBF2-AgNPs, inset the average diameter of 7.75 ± 2.26 nm (n = 70).
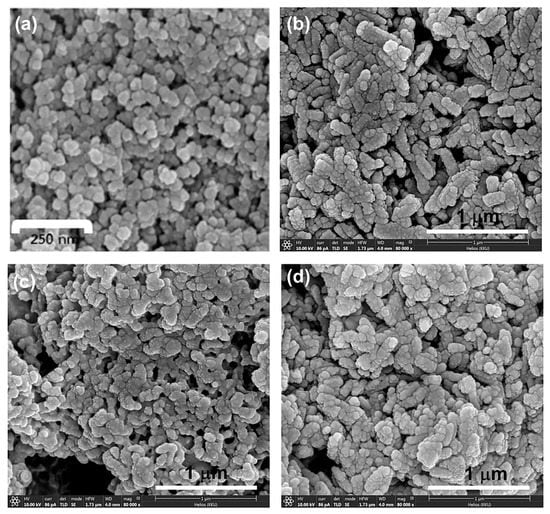
Figure 2.
(a) SEM image of ZnO NPs, (b) FIB-SEM images of ZnO NPs-GABA, (c) ZnO NPs-GABA-CurBF2, and (d) ZnO NPs-GABA/CurBF2-AgNPs.
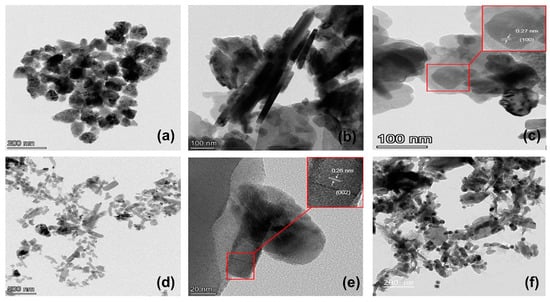
Figure 3.
(a) TEM image of ZnO NPs, (b) TEM and (c) HRTEM images of ZnO NPs-GABA, (d) TEM and (e) HRTEM images of ZnO NPs-GABA-CurBF2, and (f) TEM image of ZnO NPs-GABA/CurBF2-AgNPs.
3.2. Morphological Analysis of the ZnO Nanostructures
The SEM and TEM images (Figure 2a and Figure 3a) of the ZnO NPs confirm a spheroid morphology with a size distribution of 76.40 ± 16.31 nm (Table 1, Figure S4). The morphology of ZnO NPs-GABA, and ZnO NPs-GABA-CurBF2 (Figure 2b,c and Figure 3b,d) show a mixture of sphere and rod-like shapes. This may be due to synthesis at high pH values (pH = 11 in this experiment). Thus, ZnO can directly precipitate and prefers to grow as nanorods [42] due to the surface modification by GABA. The size distribution of the ZnO NPs-GABA spheres is 31.75 ± 10.42 nm, while the length and average diameter of the rod-shaped particles are 139.24 ± 38.72 nm and 31.36 ± 9.13 nm, respectively (Table 1 and Figure S5). Meanwhile, the dimensions of both spheres (53.46 ± 14.60 nm) and rods of ZnO NPs GABA-CurBF2 (length and average diameter: 56.54 ± 12.00 nm and 141.44 ± 47.57 nm) are larger than ZnO NPs-GABA (Table 1 and Figure S6). The results indicate that the average particles size of ZnO NPs-GABA was smaller than the size of ZnO NPs, likely because GABA inhibited the growth of ZnO NPs during the co-precipitation process. In addition, HRTEM images of ZnO NPs-GABA and ZnO NPs-GABA-CurBF2 (Figure 3c,e) show lattice fringes of d = 0.27 nm and 0.26 nm, corresponding to the (100) [43] and (002) [44] interplanar spacing of the wurtzite ZnO phase. The rod-like growth of ZnO NPs-GABA and ZnO NPs-GABA-CurBF2 is attributed to the attachment of GABA molecules that encourage the anisotropic growth of ZnO along the (001) direction, leading to nano-rod-like shapes [45]. From the above results, the size of samples determined by TEM are always larger than those determined by XRD. We note that the size extracted from XRD via the Scherrer equation correspond to the average size of the crystalline domain in the particles [46] whereas that determined by TEM is the absolute size of the ZnO NPs. This suggests that at least some particles contain more than one crystalline domain, or that aggregation of ZnO nanocrystallites occurs [47].

Table 1.
Size of ZnO NPs, ZnO NPs-GABA, and ZnO NPs-GABA-CurBF2 from the TEM technique.
3.3. Zeta Potential
Next, we evaluated the zeta potential values of the ZnO nanostructures and those decorated with GABA and CurBF2-AgNPs. The pH solutions of all samples were determined as shown in Table S1. Wurtzite-type ZnO NPs are recognized to have positive surface charges in the as-prepared state [46]. The result showed that unmodified ZnO NPs showed zeta potential of 5.3 ± 1.1 mV while ZnO NPs modified by GABA demonstrated a zeta potential value of 24.9 ± 0.3 mV. This suggests that GABA was attached to the surface of ZnO NPs. The pH of the ZnO NPs-GABA solution for zeta potential measurements was 7.47. This pH is intermediate between the pKa of the GABA carboxyl and amine groups, and thus the GABA is expected to be neutral. The fact that the zeta potential studies show that ZnO NPs-GABA is in fact positively charged is similar to the result obtained by Kim et al. [46], who functionalized ZnO NPs with L-serine and measured a zeta potential of +26.8 mV at pH 7.0. After ZnO NPs-GABA was covalently attached with CurBF2, this value shifted to −9.8 ± 0.5 mV. This may be due to the negative charge of CurBF2 influencing the charge on the ZnO NPs’ surface. Moreover, the as-prepared CurBF2-AgNPs exhibited zeta potential value of −25.5 ± 1.2 mV indicating high colloid stability [48]. Once CurBF2-AgNPs were decorated on the surface of ZnO NPs-GABA, the latter’s zeta potential value changed to −4.5 ± 1.1 mV. This confirms the successful binding of AgNPs to the ZnO NP surface.
3.4. Dynamic Light Scattering
DLS was used to determine the hydrodynamic size of all nanomaterial solutions prepared in this work and all results are collected in Table S1. The hydrodynamic sizes of the ZnO NPs (2136 ± 96 nm) were bigger than that of the ZnO NPs-GABA (926 ± 90 nm), corresponding well with the results from TEM. ZnO NPs-GABA-CurBF2 have hydrodynamic diameter of 2129 ± 86 nm, consistent with the attachment of CurBF2 on the surface of ZnO NPs-GABA. Next, we evaluated the hydrodynamic size of CurBF2-AgNPs revealing a value of 40 ± 1 nm, while the hydrodynamic size of the ZnO NPs-GABA decorated with CurBF2-AgNPs shows the value 5317 ± 579 nm. This is consistent with the imaging and Zeta potential studies suggesting that the AgNPs are indeed successfully decorated on the surface of ZnO NPs-GABA. The measured hydrodynamic sizes of all nanomaterials are larger than those measured by TEM. This may be due to the DLS experiments being undertaken in aqueous solution, where agglomeration of nanoparticles or the influence of capping molecules on the hydration sphere can be profound [37].
3.5. Crystallinity of ZnO Nanostructures
XRD analysis was used to confirm the formation of ZnO crystal phase and to investigate the influence of the surface modification on the ZnO NPs. Figure 4 shows the X-Ray diffraction (XRD) patterns of the prepared ZnO NPs, ZnO NPs-GABA, and ZnO NPs-GABA-CurBF2, and ZnO NPs-GABA/CurBF2-AgNPs. All the ZnO diffraction peaks were found at 2θ of 31.82° (h k l, 100), 34.43° (h k l, 002), 36.39° (h k l, 101), 47.65° (h k l, 102), 56.62° (h k l, 110), 62.91° (h k l, 103), 66.52° (h k l, 200), 68.13° (h k l, 112), 69.11° (h k l, 201), and 77.01° (h k l, 202). The observed peaks in the diffraction patterns of these as-prepared ZnO NPs are in good agreement with the JCPDS profile of hexagonal wurtzite zinc oxide (JCPDS #36-1451) [8,49]. The patterns of pure GABA at 2θ of 10–70° and pure CurBF2 at 2θ of 10–30°, as shown in Figure S7, are the characteristic peaks of both compounds. The additional peaks of ZnO NPs-GABA found at 16.60°, 22.04°, 24.90°, 28.14°, and 54.02° were assigned to GABA. In addition, the characteristic peaks of CurBF2 on ZnO NPs-GABA-CurBF2 at 14.76°, 15.51°, 17.32°, 18.38°, 20.60°, and 21.97° were clearly present. These results suggest firstly that the addition of GABA, CurBF2, and CurBF2-AgNPs to the reactions did not change the crystal structure of the ZnO NP cores. The sharp and intense peaks also indicate good crystallinity of ZnO NPs for all conditions. The Scherrer equation [36] can indicate the average crystallite size of zinc oxide nanoparticle samples via line broadening, estimated by the full width at half maximum (FWHM). By this model, Equation (4) was used to calculate the average crystallite sizes of our as-prepared zinc oxide nanoparticles:
where DXRD is the average crystallite size in nm, λ is the X-ray wavelength (λ = 1.5418 A), β is the FWHM of the highest intensity peak in radians, and θ is the diffraction peak half angle. The peak of the (100) plane was used to determine relative crystallite size of each prepared zinc oxide nanoparticle sample. Crystallite sizes of 19.98 nm, 25.41 nm, 26.50 nm, and 26.15 nm were estimated for ZnO NPs, ZnO NPs-GABA, ZnO NPs-GABA-CurBF2, and ZnO NPs-GABA/CurBF2-AgNPs, respectively. The crystallites of ZnO NPs-GABA being estimated to be larger than those of ZnO NPs may be due to a morphology change from spherical to rod-shaped (Figure 3b). The estimated crystalline sizes of ZnO NPs-GABA and ZnO NPs-GABA-CurBF2, ZnO NPs-GABA/CurBF2-AgNPs, and their morphology were similar.
DXRD = 0.9 λβ/cos θ
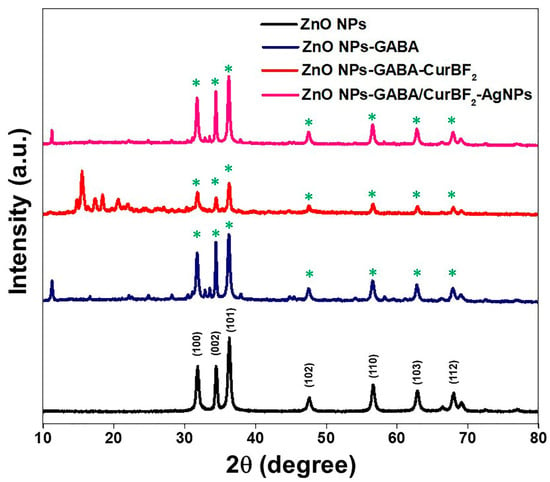
Figure 4.
XRD patterns of ZnO NPs, ZnO NPs-GABA, ZnO NPs-GABA-CurBF2 and ZnO NPs-GABA/CurBF2-AgNPs powder (the * on top of each peak in the ZnO NPs-GABA, ZnO NPs-GABA-CurBF2, and ZnO NPs-GABA/CurBF2-AgNPs diffractograms are peaks that also appear for ZnO NPs).
3.6. Surface Functional Group Analysis
The functional groups on the surface of ZnO NPs were investigated using FTIR spectroscopy. The FTIR spectra of as-prepared ZnO NPs along with free CurBF2 are shown in Figure 5. FTIR spectra of ZnO NPs clearly show a broad peak between 3000 and 3600 cm−1 attributed to O-H stretching vibration of the hydroxyl groups on the surface of ZnO [8,50]. The peak at 487 cm−1 in ZnO is shifted to 466 cm−1 in the spectrum of ZnO NPs-GABA, the latter also showing the characteristic peak of Zn-O bonds at 466 cm−1. The peak shift from 487 cm−1 to 466 cm−1 suggests the presence of altered bonding, attributed to the interaction with GABA at the NP surface. The peaks for ZnO NPs-GABA at 1623 cm−1 are assigned to stretching vibrations of carbonyl groups (C=O group) while the peak at 1505 cm−1 is characteristic of C-N bending, both indicating the presence of γ-aminobutyric acid on the surface of modified ZnO NPs. The broad peak between 3000 and 3600 cm−1 and the peak at 438 cm−1 in the spectrum of ZnO NPs-GABA-CurBF2 imply the presence of hydroxyl groups and Zn-O bonds, respectively. The peak at 1662 cm−1 may be due to carbonyl asymmetric vibration in the enol form of CurBF2 [36]. The peaks at 1493 cm−1, 1088 cm−1, 951 cm−1, 760 cm−1, and 655 cm−1 in the spectrum of ZnO NPs-GABA-CurBF2 are the characteristic peaks of C-H bending, C-O stretching, C=C bending, indicating that CurBF2 was successfully attached at the surface of ZnO NPs-GABA. The IR spectrum of ZnO NPs-GABA/CurBF2-AgNPs showed peaks characteristic of ZnO NPs-GABA. These peaks include the peaks of γ-aminobutyric acid at 1591 cm−1 and 1505 cm−1, assigned to the C=O stretch and C-N bending vibration, respectively. However, the intensity of these peaks decreased for ZnO NPs-GABA/CurBF2-AgNPs due to the binding of CurBF2-AgNPs to the ZnO NPs-GABA surface [51].
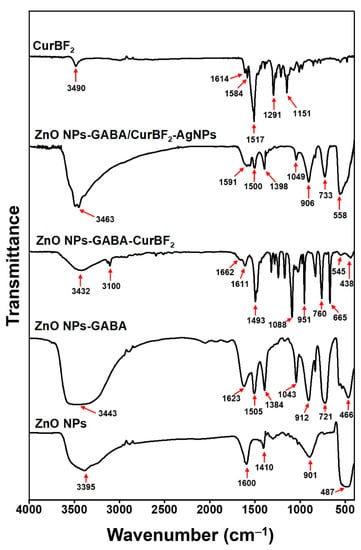
Figure 5.
FTIR spectra of CurBF2, ZnO NPs-GABA/CurBF2-AgNPs, ZnO NPs-GABA-CurBF2, ZnO NPs-GABA, and ZnO NPs.
3.7. The Photophysical Properties of ZnO Nanostructures
3.7.1. UV–VIS Diffuse Reflectance Spectra
Figure 6a depicts the UV–VIS diffuse reflectance spectra of powders of the as-prepared ZnO NPs, ZnO NPs-GABA, ZnO NPs-GABA-CurBF2, ZnO NPs-GABA/CurBF2-AgNPs, and free GABA and CurBF2. The spectra of all nanostructures show a similar broad absorption spectrum with a maximum at about 340 nm and an onset at about 400 nm, which are characteristic ZnO as a wide band gap semiconductor. For ZnO NPs-GABA-CurBF2, other strong absorption maxima were observed at around 220 nm, 480 nm, and 610 nm with absorption peak onsets at about 250 nm, 570 nm, and 680 nm, indicating the presence of both GABA and CurBF2 on the surface of the ZnO NPs. The position of GABA and CurBF2 peaks in ZnO NPs-GABA-CurBF2 is, however, redshifted compared to free GABA and CurBF2. This shift may be due to the interaction of GABA and CurBF2 with zinc ions that reduces the band gap between π–π* electronic transition of CurBF2 [36]. In addition, the absorption band of ZnO NPs-GABA/CurBF2-AgNPs occurs at 426 nm with an onset of 500 nm, further evidence of successful decoration of AgNPs on the surface of ZnO NPs-GABA.
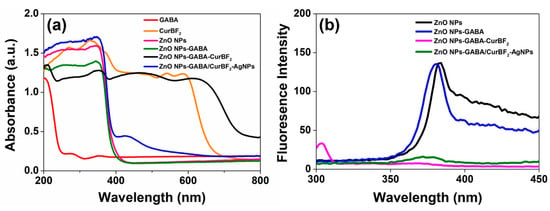
Figure 6.
(a) UV–VIS diffuse reflectance spectra of free GABA and CurBF2, and powders of ZnO NPs, ZnO NPs-GABA, ZnO NPs-GABA-CurBF2, and ZnO NPs-GABA/CurBF2-AgNPs at room temperature. (b) Fluorescence spectra of ZnO NPs, ZnO NPs-GABA, ZnO NPs-GABA-CurBF2, and ZnO NPs-GABA/CurBF2-AgNPs with slit widths at 5 nm/5 nm excited at 260 nm.
3.7.2. Fluorescence Spectra
The photoluminescence (PL) spectra of ZnO NPs, ZnO NPs-GABA, ZnO NPs-GABA-CurBF2 and ZnO NPs-GABA/CurBF2-AgNPs with excitation at 260 nm were also investigated in order to determine the influence of capping molecules/NPs. The results shown in Figure 6b indicate that only ZnO NPs and ZnO NPs-GABA exhibit an emission band at about 380 nm. This blue-violet emission band was also reported by Sengupta et al. and Mahmoud et al. [52,53] and is attributed to recombination of photo-generated excitons. After the modification of ZnO NPs-GABA with CurBF2 and combining with CurBF2-AgNPs, those emission bands were quenched, likely by charge transfer to the electron-deficient CurBF2. Since plenty of electron-rich hydroxy groups are present on the native surface of ZnO NPs, the CurBF2 may possibly interact with these electron-rich hydroxyl groups, becoming efficient surface traps for electrons [54].
3.8. Antibacterial Activity
3.8.1. Inhibition Zone Method
Antibacterial activities by the well-diffusion method for ZnO NPs, ZnO NPs-GABA and ZnO NPs-GABA-CurBF2 are shown in Figure S8. The inhibition zones of all samples were absent due to the poor solubility and poor dispersion of ZnO NPs in water leading to impermeability. However, for ZnO NPs-GABA/CurBF2-AgNPs enhanced their dispersivity in aqueous phase allowed clear inhibition zones and anti-bacterial activity to be observed (see Figure 7). ZnO NPs-GABA/CurBF2-AgNPs exhibit strong antimicrobial activity against both E. coli and S. aureus bacteria, forming zones of inhibition of diameters 10.17 ± 0.24 and 13.50 ± 0.41 mm, respectively, as shown in Table 2. In comparison, CurBF2-AgNPs showed antibacterial activity against only S. aureus with inhibition zone of 7.33 ± 0.29 mm (Table 2). These results demonstrate that the combination of ZnO NPs with CurBF2-AgNPs gives a synergistic antibacterial effect [25] as compared to AgNPs or ZnO NPs alone.
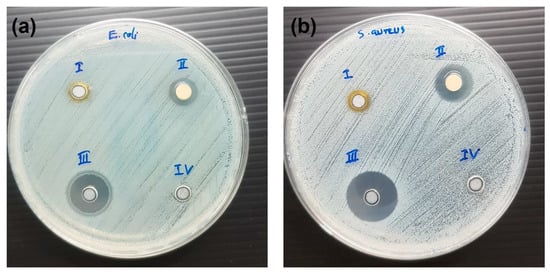
Figure 7.
Antimicrobial activity of (i) CurBF2-AgNPs (ii) ZnO-GABA/CurBF2-AgNPs, (iii) gentamicin, (iv) DI water against (a) E. coli and (b) S. aureus.

Table 2.
Zone of inhibition in millimeters of ZnO NPs-GABA/CurBF2-AgNPs, CurBF2-AgNPs, and standard drug against Gram-negative and Gram-positive bacteria.
3.8.2. Minimum Inhibitory Concentration (MIC) Determination
We then have investigated the MIC values of ZnO-NPs, ZnO NPs-GABA, ZnO NPs-GABA-CurBF2, and ZnO NPs-GABA/CurBF2-AgNPs for their antibacterial activities against Gram positive (S. aureus) and Gram negative (E. coli) bacteria as shown in Table 3. The MIC of ZnO NPs against S. aureus was found to be 100 μg/mL whilst an inhibition effect of these NPs towards E. coli was not found. Also, a bacterial inhibition of ZnO NPs-GABA-CurBF₂ against E. coli was not found, whilst MIC of ZnO NPs-GABA-CurBF₂ against S. aureus were found to be 200 μg/mL. It is clear that ZnO NPs and ZnO NPs-GABA-CurBF₂ exhibited a potent inhibition effect against Gram-positive bacteria. For ZnO NPs-GABA, MIC values against S. aureus and E. coli were observed to be 100 μg/mL and 200 μg/mL, respectively. Therefore, the synthesized ZnO NPs-GABA can be an effective nano-agent against both Gram-negative and Gram-positive bacterial cells. Conversely, treating both Grams of bacteria with GABA and CurBF2 showed no antibacterial effect (Table S2). Interestingly, the antimicrobial activity of ZnO NPs-GABA/CurBF2-AgNPs is enhanced against both S. aureus and E. coli with lower MIC values than the other ZnO NPs which are ≤6.25 µg/mL and 12.5 µg/mL, respectively. Therefore, we can confirm that AgNPs decorated ZnO NPs-GABA nanocomposite can improve the antimicrobial activity of ZnO NPs with synergistic effect.

Table 3.
Minimum inhibitory concentration (MIC) of each agent against Gram-positive and Gram-negative bacteria.
These antibacterial activity studies indicate that the bactericidal effects of all the tested agents were much more noticeable against S. aureus than against E. coli. This difference is likely due to the different structure of the cell membrane, together with the cell wall composition of Gram-positive and Gram-negative bacteria. It could also depend on the differences of cell envelope components. The cell wall of Gram-negative bacteria (E. coli) is wavy and double-layered, with the presence of an outer membrane with high amounts of lipopolysaccharides. On the other hand, Gram-positive bacteria (S. aureus) feature a smooth and single-layered cell wall and the absence of an outer membrane, as well as virtually no lipopolysaccharides. These differences may account for the greater susceptibility of Gram-positive bacteria to the antibacterial agents tested here [55]. Moreover, the sizes of ZnO NPs also play a significant role in antibacterial activities. ZnO NPs with smaller sizes can easily penetrate bacterial cell membranes, thus increasing their antibacterial activity [56,57]. In this work, ZnO NPs-GABA demonstrated the smallest size with the highest specific surface areas compared to other as-prepared ZnO NPs, and also showed the highest efficiency of antibacterial activity. Furthermore, ZnO NPs-GABA/CurBF2-AgNPs illustrated an excellent antibacterial efficiency due to a synergistic effect and better dispersion in aqueous phases.
4. Conclusions
Zinc oxide nanoparticles (ZnO NPs) were prepared by a simple method with a surface modification with gamma-amino butyric acid (GABA) and were then covalently attached with CurBF2. The nanostructures of ZnO NPs were observed to be spherical and rod-like shapes with a hexagonal crystal structure. Modifying ZnO NPs-GABA surface with CurBF2 (ZnO NPs-GABA-CurBF2) resulted in enhanced near-IR absorption. Finally, as-prepared ZnO NPs showed antibacterial activities against only Gram-positive (S. aureus) for ZnO NPs and ZnO NPs-GABA-CurBF2. Meanwhile, ZnO NPs-GABA showed good efficiency of antibacterial activity again both Gram-positive (S. aureus) and Gram-negative (E. coli) bacteria. In addition, the incorporation of CurBF2-AgNPs on the surface of ZnO NPs-GABA showed excellent antibacterial activity with synergistic effect. Thus, these nanomaterials may further have potential for biomedical applications, especially given the facile and inexpensive fabrication method developed here.
Supplementary Materials
The following are available online at https://www.mdpi.com/2079-4991/11/2/442/s1, Figure S1. 1H-NMR spectra of (a) Curcumin and (b) CurBF2; Figure S2. EDX spectrum of ZnO NPs-GABA/CurBF2-AgNPs; Figure S3. (a) EDS image of ZnO NPs-GABA/CurBF2-AgNPs and element mapping of (b) Zn, (c) O, (d) N, (e) B, (f) F, and (g) Ag; Figure S4. Histogram showing the diameter distribution of the sphere shape of ZnO NPs with the average diameter of 76.40 ± 16.31 nm (n = 50); Figure S5. (a) A histogram showing diameter distribution of the sphere shape of ZnO NPs-GABA with the average diameter of 31.75 ± 10.42 nm (n = 50); (b) a histogram showing the distribution of the rod shape of ZnO NPs-GABA with the average diameter of 31.36 ± 9.13 nm (n = 50); and (c) a histogram showing the distribution of the rod shape of ZnO-NPs-GABA with the average length of 139.24 ± 38.72 nm (n = 50); Figure S6. (a) A histogram showing thre diameter distribution of the sphere shape of ZnO NPs-GABA-CurBF2 with the average diameter of 53.46 ± 14.60 nm (n = 50); (b) a histogram showing the distribution of the rod shape of ZnO NPs-GABA-CurBF2 with the average diameter of 56.54 ± 12.00 nm (n = 50); and (c) a histogram showing the distribution of the rod shape of ZnO-NPs-GABA-Cur-BF2 with the average length of 141.44 ± 47.57 nm (n = 50); Figure S7. XRD patterns of ZnO NPs, ZnO NPs-GABA, ZnO NPs-GABA-CurBF2, free γ-aminobutyric acid (GABA), and free CurBF2 powder (the * on top of each peak in the ZnO NPs-GABA and ZnO NPs-GABA-CurBF2 diffractograms are peaks that also appear for ZnO NPs).; Figure S8. Antibacterial activity by well diffusion method: (a) S. aureus, position (i) gentamicin, (ii) ZnO NPs-GABA, (iii) ZnO NPs-GABA-CurBF2, (iv) DI water. (b) S. aureus, position (i) gentamicin, (ii) ZnO NPs, (iii) curcumin, (iv) CurBF2, (v) DI water. (c) E. coli, position (i) gentamicin, (ii) DI water, (iii) ZnO NPs-GABA, (iv) ZnO NPs-GABA-CurBF2. (d) E. coli, position (i) gentamicin, (ii) DI water, (iii) ZnO NPs, (iv) curcumin, (v) CurBF2; Table S1. pH solution, zeta potential and their hydrodynamic sizes of as-prepared nanomaterials; Table S2. Minimum inhibitory concentration (MIC) of each agent against Gram-positive and Gram-negative bacteria.
Author Contributions
Conceptualization: S.K.; methodology: C.T., W.B.; investigation: C.T., K.P. C.M., O.S., N.K.; formal analysis: C.T., K.P., C.M., O.S., N.K., and S.K.; writing—original draft preparation: C.T. and S.K.; supervision and resources: R.P., N.K., and S.K.; visualization: S.K.; project administration: S.K.; writing—reviewing and editing, and funding acquisition: S.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Research and Graduate Studies, Khon Kaen University, Faculty of Science, Khon Kaen University, the Science achievement scholarship of Thailand (SAST), and Development and Promotion of Science and Technology Talents Project (DPST). The APC was funded by Khon Kaen University.
Data Availability Statement
The research data supporting this publication are given within this paper and as supplementary material.
Acknowledgments
This research was supported by Research and Graduate Studies, Khon Kaen University. We thank the financial and laboratories support from Faculty of Science, Khon Kaen University. C.T. was supported by the Science achievement scholarship of Thailand (SAST). W.B. was supported by the Development and Promotion of Science and Technology Talents Project (DPST). O.S. was supported by The Royal Golden Jubilee Ph.D. Programme Scholarships (RGJ-Ph.D.). We thank Anuchit Ruangvittayanon for TEM and HRTEM measurements at Synchrotron Light Research Institute (SLRI), Thailand. We thank James Andell Hutchison for the fruitful discussion about the formation and surface modification of ZnO nanoparticles using organic molecules.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mirhosseini, M.; Firouzabadi, F.B. Antibacterial activity of zinc oxide nanoparticle suspensions on food-borne pathogens. Int. J. Dairy Technol. 2013, 66, 291–295. [Google Scholar] [CrossRef]
- Padmavathy, N.; Vijayaraghavan, R. Enhanced bioactivity of ZnO nanoparticles-an antimicrobial study. Sci. Technol. Adv. Mater. 2008, 9, 035004. [Google Scholar] [CrossRef] [PubMed]
- Raghupathi, K.R.; Koodali, R.T.; Manna, A.C. Size-Dependent Bacterial Growth Inhibition and Mechanism of Antibacterial Activity of Zinc Oxide Nanoparticles. Langmuir 2011, 27, 4020–4028. [Google Scholar] [CrossRef]
- Buzea, C.; Pacheco, I.I.; Robbie, K. Nanomaterials and nanoparticles: Sources and toxicity. Biointerphases 2007, 2, MR17–MR71. [Google Scholar] [CrossRef]
- Seil, J.T.; Webster, T.J. Antimicrobial applications of nanotechnology: Methods and literature. Int. J. Nanomed. 2012, 7, 2767–2781. [Google Scholar] [CrossRef]
- Xie, Y.; He, Y.; Irwin, P.L.; Jin, T.; Shi, X. Antibacterial activity and mechanism of action of zinc oxide nanoparticles against Campylobacter jejuni. Appl. Environ. Microbiol. 2011, 77, 2325–2331. [Google Scholar] [CrossRef]
- Sawai, J. Quantitative evaluation of antibacterial activities of metallic oxide powders (ZnO, MgO and CaO) by conductimetric assay. J. Microbiol. Methods 2003, 54, 177–182. [Google Scholar] [CrossRef]
- Ghaffari, S.-B.; Sarrafzadeh, M.-H.; Fakhroueian, Z.; Shahriari, S.; Khorramizadeh, M.R. Functionalization of ZnO nanoparticles by 3-mercaptopropionic acid for aqueous curcumin delivery: Synthesis, characterization, and anticancer assessment. Mater. Sci. Eng. C 2017, 79, 465–472. [Google Scholar] [CrossRef]
- Xu, C.; Yang, C.; Gu, B.; Fang, S. Nanostructured ZnO for biosensing applications. Chin. Sci. Bull. 2013, 58, 2563–2566. [Google Scholar] [CrossRef]
- Safaei-Ghomi, J.; Ghasemzadeh, M.A. Zinc oxide nanoparticles: A highly efficient and readily recyclable catalyst for the synthesis of xanthenes. Chin. Chem. Lett. 2012, 23, 1225–1229. [Google Scholar] [CrossRef]
- Nageswara Rao, T.; Apparao, K.; Murthy, S.N.V.S.; Manohara Naidu, T. Applications of Zinc oxide Nanoparticles as Catalyst in Dissipation Kinetics of S-Metolachlor Herbicide in Different pH waters UnderDirect sun light. Mater. Today Proc. 2016, 3, 3799–3804. [Google Scholar] [CrossRef]
- Kakarndee, S.; Nanan, S. SDS capped and PVA capped ZnO nanostructures with high photocatalytic performance toward photodegradation of reactive red (RR141) azo dye. J. Environ. Chem. Eng. 2018, 6, 74–94. [Google Scholar] [CrossRef]
- Chankhanittha, T.; Watcharakitti, J.; Nanan, S. PVP-assisted synthesis of rod-like ZnO photocatalyst for photodegradation of reactive red (RR141) and Congo red (CR) azo dyes. J. Mater. Sci. Mater. Electron. 2019, 30, 17804–17819. [Google Scholar] [CrossRef]
- Zhang, Z.-Y.; Xiong, H.-M. Photoluminescent ZnO nanoparticles and their biological applications. Materials 2015, 8, 3101–3127. [Google Scholar] [CrossRef]
- Yahya, N.; Daud, H.; Tajuddin, N.; Daud, H.; Shafie, A.; Puspitasari, P. Application of ZnO Nanoparticles EM Wave Detector Prepared by Sol-Gel and Self-Combustion Techniques. J. Nano Res. J. Nano Res. 2010, 11, 25–34. [Google Scholar] [CrossRef]
- Ali, A.; Phull, A.-R.; Zia, M. Elemental zinc to zinc nanoparticles: Is ZnO NPs crucial for life? Synthesis, toxicological, and environmental concerns. Nanotechnol. Rev. 2018, 7, 413–441. [Google Scholar] [CrossRef]
- Wojnarowicz, J.; Chudoba, T.; Lojkowski, W. A Review of Microwave Synthesis of Zinc Oxide Nanomaterials: Reactants, Process Parameters and Morphologies. Nanomaterials 2020, 10, 1086. [Google Scholar] [CrossRef] [PubMed]
- Chimanpure, J.; Ashtaputre, S.; Marathe, S.; Hebalkar, N.; Kharrazi, S.; Pasricha, R.; Kulkarni, S.K. Synthesis and Characterization of Mercaptoethanol Capped Zinc Oxide Nanoparticles Capped with Organic Molecules. Synth. React. Inorg. Met. Org. Nano-Met. Chem. 2006, 36, 65–69. [Google Scholar] [CrossRef]
- Saravanan, P.; Jayamoorthy, K.; Ananda Kumar, S. Switch-On fluorescence and photo-induced electron transfer of 3-aminopropyltriethoxysilane to ZnO: Dual applications in sensors and antibacterial activity. Sens. Actuators B Chem. 2015, 221, 784–791. [Google Scholar] [CrossRef]
- Liao, Y.; Hu, Z.; Gu, Q.; Xue, C. Amine-Functionalized ZnO Nanosheets for Efficient CO2 Capture and Photoreduction. Molecules 2015, 20, 18847–18855. [Google Scholar] [CrossRef]
- Fakhroueian, Z.; Harsini, F.; Chalabian, F.; Katouzian, F.; Shafiekhani, A.; Esmaeilzadeh, P. Influence of Modified ZnO Quantum Dots and Nanostructures as New Antibacterials. Adv. Nanoparticles 2013, 2, 247–258. [Google Scholar] [CrossRef][Green Version]
- Zhuang, J.; Liu, M.; Liu, H. MAA-modified and luminescence properties of ZnO quantum dots. Sci. China Ser. B-Chem. 2009, 52, 2125. [Google Scholar] [CrossRef]
- Kim, K.-E.; Kim, T.G.; Sung, Y.-M. Enzyme-conjugated ZnO nanocrystals for collisional quenching-based glucose sensing. CrystEngComm 2012, 14, 2859–2865. [Google Scholar] [CrossRef]
- Ray, P.G.; Biswas, S.; Roy, T.; Ghosh, S.; Majumder, D.; Basak, P.; Roy, S.; Dhara, S. Sonication Assisted Hierarchical Decoration of Ag-NP on Zinc Oxide Nanoflower Impregnated Eggshell Membrane: Evaluation of Antibacterial Activity and in Vitro Cytocompatibility. ACS Sustain. Chem. Eng. 2019, 7, 13717–13733. [Google Scholar] [CrossRef]
- Shakerimoghaddam, A.; Razavi, D.; Rahvar, F.; Khurshid, M.; Ostadkelayeh, S.M.; Esmaeili, S.-A.; Khaledi, A.; Eshraghi, M. Evaluate the Effect of Zinc Oxide and Silver Nanoparticles on Biofilm and icaA Gene Expression in Methicillin-Resistant Staphylococcus aureus Isolated From Burn Wound Infection. J. Burn Care Res. 2020, 41, 1253–1259. [Google Scholar] [CrossRef]
- Kinnersley, A.M.; Turano, F.J. Gamma Aminobutyric Acid (GABA) and Plant Responses to Stress. Crit. Rev. Plant Sci. 2000, 19, 479–509. [Google Scholar] [CrossRef]
- Wu, C.; Sun, D. GABA receptors in brain development, function, and injury. Metab. Brain Dis. 2015, 30, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.; Ryan, P.M.; Wiley, N.; Carafa, I.; Sherwin, E.; Moloney, G.; Franciosi, E.; Mandal, R.; Wishart, D.S.; Tuohy, K.; et al. Gamma-aminobutyric acid-producing lactobacilli positively affect metabolism and depressive-like behaviour in a mouse model of metabolic syndrome. Sci. Rep. 2019, 9, 16323. [Google Scholar] [CrossRef]
- Abdeldaiem, M.H. Use of Yellow Pigment Extracted from Turmeric (Curcuma Longa) Rhizomes Powder as Natural Food Preservative. Am. J. Food Sci. Technol. 2014, 2, 36–47. [Google Scholar] [CrossRef]
- Akbik, D.; Ghadiri, M.; Chrzanowski, W.; Rohanizadeh, R. Curcumin as a wound healing agent. Life Sci. 2014, 116, 1–7. [Google Scholar] [CrossRef]
- Mishra, S.; Palanivelu, K. The effect of curcumin (turmeric) on Alzheimer’s disease: An overview. Ann. Indian Acad. Neurol. 2008, 11, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Hodaei, H.; Adibian, M.; Nikpayam, O.; Hedayati, M.; Sohrab, G. The effect of curcumin supplementation on anthropometric indices, insulin resistance and oxidative stress in patients with type 2 diabetes: A randomized, double-blind clinical trial. Diabetol. Metab. Syndr. 2019, 11, 41. [Google Scholar] [CrossRef]
- Burge, K.; Gunasekaran, A.; Eckert, J.; Chaaban, H. Curcumin and Intestinal Inflammatory Diseases: Molecular Mechanisms of Protection. Int. J. Mol. Sci. 2019, 20, 1912. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Zhou, D.; Xu, L.; Song, X. Curcumin alleviates rheumatoid arthritis-induced inflammation and synovial hyperplasia by targeting mTOR pathway in rats. Drug Des. Dev. Ther. 2018, 12, 4095–4105. [Google Scholar] [CrossRef] [PubMed]
- Chaicham, A.; Kulchat, S.; Tumcharern, G.; Tuntulani, T.; Tomapatanaget, B. Synthesis, photophysical properties, and cyanide detection in aqueous solution of BF2-curcumin dyes. Tetrahedron 2010, 66, 6217–6223. [Google Scholar] [CrossRef]
- Moussawi, R.N.; Patra, D. Modification of nanostructured ZnO surfaces with curcumin: Fluorescence-based sensing for arsenic and improving arsenic removal by ZnO. RSC Adv. 2016, 6, 17256–17268. [Google Scholar] [CrossRef]
- Talodthaisong, C.; Boonta, W.; Thammawithan, S.; Patramanon, R.; Kamonsutthipaijit, N.; Hutchison, J.A.; Kulchat, S. Composite guar gum-silver nanoparticle hydrogels as self-healing, injectable, and antibacterial biomaterials. Mater. Today Commun. 2020, 24, 100992. [Google Scholar] [CrossRef]
- Paramelle, D.; Sadovoy, A.; Gorelik, S.; Free, P.; Hobley, J.; Fernig, D.G. A rapid method to estimate the concentration of citrate capped silver nanoparticles from UV-visible light spectra. Analyst 2014, 139, 4855–4861. [Google Scholar] [CrossRef]
- Siritongsuk, P.; Hongsing, N.; Thammawithan, S.; Daduang, S.; Klaynongsruang, S.; Tuanyok, A.; Patramanon, R. Two-Phase Bactericidal Mechanism of Silver Nanoparticles against Burkholderia pseudomallei. PLoS ONE 2016, 11, e0168098. [Google Scholar] [CrossRef]
- Kanthawong, S.; Bolscher, J.G.M.; Veerman, E.C.I.; van Marle, J.; de Soet, H.J.J.; Nazmi, K.; Wongratanacheewin, S.; Taweechaisupapong, S. Antimicrobial and antibiofilm activity of LL-37 and its truncated variants against Burkholderia pseudomallei. Int. J. Antimicrob. Agents 2012, 39, 39–44. [Google Scholar] [CrossRef]
- Saenchoopa, A.; Boonta, W.; Talodthaisong, C.; Srichaiyapol, O.; Patramanon, R.; Kulchat, S. Colorimetric detection of Hg(II) by γ-aminobutyric acid-silver nanoparticles in water and the assessment of antibacterial activities. Spectrochim. Acta Part A: Mol. Biomol. Spectrosc. 2021, 251, 119433. [Google Scholar] [CrossRef]
- Ghaffari, S.B.; Moghaddam, J. Precipitation of various shapes of nanosized zinc oxide from zinc chloride solutions by neutralization with MgO and Ca(OH)2 as non-transparent basic agents. J. Iran. Chem. Soc. 2012, 9, 687–692. [Google Scholar] [CrossRef]
- Liu, Y.; Kou, Q.; Xing, S.; Mao, C.; Naveen, K.; Han, Q.; Song, J.; Liu, H.; Liu, Y.; Yan, Y.; et al. Structural, Magnetic and Optical Properties in V Doped ZnO Nanoparticles by Sol–Gel Method. Nanosci. Nanotechnol. Lett. 2015, 7, 1–6. [Google Scholar] [CrossRef]
- Adhikari, S.; Banerjee, A.; KrishnaRao Eswar, N.; Sarkar, D.; Madras, G. Photocatalytic inactivation of E. Coli by ZnO–Ag nanoparticles under solar radiation. RSC Adv. 2015, 5, 51067–51077. [Google Scholar] [CrossRef]
- Bhushan, B.; Murty, B.S.; Mondal, K. A new approach for synthesis of ZnO nanorod flowerets and subsequent pure free-standing ZnO nanorods. Adv. Powder Technol. 2019, 30, 30–41. [Google Scholar] [CrossRef]
- Kim, K.-M.; Choi, M.-H.; Lee, J.-K.; Jeong, J.; Kim, Y.-R.; Kim, M.-K.; Paek, S.-M.; Oh, J.-M. Physicochemical properties of surface charge-modified ZnO nanoparticles with different particle sizes. Int. J. Nanomed. 2014, 9 (Suppl. 2), 41–56. [Google Scholar] [CrossRef]
- Becker, J.; Raghupathi, K.R.; St Pierre, J.; Zhao, D.; Koodali, R.T. Tuning of the Crystallite and Particle Sizes of ZnO Nanocrystalline Materials in Solvothermal Synthesis and Their Photocatalytic Activity for Dye Degradation. J. Phys. Chem. C 2011, 115, 13844–13850. [Google Scholar] [CrossRef]
- Mukherjee, S.; Chowdhury, D.; Kotcherlakota, R.; Patra, S.; Vinothkumar, B.; Bhadra, M.P.; Sreedhar, B.; Patra, C.R. Potential theranostics application of bio-synthesized silver nanoparticles (4-in-1 system). Theranostics 2014, 4, 316–335. [Google Scholar] [CrossRef]
- Mishra, D.S.; Srivastava, R.; Prakash, S. Photoluminescence and Photoconductivity Studies of ZnO Nanoparticles Prepared by Solid State Reaction Method. J. Mater. Sci. Mater. Electron. 2012, 24, 125–134. [Google Scholar] [CrossRef]
- Šimšíková, M.; Antalík, M.; Kaňuchová, M.; Škvarla, J. Anionic 11-mercaptoundecanoic acid capped ZnO nanoparticles. Appl. Surf. Sci. 2013, 282, 342–347. [Google Scholar] [CrossRef]
- Nagaraju, G.; Udayabhanu, S.; Prashanth, S.A.; Shastri, M.; Yathish, K.V.; Anupama, C.; Rangappa, D. Electrochemical heavy metal detection, photocatalytic, photoluminescence, biodiesel production and antibacterial activities of Ag–ZnO nanomaterial. Mater. Res. Bull. 2017, 94, 54–63. [Google Scholar] [CrossRef]
- Saikia, L.; Bhuyan, D.; Saikia, M.; Malakar, B.; Dutta, D.K.; Sengupta, P. Photocatalytic performance of ZnO nanomaterials for self sensitized degradation of malachite green dye under solar light. Appl. Catal. A Gen. 2015, 490, 42–49. [Google Scholar] [CrossRef]
- Musa, I.; Qamhieh, N.; Mahmoud, S.T. Synthesis and length dependent photoluminescence property of zinc oxide nanorods. Results Phys. 2017, 7, 3552–3556. [Google Scholar] [CrossRef]
- Venkatesan, G.; Vijayaraghavan, R.; Chakravarthula, S.N.; Sathiyan, G. Fluorescent zinc oxide nanoparticles of Boswellia ovalifoliolata for selective detection of picric acid. Front. Res. Today 2019, 2, 2002. [Google Scholar] [CrossRef]
- Ferreyra Maillard, A.P.V.; Gonçalves, S.; Santos, N.C.; López de Mishima, B.A.; Dalmasso, P.R.; Hollmann, A. Studies on interaction of green silver nanoparticles with whole bacteria by surface characterization techniques. Biochim. Biophys. Acta Biomembr. 2019, 1861, 1086–1092. [Google Scholar] [CrossRef]
- Yamamoto, O. Influence of particle size on the antibacterial activity of zinc oxide. Int. J. Inorg. Mater. 2001, 3, 643–646. [Google Scholar] [CrossRef]
- Sawai, J.; Kawada, E.; Kanou, F.; Igarashi, H.; Hashimoto, A.; Kokugan, T.; Shimizu, M. Detection of active oxygen generated from ceramic powders having antibacterial activity. J. Chem. Eng. Jpn. 1996, 29, 627–633. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).