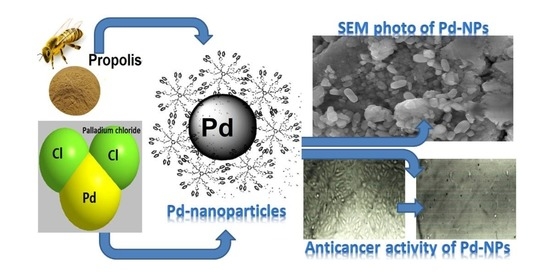
Characterization, Antimicrobial and Anticancer Properties of Palladium Nanoparticles Biosynthesized Optimally Using Saudi Propolis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Propolis Extract Preparation
2.3. Optimal Biosynthesis of Palladium NPs
2.3.1. Ratio and Concentration Effects
2.3.2. Effect of pH
2.3.3. Temperature Effect
2.3.4. Effect of Reaction Time
2.4. Characterization Techniques
2.5. Antimicrobial Experiment Design
2.6. Cytotoxicity Assay on MCF-7 Cells
2.6.1. Cell Culture
2.6.2. MTT Protocol
2.6.3. Statistical Analysis
3. Results
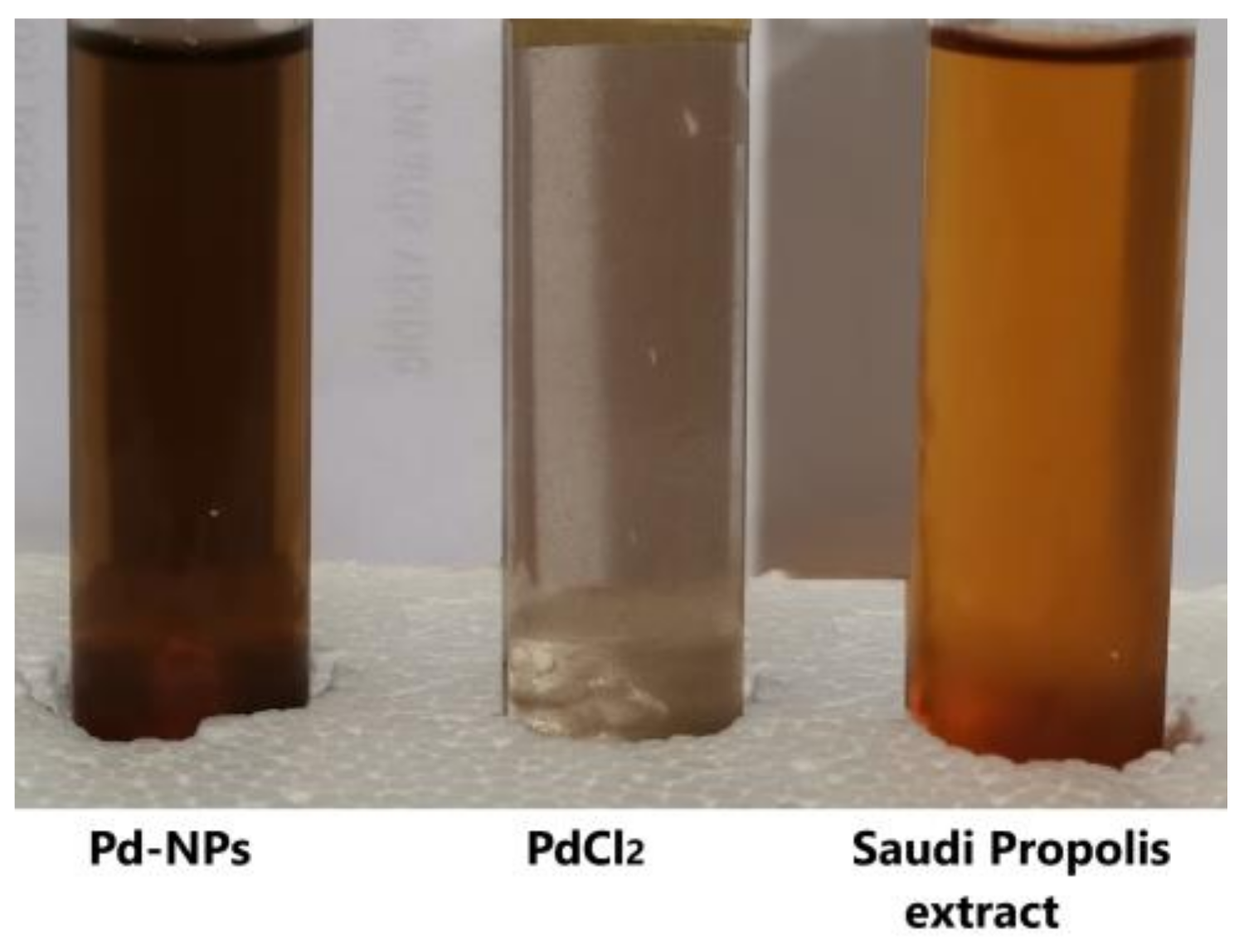
3.1. UV Spectrometer Results
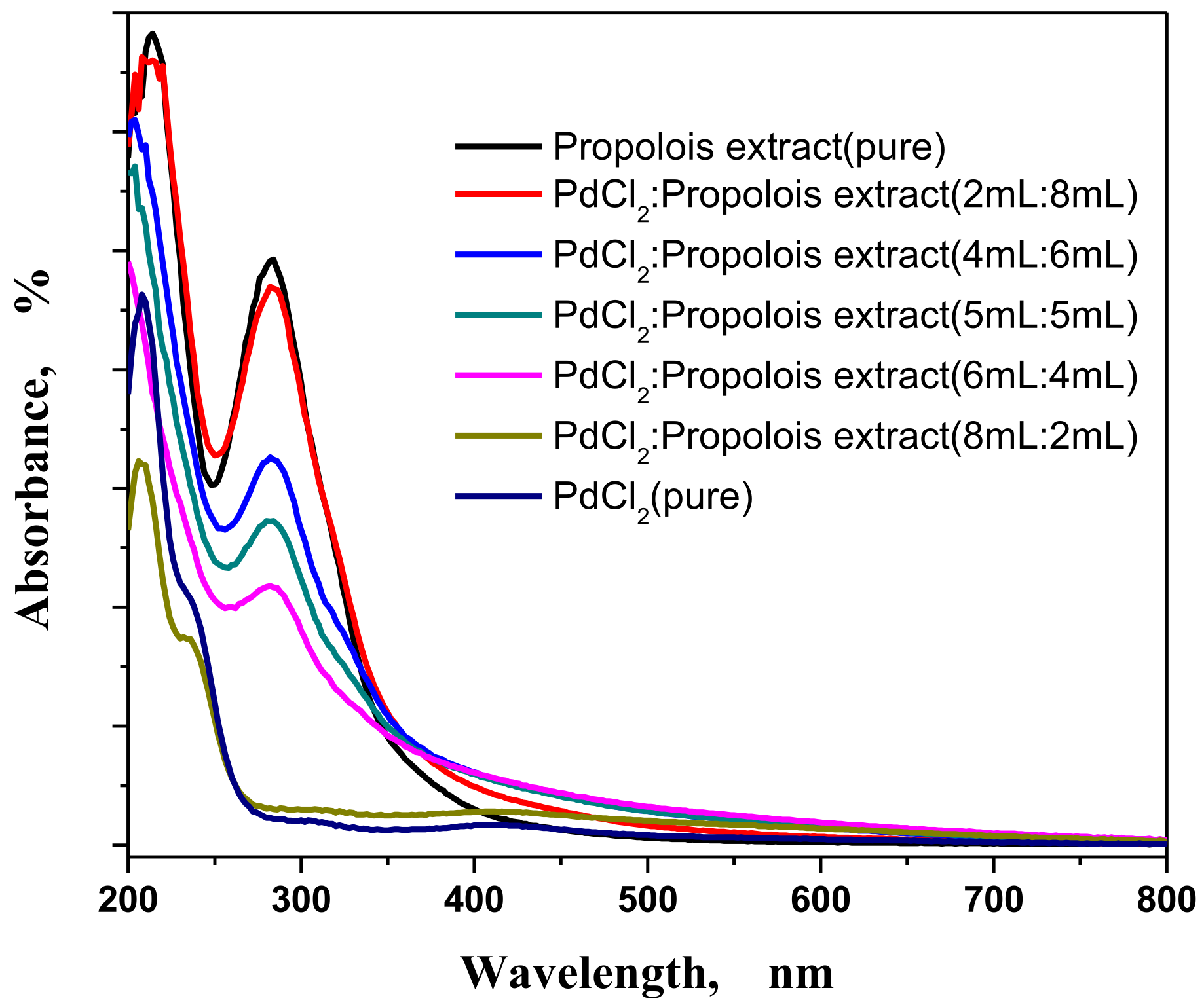
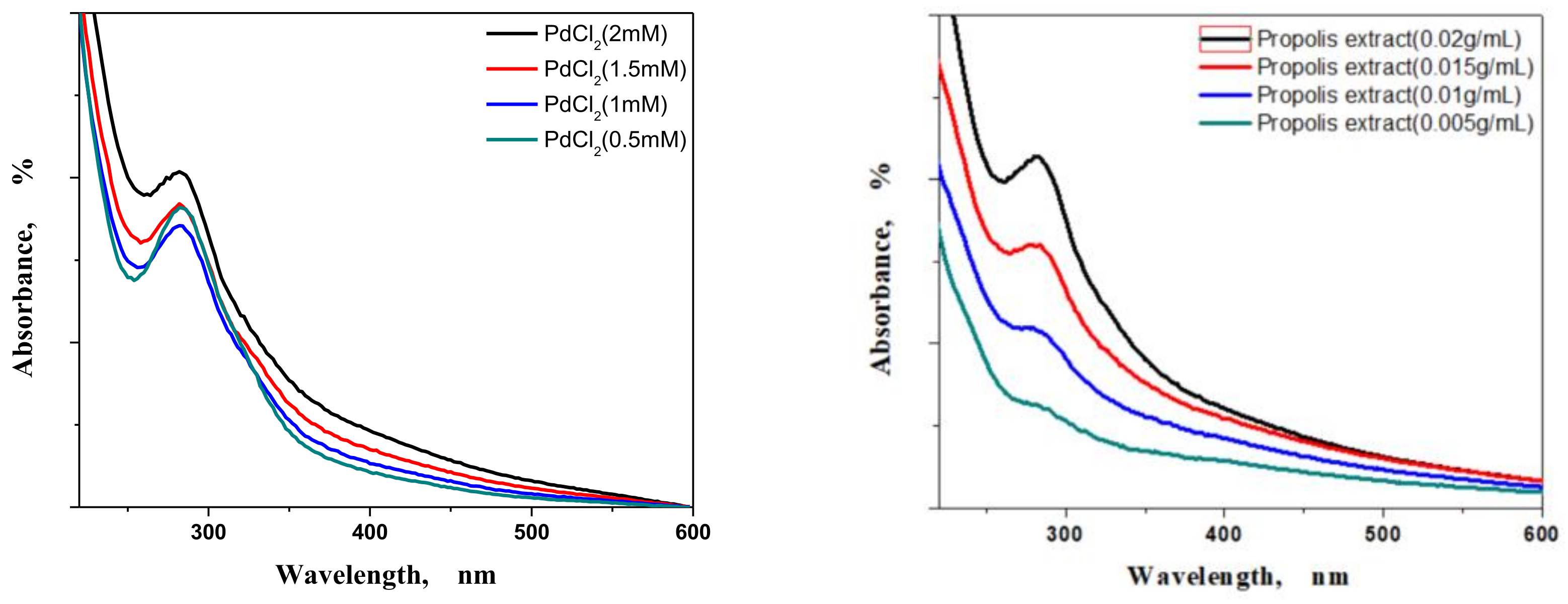
3.1.1. Concentrations and Ratio Effects
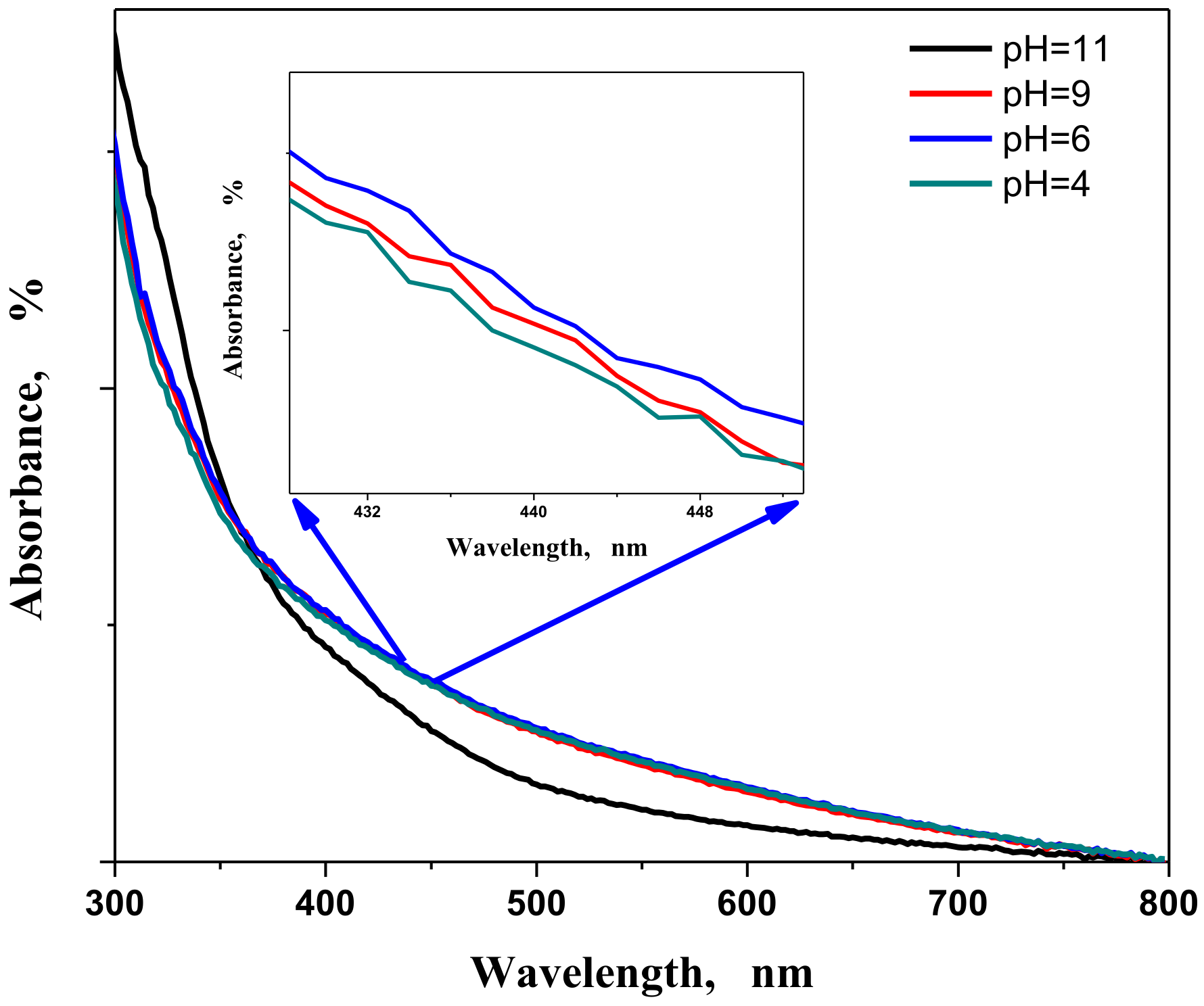
3.1.2. pH Factor
3.1.3. Temperature Factor
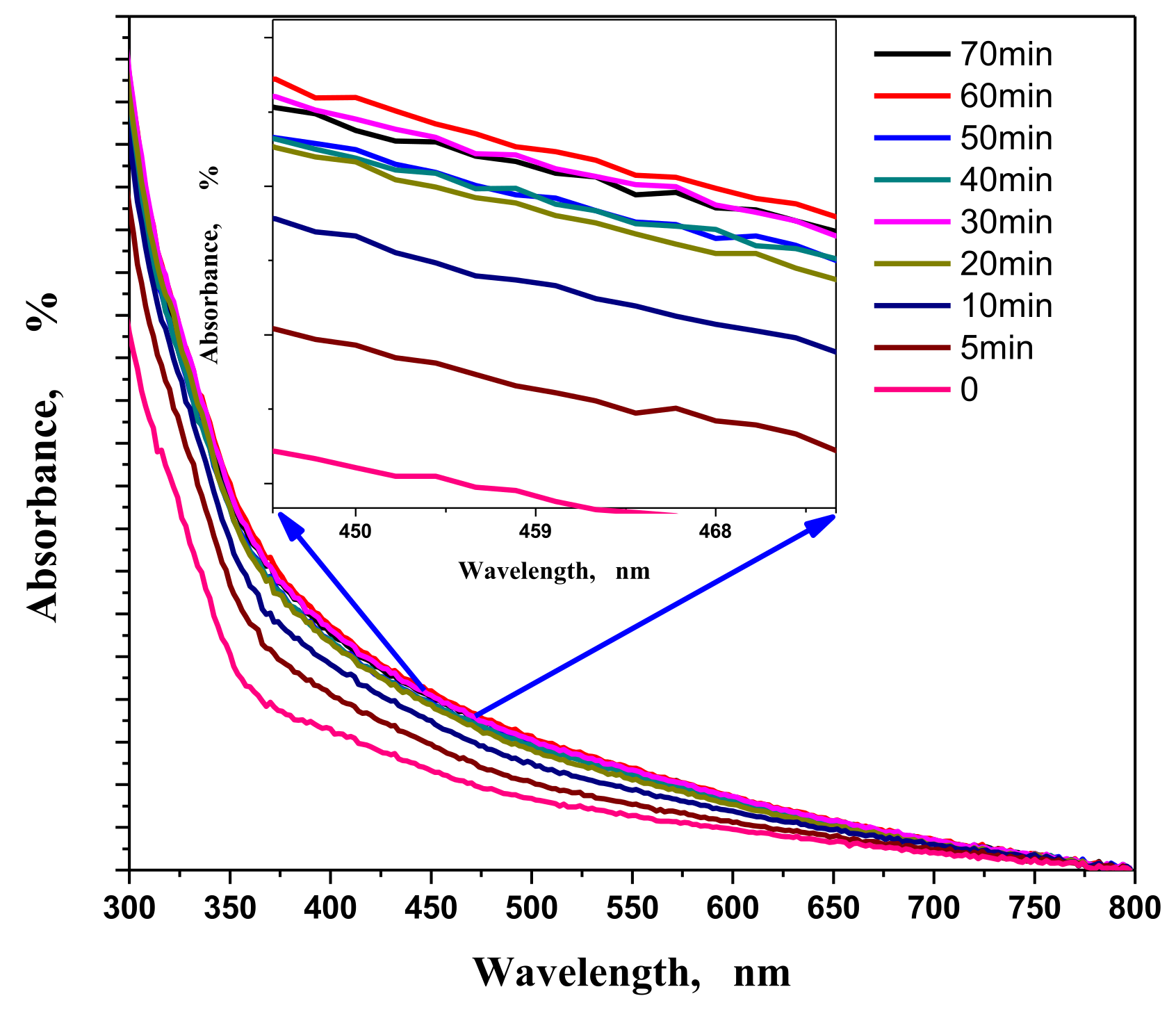
3.1.4. Contact Time Effect
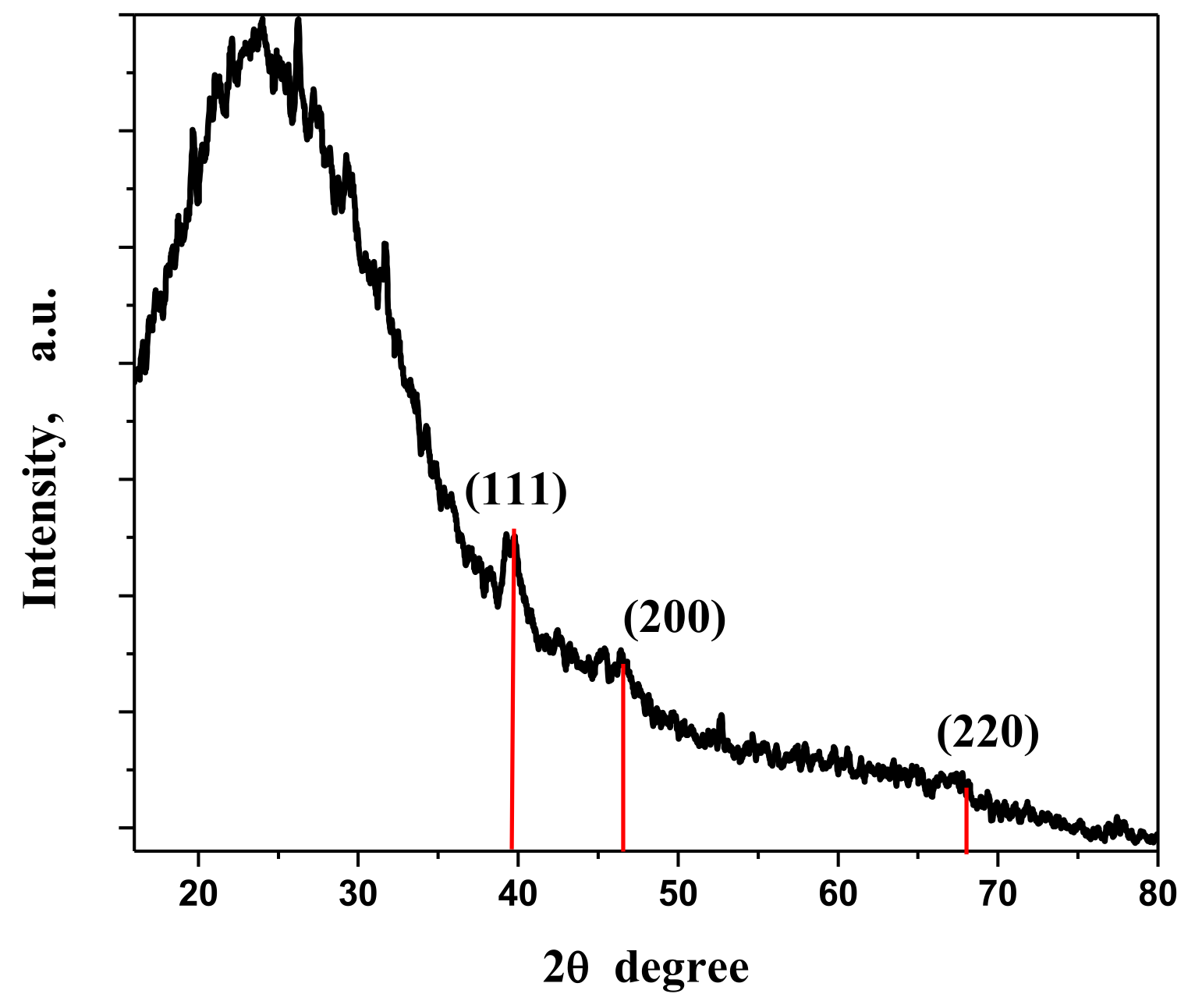
3.2. XRD Analysis
3.3. FT-IR Analysis
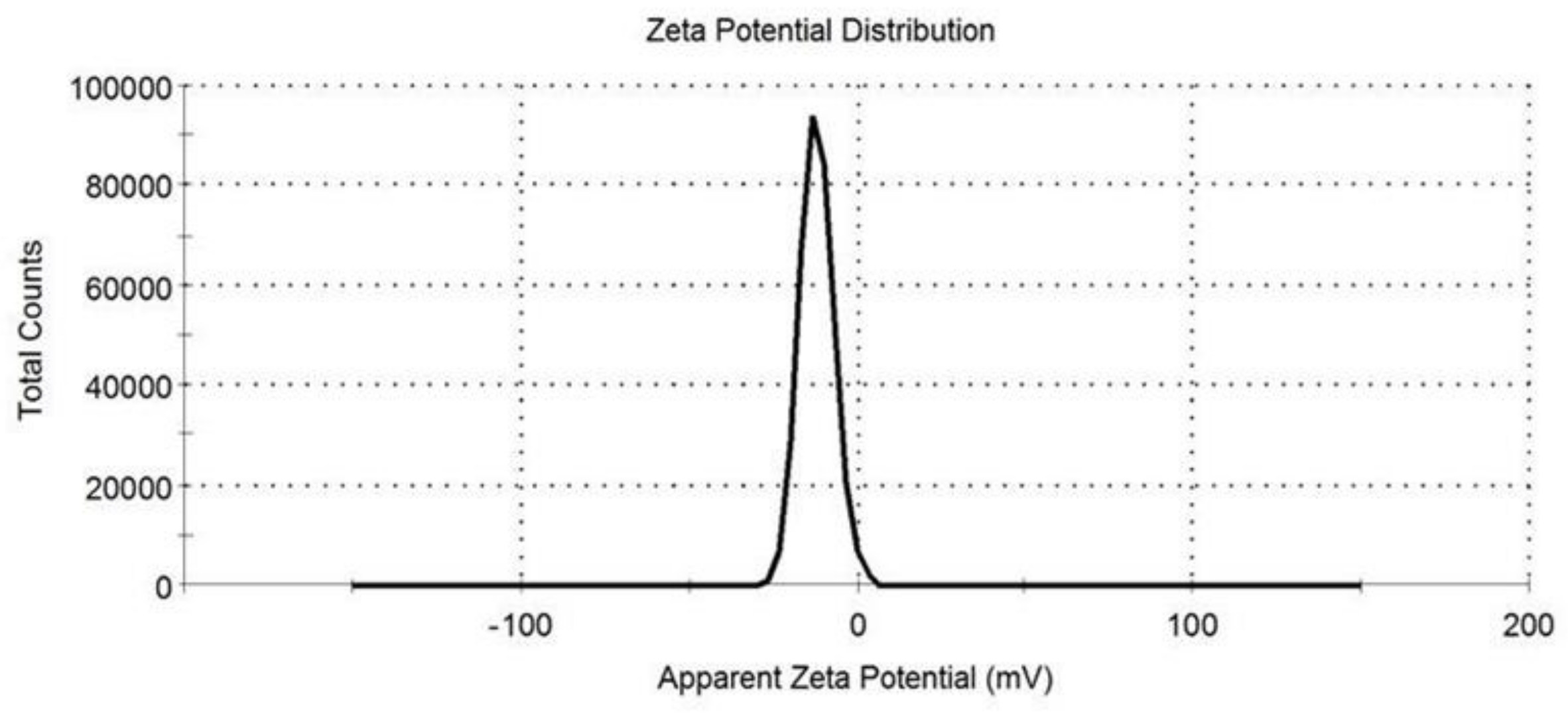
3.4. Zeta Potential
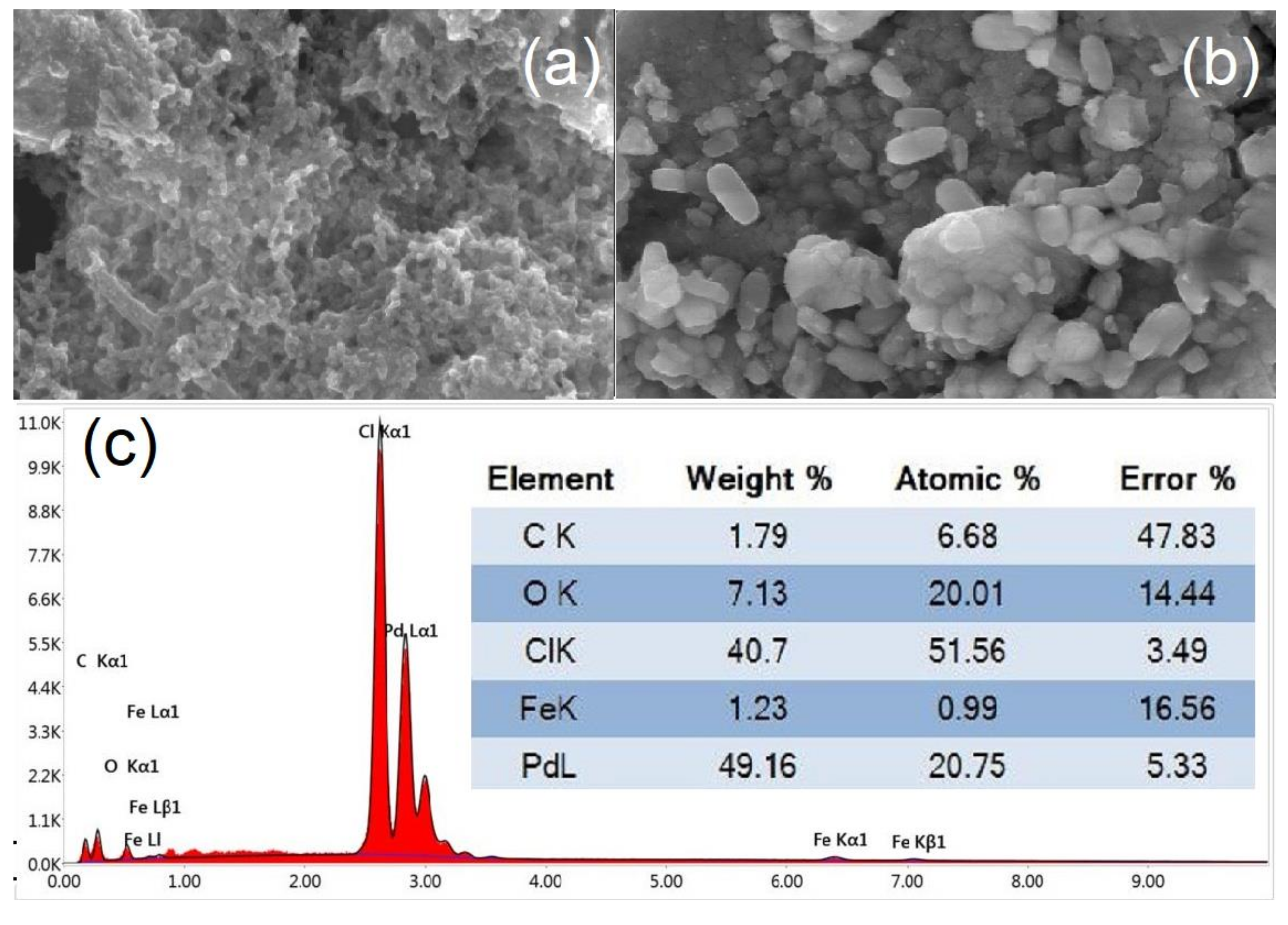
3.5. TEM, SEM, and EDX Measurements
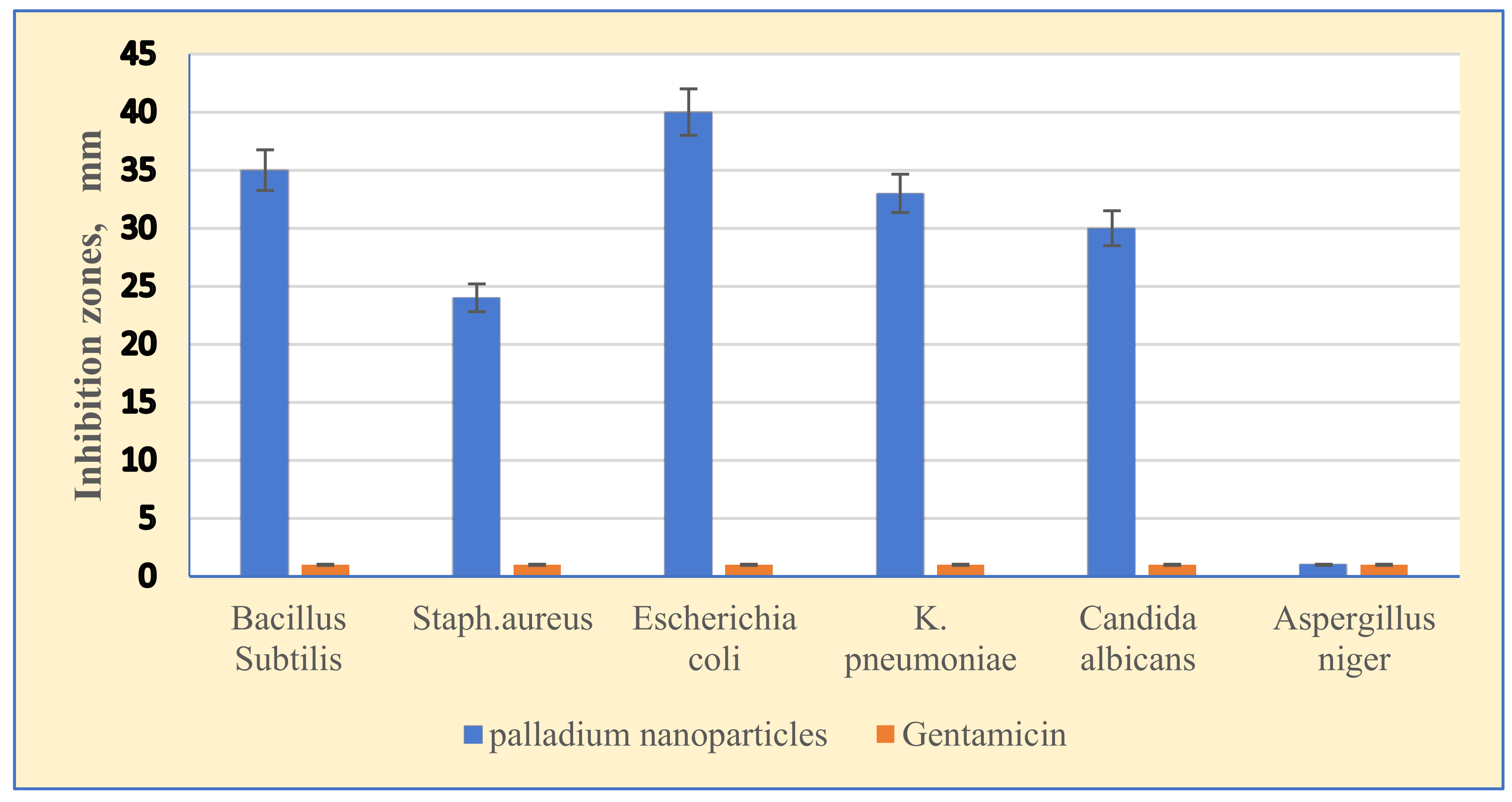
3.6. Antimicrobial Activity of Palladium Nanoparticles
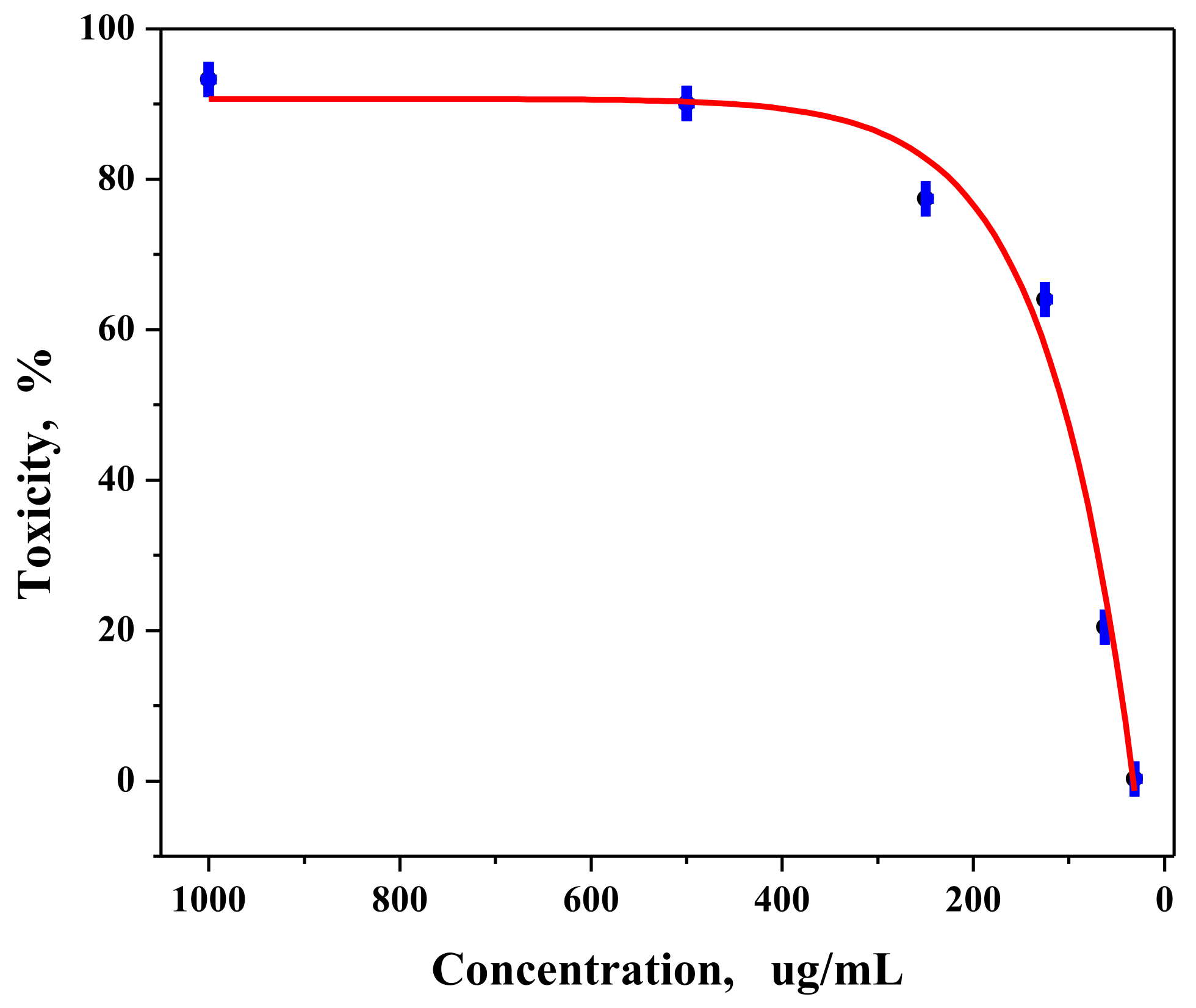
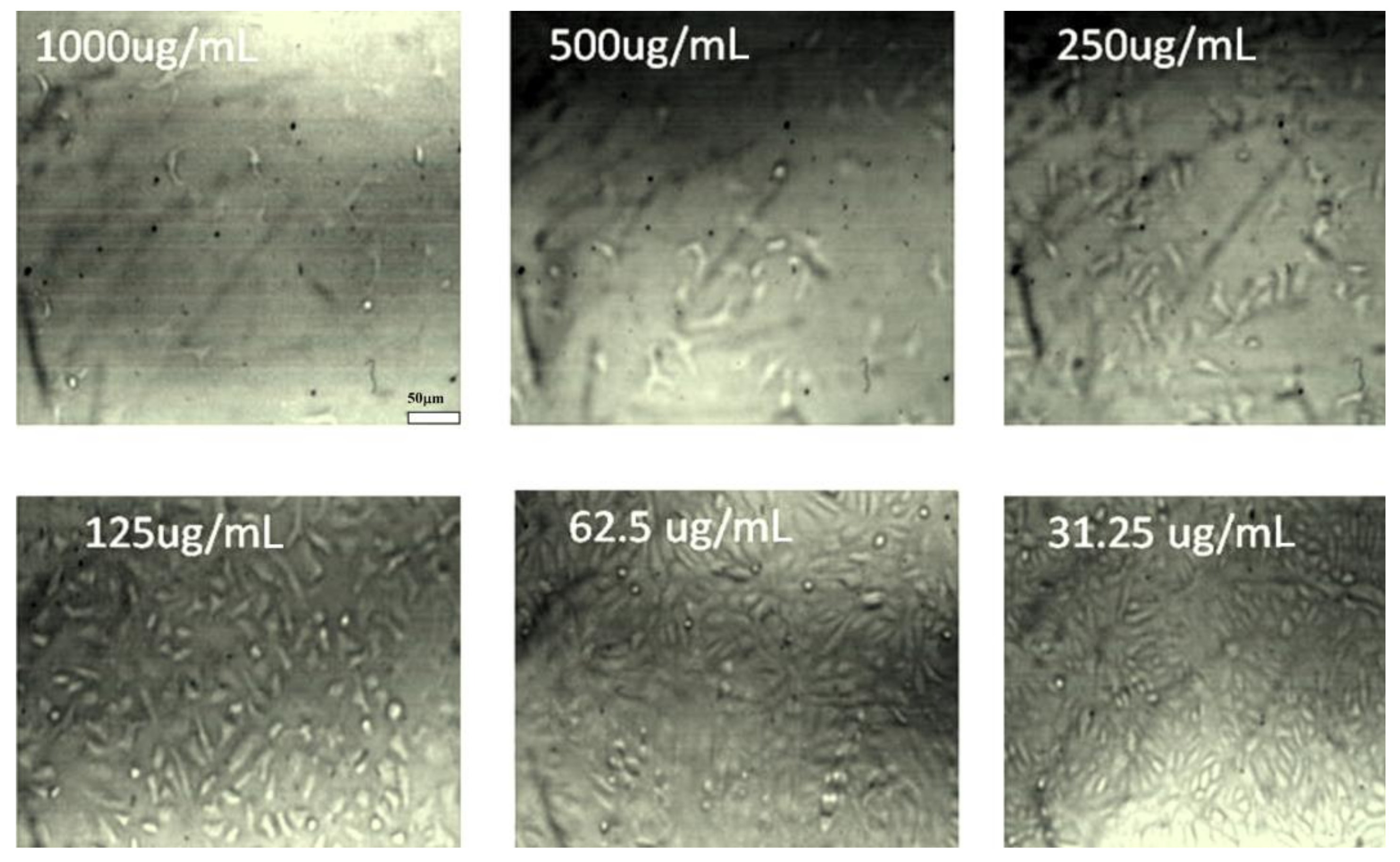
3.7. In Vitro Anticancer Activity of Palladium Nanoparticles against MCF-7 Cell Line
4. Discussion
4.1. Characterization of Pd-NPS
4.2. Antimicrobial and Anticancer Mechanisms
4.2.1. Antimicrobial Activity of Palladium Nanoparticles
4.2.2. Anticancer Activity of Palladium Nanoparticles against MCF-7 Cell Line
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sforcin, J.M. Biological Properties and Therapeutic Applications of Propolis. Phytother. Res. 2016, 30, 894–905.4. [Google Scholar] [CrossRef] [PubMed]
- Alanazi, S.; Alenzi, N.; Alenazi, F.; Tabassum, H.; Watson, D. Chemical characterization of Saudi propolis and its antiparasitic and anticancer properties. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Khan, I.; Saeed, K.; Khan, I. Nanoparticles: Properties, applications and toxicities. Arab. J. Chem. 2019, 12, 908–931. [Google Scholar] [CrossRef]
- Stark, W.J.; Stoessel, P.R.; Wohlleben, W.; Hafner, A. Industrial applications of nanoparticles. Chem. Soc. Rev. 2015, 44, 5793–5805. [Google Scholar] [CrossRef] [Green Version]
- Mohanpuria, P.; Rana, N.K.; Yadav, S.K. Biosynthesis of nanoparticles: Technological concepts and future applications. J. Nanopart. Res. 2008, 10, 507–517. [Google Scholar] [CrossRef]
- Hazarika, M.; Borah, D.; Bora, P.; Silva, A.R.; Das, P. Biogenic synthesis of palladium nanoparticles and their applications as catalyst and antimicrobial agent. PLoS ONE 2017, 12, e0184936. [Google Scholar] [CrossRef]
- Phan, T.T.V.; Huynh, T.-C.; Manivasagan, P.; Mondal, S.; Oh, J. An up-to-date review on biomedical applications of palladium nanoparticles. Nanomaterials 2020, 10, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saldan, I.; Semenyuk, Y.; Marchuk, I.; Reshetnyak, O. Chemical synthesis and application of palladium nanoparticles. J. Mater. Sci. 2015, 50, 2337–2354. [Google Scholar] [CrossRef]
- Chen, H.; Wei, G.; Ispas, A.; Hickey, S.G.; Eychmüller, A. Synthesis of palladium nanoparticles and their applications for surface-enhanced Raman scattering and electrocatalysis. J. Phys. Chem. C 2010, 114, 21976–21981. [Google Scholar] [CrossRef]
- Anand, K.; Tiloke, C.; Phulukdaree, A.; Ranjan, B.; Chuturgoon, A.; Singh, S.; Gengan, R. Biosynthesis of palladium nanoparticles by using Moringa oleifera flower extract and their catalytic and biological properties. J. Photochem. Photobiol. B Biol. 2016, 165, 87–95. [Google Scholar] [CrossRef]
- Fahmy, A.H.; Preis, E.; Bakowsky, U.; Azzazy, H.M.E.-S. Palladium Nanoparticles Fabricated by Green Chemistry: Promising Chemotherapeutic, Antioxidant and Antimicrobial Agents. Materials 2020, 13, 3661. [Google Scholar] [CrossRef]
- Rokade, S.S.; Joshi, K.A.; Mahajan, K.; Tomar, G. Novel anticancer platinum and palladium nanoparticles from Barleria prionitis. Glob. J. Nanomedicine 2017, 2, 555600. [Google Scholar]
- Gurunathan, S.; Kim, E.; Han, J.W.; Park, J.H.; Kim, J.H. Green chemistry approach for synthesis of effective anticancer palladium nanoparticles. Molecules 2015, 20, 22476–22498. [Google Scholar] [CrossRef]
- Adams, C.P.; Walker, K.A.; Obare, S.O.; Docherty, K.M. Size-dependent antimicrobial effects of novel palladium nanoparticles. PLoS ONE 2014, 9, e85981. [Google Scholar] [CrossRef] [Green Version]
- Manikandan, V.; Velmurugan, P.; Park, J.-H.; Lovanh, N.; Seo, S.-K.; Jayanthi, P.; Palrk, Y.-J.; Cho, M.; Oh, B.-T. Synthesis and antimicrobial activity of palladium nanoparticles from Prunus× yedoensis leaf extract. Mater. Lett. 2016, 185, 335–338. [Google Scholar] [CrossRef]
- Osonga, F.J.; Kalra, S.; Miller, R.M.; Isika, D.; Sadik, O.A. Synthesis, characterization and antifungal activities of eco-friendly palladium nanoparticles. RSC Adv. 2020, 10, 5894–5904. [Google Scholar] [CrossRef]
- Azizi, S.; Shahri, M.M.; Rahman, H.S.; Rahim, R.A.; Rasetdee, A.; Mohamad, R. Green synthesis palladium nanoparticles mediated by white tea (Camellia sinensis) extract with antioxidant, antibacterial, and antiproliferative activities toward the human leukemia (MOLT-4) cell line. Int. J. Nanomed. 2017, 12, 8841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Indrigo, E.; Clavadetscher, J.; Chankeshwara, S.V.; Lilienkampf, A.; Bradley, M. Palladium-mediated in situ synthesis of an anticancer agent. Chem. Commun. 2016, 52, 14212–14214. [Google Scholar] [CrossRef] [Green Version]
- Gnanasekar, S.; Murugaraj, J.; Dhivyabharathi, B.; Krishnamoorthy, V.; Jha, P.; Seetharaman, P.; Vilwanathan, R.; Sivaperumal, S. Antibacterial and cytotoxicity effects of biogenic palladium nanoparticles synthesized using fruit extract of Couroupita guianensis Aubl. J. Appl. Biomed. 2018, 16, 59–65. [Google Scholar] [CrossRef]
- Malik, P.; Shankar, R.; Malik, V.; Sharma, N.; Mukherjee, T.K. Green chemistry based benign routes for nanoparticle synthesis. J. Nanopart. 2014, 2014, 302429. [Google Scholar] [CrossRef] [Green Version]
- Akhtar, M.S.; Panwar, J.; Yun, Y.-S. Biogenic synthesis of metallic nanoparticles by plant extracts. ACS Sustain. Chem. Eng. 2013, 1, 591–602. [Google Scholar] [CrossRef]
- Castro, L.; Blázquez, M.L.; González, F.G.; Ballester, A. Mechanism and applications of metal nanoparticles prepared by bio-mediated process. Rev. Adv. Sci. Eng. 2014, 3, 199–216. [Google Scholar] [CrossRef]
- Singh, J.; Dutta, T.; Kim, K.-H.; Rawat, M.; Samddar, P.; Kumar, P. “Green” synthesis of metals and their oxide nanoparticles: Applications for environmental remediation. J. Nanobiotechnol. 2018, 16, 84. [Google Scholar] [CrossRef]
- Bankova, V. Recent trends and important developments in propolis research. Evidence-based Complement. Altern. Med. 2005, 2, 29–32. [Google Scholar]
- Ali, A.M.; Kunugi, H. Propolis, bee honey, and their components protect against coronavirus disease 2019 (Covid-19): A review of in silico, in vitro, and clinical studies. Molecules 2021, 26, 1232. [Google Scholar] [CrossRef] [PubMed]
- Castaldo, S.; Capasso, F. Propolis, an old remedy used in modern medicine. Fitoterapia 2002, 73, S1–S6. [Google Scholar] [CrossRef]
- Bouzahouane, H.; Ayari, A.; Guehria, I.; Riah, O. The Propolis: Antimicrobial activity and chemical composition analysis: Properties of propolis. J. Microbiol. Biotechnol. Food Sci. 2021, e3211. [Google Scholar]
- Amarnath, K.; Kumar, J.; Reddy, T.; Mahesh, V.; Ayyappan, S.R.; Nellore, J. Synthesis and characterization of chitosan and grape polyphenols stabilized palladium nanoparticles and their antibacterial activity. Colloids Surf. B Biointerfaces 2011, 92, 254–261. [Google Scholar] [CrossRef]
- Wikler, M.A. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically: Approved standard. CLSI 2006, 26, M7-A7. [Google Scholar]
- Slater, T.F.; Sawyer, B.; Sträuli, U. Studies on succinate-tetrazolium reductase systems: III. Points of coupling of four different tetrazolium salts III. Points of coupling of four different tetrazolium salts. Biochim. Biophys. Acta 1963, 77, 383–393. [Google Scholar] [CrossRef]
- Van de Loosdrecht, A.A.; Beelen, R.H.J.; Ossenkoppele, G.J.; Broekhoven, M.G.; Langenhuijsen, M. A tetrazolium-based colorimetric MTT assay to quantitate human monocyte mediated cytotoxicity against leukemic cells from cell lines and patients with acute myeloid leukemia. J. Immunol. Methods 1994, 174, 311–320. [Google Scholar] [CrossRef]
- Alley, M.C.; Scudiero, D.A.; Monks, A.; Hursey, M.L.; Czerwinski, M.J.; Fine, D.L.; Abbott, B.J.; Mayo, J.G.; Shoemaker, R.H.; Boyd, M.R. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 1988, 48, 589–601. [Google Scholar]
- Sheny, D.S.; Philip, D.; Mathew, J. Rapid green synthesis of palladium nanoparticles using the dried leaf of Anacardium occidentale. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2012, 91, 35–38. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, X. Carbon-supported PdSn nanoparticles as catalysts for formic acid oxidation. Electrochem. Commun. 2009, 11, 1667–1670. [Google Scholar] [CrossRef]
- Cheng, H.; Yang, N.; Liu, G.; Ge, Y.; Huang, J.; Yun, Q.; Du, Y.; Sun, C.-J.; Chen, B.; Liu, J.; et al. Ligand-Exchange-Induced Amorphization of Pd Nanomaterials for Highly Efficient Electrocatalytic Hydrogen Evolution Reaction. Adv. Mater. 2020, 32, 1902964. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Liu, J.; Roberts, C.B.; Zhao, D. One-step “green” synthesis of Pd nanoparticles of controlled size and their catalytic activity for trichloroethene hydrodechlorination. Ind. Eng. Chem. Res. 2009, 48, 6550–6557. [Google Scholar] [CrossRef]
- Debye, P.; Scherrer, P. Interferenzen an regellos orientierten Teilchen im Röntgenlicht. I. Nachrichten von der Gesellschaft der Wissenschaften zu Göttingen. Math. Klasse 1916, 1916, 1–15. [Google Scholar]
- Basavegowda, N.; Mishra, K.; Lee, Y.R. Ultrasonic-assisted green synthesis of palladium nanoparticles and their nanocatalytic application in multicomponent reaction. New J. Chem. 2015, 39, 972–977. [Google Scholar] [CrossRef]
- Rehbock, C.; Merk, V.; Gamrad, L.; Streubel, R.; Barcikowski, S. Size control of laser-fabricated surfactant-free gold nanoparticles with highly diluted electrolytes and their subsequent bioconjugation. Phys. Chem. Chem. Phys. 2013, 15, 3057–3067. [Google Scholar] [CrossRef] [Green Version]
- Sathishkumar, M.; Sneha, K.; Kwak, I.S.; Mao, J.; Tripathy, S.; Yun, Y.-S. Phyto-crystallization of palladium through reduction process using Cinnamom zeylanicum bark extract. J. Hazard. Mater. 2009, 171, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Marzun, G.; Nakamura, J.; Zhang, X.; Barcikowski, S.; Wagener, P. Size control and supporting of palladium nanoparticles made by laser ablation in saline solution as a facile route to heterogeneous catalysts. Appl. Surf. Sci. 2015, 348, 75–84. [Google Scholar] [CrossRef]
- Wisam, J.A.; Haneen, A.J. A novel study of pH influence on Ag nanoparticles size with antibacterial and antifungal activity using green synthesis. World Sci. News 2018, 97, 139–152. [Google Scholar]
- Narain, R. Polymer Science and Nanotechnology: Fundamentals and Applications; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Lu, W.; Wang, B.; Wang, K.; Wang, X.; Hou, J.G. Synthesis and characterization of crystalline and amorphous palladium nanoparticles. Langmuir 2003, 19, 5887–5891. [Google Scholar] [CrossRef]
- Venkatesham, M.; Ayodhya, D.; Veerabhadram, G. Green synthesis, characterization and catalytic activity of palladium nanoparticles by xanthan gum. Appl. Nanosci. 2015, 5, 315–320. [Google Scholar]
- Hussain, M.; Raja, N.I.; Iqbal, M.; Aslam, S. Applications of plant flavonoids in the green synthesis of colloidal silver nanoparticles and impacts on human health. Iran. J. Sci. Technol. Trans. A Sci. 2019, 43, 1381–1392. [Google Scholar] [CrossRef]
- Shanthi, K.; Sreevani, V.; Vimala, K.; Kannan, S. Cytotoxic effect of palladium nanoparticles synthesized from Syzygium aromaticum aqueous extracts and induction of apoptosis in cervical carcinoma. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2017, 87, 1101–1112. [Google Scholar] [CrossRef]
- Tuan, T.Q.; Van Hao, P.; Quynh, L.M.; Luong, N.H.; Hai, N.H. Preparation and properties of silver nanoparticles by heat-combined electrochemical method. VNU J. Sci. Math. 2015, 31, 36–44. [Google Scholar]
- Murugesan, B.; Pandiyan, N.; Arumugam, M.; Sonamuthu, J.; Samayanan, S.; Yurong, C.; Juming, Y.; Mahalingam, S. Fabrication of palladium nanoparticles anchored polypyrrole functionalized reduced graphene oxide nanocomposite for antibiofilm associated orthopedic tissue engineering. Appl. Surf. Sci. 2020, 510, 145403. [Google Scholar] [CrossRef]
- Rai, M.; Deshmukh, S.; Gade, A. Strategic nanoparticle-mediated gene transfer in plants and animals-a novel approach. Curr. Nanosci. 2012, 8, 170–179. [Google Scholar] [CrossRef]
- Li, Q.; Mahendra, S.; Lyon, D.Y.; Brunet, L.; Liga, M.V.; Li, D.; Alvarez, P.J. Antimicrobial nanomaterials for water disinfection and microbial control: Potential applications and implications. Water Res. 2008, 42, 4591–4602. [Google Scholar] [CrossRef]
- Pal, S.; Tak, Y.K.; Song, J.M. Does the antibacterial activity of silver nanoparticles depend on the shape of the nanoparticle? A study of the gram-negative bacterium Escherichia coli. Appl. Environ. Microbiol. 2007, 73, 1712–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morones, J.R.; Elechiguerra, J.L.; Camacho, A.; Holt, K.; Kouri, J.B.; Ramírez, J.T.; Yacaman, M.J. The bactericidal effect of silver nanoparticles. Nanotechnology 2005, 16, 2346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keat, C.L.; Abdul-Aziz, A.; Eid, A.M.; Elmarzugi, N.A. Elmarzugi. Biosynthesis of nanoparticles and silver nanoparticles. Bioresour. Bioprocess. 2015, 2, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Siddiqi, K.S.; Husen, A.; Rao, R.A.K. A review on biosynthesis of silver nanoparticles and their biocidal properties. J. Nanobiotechnol. 2018, 16, 1–28. [Google Scholar] [CrossRef]
- Vega-Baudrit, J.; Gamboa, S.M.; Rojas, E.R.; Martinez, V.V. Synthesis and characterization of silver nanoparticles and their application as an antibacterial agent. Int. J. Biosen. Bioelectron. 2019, 5, 166–173. [Google Scholar] [CrossRef] [Green Version]
- Kocot, J.; Kiełczykowska, M.; Luchowska-Kocot, D.; Kurzepa, J.; Musik, I. Antioxidant potential of propolis, bee pollen, and royal jelly: Possible medical application. Oxid. Med. Cell. Longev. 2018, 2018, 7074209. [Google Scholar] [CrossRef]
- Sharmila, G.; Haries, S.; Fathima, M.F.; Geetha, S.; Kumar, N.M.; Muthukumaran, C. Enhanced catalytic and antibacterial activities of phytosynthesized palladium nanoparticles using Santalum album leaf extract. Powder Technol. 2017, 320, 22–26. [Google Scholar] [CrossRef]
- Tahir, K.; Nazir, S.; Li, B.; Ahmad, A.; Nasir, T.; Khan, A.U.; Shah, S.A.; Khan, Z.U.; Yasin, G.; Hameed, M.U. Sapium sebiferum leaf extract mediated synthesis of palladium nanoparticles and in vitro investigation of their bacterial and photocatalytic activities. J. Photochem. Photobiol. B Biol. 2016, 164, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Vaghela, H.; Shah, R.; Pathan, A. Palladium nanoparticles mediated through bauhinia variegata: Potent in vitro anticancer activity against mcf-7 cell lines and antimicrobial assay. Curr. Nanomater. 2018, 3, 168–177. [Google Scholar] [CrossRef]
- Bendale, Y.; Bendale, V.; Paul, S. Evaluation of cytotoxic activity of platinum nanoparticles against normal and cancer cells and its anticancer potential through induction of apoptosis. Integr. Med. Res. 2017, 6, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Fahmy, S.A.; Fawzy, I.M.; Saleh, B.M.; Issa, M.Y.; Bakowsky, U.; Azzazy, H.M.E. Green Synthesis of Platinum and Palladium Nanoparticles Using Peganum harmala L. Seed Alkaloids: Biological and Computational Studies. Nanomaterials 2021, 11, 965. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Fakeh, M.S.; Osman, S.O.M.; Gassoumi, M.; Rabhi, M.; Omer, M. Characterization, Antimicrobial and Anticancer Properties of Palladium Nanoparticles Biosynthesized Optimally Using Saudi Propolis. Nanomaterials 2021, 11, 2666. https://doi.org/10.3390/nano11102666
Al-Fakeh MS, Osman SOM, Gassoumi M, Rabhi M, Omer M. Characterization, Antimicrobial and Anticancer Properties of Palladium Nanoparticles Biosynthesized Optimally Using Saudi Propolis. Nanomaterials. 2021; 11(10):2666. https://doi.org/10.3390/nano11102666
Chicago/Turabian StyleAl-Fakeh, Maged S., Samir Osman Mohammed Osman, Malek Gassoumi, Mokded Rabhi, and Mohamed Omer. 2021. "Characterization, Antimicrobial and Anticancer Properties of Palladium Nanoparticles Biosynthesized Optimally Using Saudi Propolis" Nanomaterials 11, no. 10: 2666. https://doi.org/10.3390/nano11102666
APA StyleAl-Fakeh, M. S., Osman, S. O. M., Gassoumi, M., Rabhi, M., & Omer, M. (2021). Characterization, Antimicrobial and Anticancer Properties of Palladium Nanoparticles Biosynthesized Optimally Using Saudi Propolis. Nanomaterials, 11(10), 2666. https://doi.org/10.3390/nano11102666