Nanomaterials in the Management of Gram-Negative Bacterial Infections
Abstract
:1. Introduction
2. Detection of E. coli Infection
2.1. Nanotechnology Approaches for E. coli Detection
2.1.1. Gold NPs
2.1.2. Silver NPs
2.1.3. QDs
2.1.4. Carbon Nanomaterials
2.1.5. Metal-Organic Frameworks (MOFs)
2.1.6. Silica NPs
2.1.7. Magnetic NPs
2.1.8. ZnO NPs
3. Nanomaterials for Treatment of E. coli Infections
3.1. Polymeric Nanocarriers
3.2. Lipidic Nanocarriers
3.3. Metallic Nanocarriers
3.4. Other Nanocarriers
4. Point of Care (POC) Devices for Clinical Applications
5. Regulatory Landscape of Nanotechnology in Biomedical Applications
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weiner, L.M.; Webb, A.K.; Limbago, B.; Dudeck, M.A.; Patel, J.; Kallen, A.J.; Edwards, J.R.; Sievert, D.M. Antimicrobial-resistant pathogens associated with healthcare-associated infections: Summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect. Control. Hosp. Epidemiol. 2016, 37, 1288–1301. [Google Scholar] [CrossRef]
- Al-Hasan, M.N.; Lahr, B.D.; Eckel-Passow, J.E.; Baddour, L.M. Antimicrobial resistance trends of Escherichia coli bloodstream isolates: A population-based study, 1998–2007. J. Antimicrob. Chemother. 2009, 64, 169–174. [Google Scholar] [CrossRef]
- Søgaard, M.; Nørgaard, M.; Dethlefsen, C.; Schønheyder, H.C. Temporal changes in the incidence and 30-day mortality associated with bacteremia in hospitalized patients from 1992 through 2006: A population-based cohort study. Clin. Infect. Dis. 2011, 52, 61–69. [Google Scholar] [CrossRef]
- Tavafi, H.; Sadrzadeh-Afshar, M.; Niroomand, S. In Vitro Effectiveness of Antimicrobial Properties of Propolis and Chlorhexidine on Oral Pathogens: A Comparative Study: Effectiveness of Antimicrobial Properties of Propolis and Chlorhexidine on Oral Pathogens. Biosis Biol. Syst. 2020, 1, 116–125. [Google Scholar] [CrossRef]
- Opal, S.M.; Garber, G.E.; LaRosa, S.P.; Maki, D.G.; Freebairn, R.C.; Kinasewitz, G.T.; Dhainaut, J.-F.; Yan, S.B.; Williams, M.D.; Graham, D.E. Systemic host responses in severe sepsis analyzed by causative microorganism and treatment effects of drotrecogin alfa (activated). Clin. Infect. Dis. 2003, 37, 50–58. [Google Scholar] [CrossRef] [PubMed]
- Ranieri, V.M.; Thompson, B.T.; Barie, P.S.; Dhainaut, J.-F.; Douglas, I.S.; Finfer, S.; Gårdlund, B.; Marshall, J.C.; Rhodes, A.; Artigas, A. Drotrecogin alfa (activated) in adults with septic shock. N. Engl. J. Med. 2012, 366, 2055–2064. [Google Scholar] [CrossRef] [PubMed]
- Hauser, A. Cell Envelope. Antibiotic Basic for Clinicians; Wolters Kluwer India Pvt Ltd.: Gurugram, India, 2015. [Google Scholar]
- Dasaraju, P.V.; Liu, C. Infections of the respiratory system. In Medical Microbiology, 4th ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300. [Google Scholar] [CrossRef]
- Pang, X.; Gong, K.; Zhang, X.; Wu, S.; Cui, Y.; Qian, B.-Z. Osteopontin as a multifaceted driver of bone metastasis and drug resistance. Pharmacol. Res. 2019, 144, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.I. Antibiotic resistance and regulation of the gram-negative bacterial outer membrane barrier by host innate immune molecules. MBio 2016, 7, e01541-16. [Google Scholar] [CrossRef]
- Gupta, V.; Datta, P. Next-generation strategy for treating drug resistant bacteria: Antibiotic hybrids. Indian J. Med. Res. 2019, 149, 97. [Google Scholar]
- Pandey, V.K.; Srivastava, K.R.; Ajmal, G.; Thakur, V.K.; Gupta, V.K.; Upadhyay, S.N.; Mishra, P.K. Differential Susceptibility of Catheter Biomaterials to Biofilm-Associated Infections and Their Remedy by Drug-Encapsulated Eudragit RL100 Nanoparticles. Int. J. Mol. Sci. 2019, 20, 5110. [Google Scholar] [CrossRef]
- Bansal, K.K.; Bhardwaj, J.K.; Saraf, P.; Thakur, V.K.; Sharma, P.C. Synthesis of Thiazole Clubbed Pyrazole Derivatives as Apoptosis Inducers and Anti-Infective Agents. Mater. Today Chem. 2020, 17, 100335. [Google Scholar] [CrossRef]
- Tavafi, H. An Investigation of Antibacterial Resistance Patterns in Isolated Bacteria from Contaminated Water Samples in Poultry Slaughterhouses. Biosis Biol. Syst. 2020, 1, 85–90. [Google Scholar] [CrossRef]
- Mohsin, M.; Haque, A.; Ali, A.; Sarwar, Y.; Bashir, S.; Tariq, A.; Afzal, A.; Iftikhar, T.; Saeed, M.A. Effects of ampicillin, gentamicin, and cefotaxime on the release of Shiga toxins from Shiga toxin–producing Escherichia coli isolated during a diarrhea episode in Faisalabad, Pakistan. Foodborne Pathog. Dis. 2010, 7, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.W.; Henderson, T.D.; Patfield, S.; Stanker, L.H.; He, X. Mouse in vivo neutralization of Escherichia coli Shiga toxin 2 with monoclonal antibodies. Toxins 2013, 5, 1845–1858. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.D.; Gaudet, R.G. Antibodies in infectious diseases: Polyclonals, monoclonals and niche biotechnology. New Biotechnol. 2011, 28, 489–501. [Google Scholar] [CrossRef]
- Saeedi, P.; Yazdanparast, M.; Behzadi, E.; Salmanian, A.H.; Mousavi, S.L.; Nazarian, S.; Amani, J. A review on strategies for decreasing E. coli O157: H7 risk in animals. Microb. Pathog. 2017, 103, 186–195. [Google Scholar] [CrossRef]
- Ling, L.L.; Schneider, T.; Peoples, A.J.; Spoering, A.L.; Engels, I.; Conlon, B.P.; Mueller, A.; Schäberle, T.F.; Hughes, D.E.; Epstein, S. A new antibiotic kills pathogens without detectable resistance. Nature 2015, 517, 455–459. [Google Scholar] [CrossRef]
- Kumar, M.; Ghosh, S.; Nayak, S.; Das, A. Recent advances in biosensor based diagnosis of urinary tract infection. Biosens. Bioelectron. 2016, 80, 497–510. [Google Scholar] [CrossRef]
- Van Nostrand, J.D.; Junkins, A.D.; Bartholdi, R.K. Poor predictive ability of urinalysis and microscopic examination to detect urinary tract infection. Am. J. Clin. Pathol. 2000, 113, 709–713. [Google Scholar] [CrossRef]
- Graham, J.; Galloway, A. ACP Best Practice No 167: The laboratory diagnosis of urinary tract infection. J. Clin. Pathol. 2001, 54, 911–919. [Google Scholar] [CrossRef]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L. The real-time polymerase chain reaction. Mol. Asp. Med. 2006, 27, 95–125. [Google Scholar] [CrossRef]
- Padmavathy, á.; Kumar, R.V.; Patel, A.; Swarnam, S.D.; Vaidehi, T.; Ali, B.J. Rapid and sensitive detection of major uropathogens in a single-pot multiplex PCR assay. Curr. Microbiol. 2012, 65, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Mohanan, P.V.; Banerjee, S.; Geetha, C.S. Detection of pyrogenicity on medical grade polymer materials using rabbit pyrogen, LAL and ELISA method. J. Pharm. Biomed. Anal. 2011, 55, 1170–1174. [Google Scholar] [CrossRef] [PubMed]
- Ivnitski, D.; Abdel-Hamid, I.; Atanasov, P.; Wilkins, E. Biosensors for detection of pathogenic bacteria. Biosens. Bioelectron. 1999, 14, 599–624. [Google Scholar] [CrossRef]
- Skottrup, P.D.; Nicolaisen, M.; Justesen, A.F. Towards on-site pathogen detection using antibody-based sensors. Biosens. Bioelectron. 2008, 24, 339–348. [Google Scholar] [CrossRef]
- Carvalho, F.; George, J.; Sheikh, H.M.A.; Selvin, R. Advances in screening, detection and enumeration of Escherichia coli using nanotechnology-based methods: A review. J. Biomed. Nanotechnol. 2018, 14, 829–846. [Google Scholar] [CrossRef]
- Ates, B.; Koytepe, S.; Ulu, A.; Gurses, C.; Thakur, V.K. Chemistry, Structures, and Advanced Applications of Nanocomposites from Biorenewable Resources. Chem. Rev. 2020, 120, 9304–9362. [Google Scholar] [CrossRef]
- Morsy, M.K.; Khalaf, H.H.; Sharoba, A.M.; El-Tanahi, H.H.; Cutter, C.N. Incorporation of essential oils and nanoparticles in pullulan films to control foodborne pathogens on meat and poultry products. J. Food Sci. 2014, 79, M675–M684. [Google Scholar] [CrossRef]
- Paredes, D.; Ortiz, C.; Torres, R. Synthesis, characterization, and evaluation of antibacterial effect of Ag nanoparticles against Escherichia coli O157: H7 and methicillin-resistant Staphylococcus aureus (MRSA). Int. J. Nanomed. 2014, 9, 1717. [Google Scholar]
- Smekalova, M.; Aragon, V.; Panacek, A.; Prucek, R.; Zboril, R.; Kvitek, L. Enhanced antibacterial effect of antibiotics in combination with silver nanoparticles against animal pathogens. Vet. J. 2016, 209, 174–179. [Google Scholar] [CrossRef]
- Hari, N.; Thomas, T.K.; Nair, A.J. Comparative Study on the Synergistic Action of Garlic Synthesized and Citrate Capped Silver Nanoparticles with β-Penem Antibiotics. Int. Sch. Res. Not. 2013, 2013, 792105. [Google Scholar] [CrossRef]
- Fang, J.; Zhong, C.; Mu, R. The study of deposited silver particulate films by simple method for efficient SERS. Chem. Phys. Lett. 2005, 401, 271–275. [Google Scholar] [CrossRef]
- Abdel-Azeem, A.; Nada, A.A.; O’donovan, A.; Thakur, V.K.; Elkelish, A. Mycogenic Silver Nanoparticles From Endophytic Trichoderma Atroviride with Antimicrobial Activity. J. Renew. Mater. 2020, 8, 171–185. [Google Scholar] [CrossRef]
- Shakeri, S.; Ashrafizadeh, M.; Zarrabi, A.; Roghanian, R.; Afshar, E.G.; Pardakhty, A.; Mohammadinejad, R.; Kumar, A.; Thakur, V.K. Multifunctional Polymeric Nanoplatforms for Brain Diseases Diagnosis, Therapy and Theranostics. Biomedicines 2020, 8, 13. [Google Scholar] [CrossRef]
- Li, P.; Li, J.; Wu, C.; Wu, Q.; Li, J. Synergistic antibacterial effects of β-lactam antibiotic combined with silver nanoparticles. Nanotechnology 2005, 16, 1912. [Google Scholar] [CrossRef]
- Oprea, O.; Andronescu, E.; Ficai, D.; Ficai, A.; N Oktar, F.; Yetmez, M. ZnO applications and challenges. Curr. Org. Chem. 2014, 18, 192–203. [Google Scholar] [CrossRef]
- Yang, S.-C.; Aljuffali, I.A.; Sung, C.T.; Lin, C.-F.; Fang, J.-Y. Antimicrobial activity of topically-applied soyaethyl morpholinium ethosulfate micelles against Staphylococcus species. Nanomedicine 2016, 11, 657–671. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Xia, X.-X.; Zhou, H.; Jin, S.-Q.; Lu, Y.-Y.; Liu, H.; Gao, M.-L.; Jin, Z.-B. COCO enhances the efficiency of photoreceptor precursor differentiation in early human embryonic stem cell-derived retinal organoids. Stem Cell Res. Ther. 2020, 11, 366. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Shandilya, P.; Saini, N.K.; Singh, P.; Thakur, V.K.; Saini, R.V.; Mittal, D.; Chandan, G.; Saini, V.; Saini, A.K. Insights into the Synthesis and Mechanism of Green Synthesized Antimicrobial Nanoparticles, Answer to the Multidrug Resistance. Mater. Today Chem. 2021, 19, 100391. [Google Scholar] [CrossRef]
- Sudhaik, A.; Raizada, P.; Thakur, S.; Saini, R.V.; Saini, A.K.; Singh, P.; Kumar Thakur, V.; Nguyen, V.-H.; Khan, A.A.P.; Asiri, A.M. Synergistic Photocatalytic Mitigation of Imidacloprid Pesticide and Antibacterial Activity Using Carbon Nanotube Decorated Phosphorus Doped Graphitic Carbon Nitride Photocatalyst. J. Taiwan Inst. Chem. Eng. 2020, 113, 142–154. [Google Scholar] [CrossRef]
- Nazaripour, E.; Mousazadeh, F.; Moghadam, M.D.; Najafi, K.; Borhani, F.; Sarani, M.; Ghasemi, M.; Rahdar, A.; Iravani, S.; Khatami, M. Biosynthesis of lead oxide and cerium oxide nanoparticles and their cytotoxic activities against colon cancer cell line. Inorg. Chem. Commun. 2021, 131, 108800. [Google Scholar] [CrossRef]
- M Balasubramaniam, B.; Prateek; Ranjan, S.; Saraf, M.; Kar, P.; Singh, S.P.; Thakur, V.K.; Singh, A.; Gupta, R.K. Antibacterial and Antiviral Functional Materials: Chemistry and Biological Activity toward Tackling COVID-19-like Pandemics. ACS Pharmacol. Transl. Sci. 2021, 4, 8–54. [Google Scholar] [CrossRef]
- Siwal, S.S.; Zhang, Q.; Saini, A.K.; Thakur, V.K. Antimicrobial Materials: New Strategies to Tackle Various Pandemics. J. Renew. Mater. 2020, 8, 1543–1563. [Google Scholar] [CrossRef]
- Arshad, R.; Pal, K.; Sabir, F.; Rahdar, A.; Bilal, M.; Shahnaz, G.; Kyzas, G.Z. A review of the nanomaterials use for the diagnosis and therapy of salmonella typhi. J. Mol. Struct. 2021, 1230, 129928. [Google Scholar] [CrossRef]
- Hakami, T.M.; Davarpanah, A.; Rahdar, A.; Barrett, S. Structural and magnetic study and cytotoxicity evaluation of tetra-metallic nanoparticles of Co0. 5Ni0. 5CrxFe2-xO4 prepared by co-precipitation. J. Mol. Struct. 2018, 1165, 344–348. [Google Scholar] [CrossRef]
- Hasanein, P.; Rahdar, A.; Bahabadi, S.E.; Kumar, A.; Kyzas, G.Z. Manganese/cerium nanoferrites: Synthesis and toxicological effects by intraperitoneal administration in rats. Inorg. Chem. Commun. 2021, 125, 108433. [Google Scholar] [CrossRef]
- Mohammadi, L.; Pal, K.; Bilal, M.; Rahdar, A.; Fytianos, G.; Kyzas, G.Z. Green nanoparticles to treat patients from Malaria disease: An overview. J. Mol. Struct. 2021, 1299, 129857. [Google Scholar] [CrossRef]
- Nikazar, S.; Sivasankarapillai, V.S.; Rahdar, A.; Gasmi, S.; Anumol, P.; Shanavas, M.S. Revisiting the cytotoxicity of quantum dots: An in-depth overview. Biophys. Rev. 2020, 12, 703–718. [Google Scholar] [CrossRef] [PubMed]
- Pillai, A.M.; Sivasankarapillai, V.S.; Rahdar, A.; Joseph, J.; Sadeghfar, F.; Rajesh, K.; Kyzas, G.Z. Green synthesis and characterization of zinc oxide nanoparticles with antibacterial and antifungal activity. J. Mol. Struct. 2020, 1211, 128107. [Google Scholar] [CrossRef]
- Rahdar, A.; Aliahmad, M.; Samani, M.; HeidariMajd, M.; Susan, M.A.B.H. Synthesis and characterization of highly efficacious Fe-doped ceria nanoparticles for cytotoxic and antifungal activity. Ceram. Int. 2019, 45, 7950–7955. [Google Scholar] [CrossRef]
- Rahdar, A.; Beyzaei, H.; Askari, F.; Kyzas, G.Z. Gum-based cerium oxide nanoparticles for antimicrobial assay. Appl. Phys. A 2020, 126, 324. [Google Scholar] [CrossRef]
- Rahdar, A.; Hajinezhad, M.R.; Hamishekar, H.; Ghamkhari, A.; Kyzas, G.Z. Copolymer/graphene oxide nanocomposites as potential anticancer agents. Polym. Bull. 2020, 78, 4877–4898. [Google Scholar] [CrossRef]
- Rahdar, A.; Taboada, P.; Aliahmad, M.; Hajinezhad, M.R.; Sadeghfar, F. Iron oxide nanoparticles: Synthesis, physical characterization, and intraperitoneal biochemical studies in Rattus norvegicus. J. Mol. Struct. 2018, 1173, 240–245. [Google Scholar] [CrossRef]
- Taimoory, S.M.; Rahdar, A.; Aliahmad, M.; Sadeghfar, F.; Hajinezhad, M.R.; Jahantigh, M.; Shahbazi, P.; Trant, J.F. The synthesis and characterization of a magnetite nanoparticle with potent antibacterial activity and low mammalian toxicity. J. Mol. Liq. 2018, 265, 96–104. [Google Scholar] [CrossRef]
- Naravaneni, R.; Jamil, K. Rapid detection of food-borne pathogens by using molecular techniques. J. Med. Microbiol. 2005, 54, 51–54. [Google Scholar] [CrossRef]
- Mukherjee, S.; Sau, S.; Madhuri, D.; Bollu, V.S.; Madhusudana, K.; Sreedhar, B.; Banerjee, R.; Patra, C.R. Green synthesis and characterization of monodispersed gold nanoparticles: Toxicity study, delivery of doxorubicin and its bio-distribution in mouse model. J. Biomed. Nanotechnol. 2016, 12, 165–181. [Google Scholar] [CrossRef]
- Bhatt, K.D.; Vyas, D.J.; Makwana, B.A.; Darjee, S.M.; Jain, V.K.; Shah, H. Turn-on fluorescence probe for selective detection of Hg (II) by calixpyrrole hydrazide reduced silver nanoparticle: Application to real water sample. Chin. Chem. Lett. 2016, 27, 731–737. [Google Scholar] [CrossRef]
- Zou, Q.; Xing, P.; Wei, L.; Liu, B. Gene2vec: Gene subsequence embedding for prediction of mammalian N6-methyladenosine sites from mRNA. RNA 2019, 25, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Xu, T.; Huang, G.; Li, Q.; Luo, L.; Zhao, Y.; Wu, Z.; Ou-Yang, J.; Yang, X.; Xie, M. Design and Fabrication of a Novel Dual-Frequency Confocal Ultrasound Transducer for Microvessels Super-Harmonic Imaging. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2020, 68, 1272–1277. [Google Scholar] [CrossRef]
- Badoei-Dalfard, A.; Sohrabi, N.; Karami, Z.; Sargazi, G. Fabrication of an efficient and sensitive colorimetric biosensor based on Uricase/Th-MOF for uric acid sensing in biological samples. Biosens. Bioelectron. 2019, 141, 111420. [Google Scholar] [CrossRef]
- Heo, J.; Hua, S.Z. An overview of recent strategies in pathogen sensing. Sensors 2009, 9, 4483–4502. [Google Scholar] [CrossRef] [PubMed]
- Nugen, S.; Baeumner, A. Trends and opportunities in food pathogen detection. Anal. Bioanal. Chem. 2008, 391, 451–454. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Raizada, P.; Verma, N.; Hosseini-Bandegharaei, A.; Thakur, V.K.; Le, Q.V.; Nguyen, V.-H.; Selvasembian, R.; Singh, P. Recent Advances on Water Disinfection Using Bismuth Based Modified Photocatalysts: Strategies and Challenges. J. Clean. Prod. 2021, 297, 126617. [Google Scholar] [CrossRef]
- Sharma, P.C.; Sharma, D.; Sharma, A.; Saini, N.; Goyal, R.; Ola, M.; Chawla, R.; Thakur, V.K. Hydrazone Comprising Compounds as Promising Anti-Infective Agents: Chemistry and Structure-Property Relationship. Mater. Today Chem. 2020, 18, 100349. [Google Scholar] [CrossRef]
- Belgrader, P.; Benett, W.; Hadley, D.; Richards, J.; Stratton, P.; Mariella, R.; Milanovich, F. PCR detection of bacteria in seven minutes. Science 1999, 284, 449–450. [Google Scholar] [CrossRef]
- Váradi, L.; Luo, J.L.; Hibbs, D.E.; Perry, J.D.; Anderson, R.J.; Orenga, S.; Groundwater, P.W. Methods for the detection and identification of pathogenic bacteria: Past, present, and future. Chem. Soc. Rev. 2017, 46, 4818–4832. [Google Scholar] [CrossRef]
- Hameed, S.; Xie, L.; Ying, Y. Conventional and emerging detection techniques for pathogenic bacteria in food science: A review. Trends Food Sci. Technol. 2018, 81, 61–73. [Google Scholar] [CrossRef]
- Ahmed, A.; Rushworth, J.V.; Hirst, N.A.; Millner, P.A. Biosensors for whole-cell bacterial detection. Clin. Microbiol. Rev. 2014, 27, 631. [Google Scholar] [CrossRef] [PubMed]
- Daminabo, S.C.; Goel, S.; Grammatikos, S.A.; Nezhad, H.Y.; Thakur, V.K. Fused Deposition Modeling-Based Additive Manufacturing (3D Printing): Techniques for Polymer Material Systems. Mater. Today Chem. 2020, 16, 100248. [Google Scholar] [CrossRef]
- Zhao, V.X.T.; Wong, T.I.; Zheng, X.T.; Tan, Y.N.; Zhou, X. Colorimetric biosensors for point-of-care virus detections. Mater. Sci. Energy Technol. 2020, 3, 237–249. [Google Scholar] [CrossRef]
- Stringer, R.C.; Schommer, S.; Hoehn, D.; Grant, S.A. Development of an optical biosensor using gold nanoparticles and quantum dots for the detection of porcine reproductive and respiratory syndrome virus. Sens. Actuators B Chem. 2008, 134, 427–431. [Google Scholar] [CrossRef]
- Li, F.; Zhao, Q.; Wang, C.; Lu, X.; Li, X.-F.; Le, X.C. Detection of Escherichia coli O157: H7 using gold nanoparticle labeling and inductively coupled plasma mass spectrometry. Anal. Chem. 2010, 82, 3399–3403. [Google Scholar] [CrossRef]
- Wang, L.; Wei, Q.; Wu, C.; Ji, J.; Liu, Q.; Yang, M.; Wang, P. Detection of E. coli O157: H7 DNA by a novel QCM biosensor coupled with gold nanoparticles amplification. In Proceedings of the 2007 7th IEEE Conference on Nanotechnology (IEEE NANO), Hong Kong, China, 2–5 August 2007; pp. 330–333. [Google Scholar]
- Peng, H.; Chen, I.A. Rapid colorimetric detection of bacterial species through the capture of gold nanoparticles by chimeric phages. ACS Nano 2018, 13, 1244–1252. [Google Scholar] [CrossRef] [PubMed]
- Raj, P.; Oh, M.H.; Han, K.; Lee, T.Y. Label-Free Electrochemical Biosensor Based on Au@ MoS₂–PANI for Escherichia coli Detection. Chemosensors 2021, 9, 49. [Google Scholar] [CrossRef]
- Ropero-Vega, J.L.; Redondo-Ortega, J.F.; Galvis-Curubo, Y.J.; Rondón-Villarreal, P.; Flórez-Castillo, J.M. A Bioinspired Peptide in TIR Protein as Recognition Molecule on Electrochemical Biosensors for the Detection of E. coli O157: H7 in an Aqueous Matrix. Molecules 2021, 26, 2559. [Google Scholar] [CrossRef]
- Pao, Y.-P.; Yu, C.-C.; Lin, Y.-Z.; Chatterjee, S.; Saha, S.; Tiwari, N.; Huang, Y.-T.; Wu, C.-C.; Choi, D.; Lin, Z.-H. Carbohydrate-protein interactions studied by solid-liquid contact electrification and its use for label-free bacterial detection. Nano Energy 2021, 85, 106008. [Google Scholar] [CrossRef]
- Hwang, C.S.H.; Ahn, M.-s.; Jeong, K.-H. Extraordinary sensitivity enhancement of Ag-Au alloy nanohole arrays for label-free detection of Escherichia coli. Biomed. Opt. Express 2021, 12, 2734–2743. [Google Scholar] [CrossRef]
- Imran, M.; Ehrhardt, C.J.; Bertino, M.F.; Shah, M.R.; Yadavalli, V.K. Chitosan Stabilized Silver Nanoparticles for the Electrochemical Detection of Lipopolysaccharide: A Facile Biosensing Approach for Gram-Negative Bacteria. Micromachines 2020, 11, 413. [Google Scholar] [CrossRef]
- Wu, D.; Wang, D.; Ye, X.; Yuan, K.; Xie, Y.; Li, B.; Huang, C.; Kuang, T.; Yu, Z.; Chen, Z. Fluorescence detection of Escherichia coli on mannose modified ZnTe quantum dots. Chin. Chem. Lett. 2020, 31, 1504–1507. [Google Scholar] [CrossRef]
- Lee, I.; Jun, S. Simultaneous detection of E. coli K12 and S. aureus Using a Continuous Flow Multijunction Biosensor. J. Food Sci. 2016, 81, N1530–N1536. [Google Scholar] [CrossRef]
- Bruce, J.A.; Clapper, J.C. Conjugation of Carboxylated Graphene Quantum Dots with Cecropin P1 for Bacterial Biosensing Applications. ACS Omega 2020, 5, 26583–26591. [Google Scholar] [CrossRef]
- Muniandy, S.; Teh, S.J.; Thong, K.L.; Thiha, A.; Dinshaw, I.J.; Lai, C.W.; Ibrahim, F.; Leo, B.F. Carbon nanomaterial-based electrochemical biosensors for foodborne bacterial detection. Crit. Rev. Anal. Chem. 2019, 49, 510–533. [Google Scholar] [CrossRef]
- Nißler, R.; Bader, O.; Dohmen, M.; Walter, S.G.; Noll, C.; Selvaggio, G.; Groß, U.; Kruss, S. Remote near infrared identification of pathogens with multiplexed nanosensors. Nat. Commun. 2020, 11, 5995. [Google Scholar] [CrossRef]
- Kaur, H.; Shorie, M.; Sabherwal, P. Electrochemical aptasensor using boron-carbon nanorods decorated by nickel nanoparticles for detection of E. coli O157: H7. Microchim. Acta 2020, 187, 461. [Google Scholar] [CrossRef]
- Kreno, L.E.; Leong, K.; Farha, O.K.; Allendorf, M.; Van Duyne, R.P.; Hupp, J.T. Metal–organic framework materials as chemical sensors. Chem. Rev. 2012, 112, 1105–1125. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Bhardwaj, S.K.; Sharma, A.L.; Kim, K.-H.; Deep, A. Development of an advanced electrochemical biosensing platform for E. coli using hybrid metal-organic framework/polyaniline composite. Environ. Res. 2019, 171, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Jenie, S.; Kusumastuti, Y.; Krismastuti, F.S.; Untoro, Y.M.; Dewi, R.T.; Udin, L.Z.; Artanti, N. Rapid Fluorescence Quenching Detection of Escherichia coli Using Natural Silica-Based Nanoparticles. Sensors 2021, 21, 881. [Google Scholar] [CrossRef] [PubMed]
- Yuhana Ariffin, E.; Heng, L.Y.; Tan, L.L.; Abd Karim, N.H.; Hasbullah, S.A. A highly sensitive impedimetric DNA biosensor based on hollow silica microspheres for label-free determination of E. coli. Sensors 2020, 20, 1279. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Lum, J.; Callaway, Z.; Lin, J.; Bottje, W.; Li, Y. A label-free impedance immunosensor using screen-printed interdigitated electrodes and magnetic nanobeads for the detection of E. coli O157: H7. Biosensors 2015, 5, 791–803. [Google Scholar] [CrossRef]
- Varshney, M.; Li, Y. Interdigitated array microelectrode based impedance biosensor coupled with magnetic nanoparticle–antibody conjugates for detection of Escherichia coli O157: H7 in food samples. Biosens. Bioelectron. 2007, 22, 2408–2414. [Google Scholar] [CrossRef]
- Lee, N.; Choi, S.-W.; Chang, H.-J.; Chun, H.S. Rapid Detection of Escherichia coli O157: H7 in Fresh Lettuce Based on Localized Surface Plasmon Resonance Combined with Immunomagnetic Separation. J. Food Prot. 2018, 81, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Wan, Y.; Qi, P.; Sun, Y.; Zhang, D.; Yu, L. Lectin functionalized ZnO nanoarrays as a 3D nano-biointerface for bacterial detection. Talanta 2017, 167, 600–606. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Liang, T.; Zhu, P.; Chen, Y.; Chen, W.; Du, L.; Wu, C.; Wang, P. Label-Free Detection of E. coli O157: H7 DNA Using Light-Addressable Potentiometric Sensors with Highly Oriented ZnO Nanorod Arrays. Sensors 2019, 19, 5473. [Google Scholar] [CrossRef] [PubMed]
- Yadav, N.; Chhillar, A.K.; Rana, J.S. Detection of pathogenic bacteria with special emphasis to biosensors integrated with gold nanoparticles. Sens. Int. 2020, 1, 100028. [Google Scholar] [CrossRef]
- Naidoo, R.; Singh, A.; Arya, S.K.; Beadle, B.; Glass, N.; Tanha, J.; Szymanski, C.M.; Evoy, S. Surface-immobilization of chromatographically purified bacteriophages for the optimized capture of bacteria. Bacteriophage 2012, 2, 15–24. [Google Scholar] [CrossRef]
- Haes, A.J.; Van Duyne, R.P. A nanoscale optical biosensor: Sensitivity and selectivity of an approach based on the localized surface plasmon resonance spectroscopy of triangular silver nanoparticles. J. Am. Chem. Soc. 2002, 124, 10596–10604. [Google Scholar] [CrossRef]
- Pangajam, A.; Theyagarajan, K.; Dinakaran, K. Highly sensitive electrochemical detection of E. coli O157: H7 using conductive Carbon dot/ZnO nanorod/PANI composite electrode. Sens. Bio-Sens. Res. 2020, 29, 100317. [Google Scholar] [CrossRef]
- Gupta, A.; Garg, M.; Singh, S.; Deep, A.; Sharma, A.L. Highly Sensitive Optical Detection of Escherichia coli Using Terbium-Based Metal–Organic Framework. ACS Appl. Mater. Interfaces 2020, 12, 48198–48205. [Google Scholar] [CrossRef]
- Ji, X.; Wang, H.; Song, B.; Chu, B.; He, Y. Silicon nanomaterials for biosensing and bioimaging analysis. Front. Chem. 2018, 6, 38. [Google Scholar] [CrossRef]
- Liu, F.; Li, Y.; Su, X.-L.; Slavik, M.F.; Ying, Y.; Wang, J. QCM immunosensor with nanoparticle amplification for detection of Escherichia coli O157: H7. Sens. Instrum. Food Qual. Saf. 2007, 1, 161–168. [Google Scholar] [CrossRef]
- Chawich, J.; Hassen, W.M.; Elie-Caille, C.; Leblois, T.; Dubowski, J.J. Regenerable ZnO/GaAs Bulk Acoustic Wave Biosensor for Detection of Escherichia coli in “Complex” Biological Medium. Biosensors 2021, 11, 145. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Chen, Y.; Zhang, Y.; Zhang, Q.; Zhang, L. Nanoparticle-based local antimicrobial drug delivery. Adv. Drug Deliv. Rev. 2018, 127, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Bagheri, R.; Ariaii, P.; Motamedzadegan, A. Characterization, antioxidant and antibacterial activities of chitosan nanoparticles loaded with nettle essential oil. J. Food Meas. Charact. 2021, 15, 1395–1402. [Google Scholar] [CrossRef]
- Alfaro-Viquez, E.; Esquivel-Alvarado, D.; Madrigal-Carballo, S.; Krueger, C.G.; Reed, J.D. Proanthocyanidin-chitosan composite nanoparticles prevent bacterial invasion and colonization of gut epithelial cells by extra-intestinal pathogenic Escherichia coli. Int. J. Biol. Macromol. 2019, 135, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Ejaz, S.; Ihsan, A.; Noor, T.; Shabbir, S.; Imran, M. Mannose functionalized chitosan nanosystems for enhanced antimicrobial activity against multidrug resistant pathogens. Polym. Test. 2020, 91, 106814. [Google Scholar] [CrossRef]
- Liu, S.; Qiao, S.; Li, L.; Qi, G.; Lin, Y.; Qiao, Z.; Wang, H.; Shao, C. Surface charge-conversion polymeric nanoparticles for photodynamic treatment of urinary tract bacterial infections. Nanotechnology 2015, 26, 495602. [Google Scholar] [CrossRef]
- Durak, S.; Arasoglu, T.; Ates, S.C.; Derman, S. Enhanced antibacterial and antiparasitic activity of multifunctional polymeric nanoparticles. Nanotechnology 2020, 31, 175705. [Google Scholar] [CrossRef]
- Li, X.; Wang, B.; Liang, T.; Wang, R.; Song, P.; He, Y. Synthesis of cationic acrylate copolyvidone-iodine nanoparticles with double active centers and their antibacterial application. Nanoscale 2020, 12, 21940–21950. [Google Scholar] [CrossRef]
- Srisang, S.; Nasongkla, N. Spray coating of foley urinary catheter by chlorhexidine-loadedpoly (ε-caprolactone) nanospheres: Effect of lyoprotectants, characteristics, and antibacterial activity evaluation. Pharm. Dev. Technol. 2019, 24, 402–409. [Google Scholar] [CrossRef]
- Ebrahimi, S.; Farhadian, N.; Karimi, M.; Ebrahimi, M. Enhanced bactericidal effect of ceftriaxone drug encapsulated in nanostructured lipid carrier against gram-negative Escherichia coli bacteria: Drug formulation, optimization, and cell culture study. Antimicrob. Resist. Infect. Control 2020, 9, 28. [Google Scholar] [CrossRef]
- He, J.; Huang, S.; Sun, X.; Han, L.; Chang, C.; Zhang, W.; Zhong, Q. Carvacrol loaded solid lipid nanoparticles of propylene glycol monopalmitate and glyceryl monostearate: Preparation, characterization, and synergistic antimicrobial activity. Nanomaterials 2019, 9, 1162. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Yang, E.; Chang, P.-S.; Ryu, S. Preparation and characterization of endolysin-containing liposomes and evaluation of their antimicrobial activities against gram-negative bacteria. Enzym. Microb. Technol. 2019, 128, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Ragioto, D.A.; Carrasco, L.D.; Carmona-Ribeiro, A.M. Novel gramicidin formulations in cationic lipid as broad-spectrum microbicidal agents. Int. J. Nanomed. 2014, 9, 3183. [Google Scholar]
- Farhadian, N.; Karimi, M.; Porozan, S. Ceftriaxone sodium loaded onto polymer-lipid hybrid nanoparticles enhances antibacterial effect on gram-negative and gram-positive bacteria: Effects of lipid-polymer ratio on particles size, characteristics, in vitro drug release and antibacterial drug efficacy. J. Drug Deliv. Sci. Technol. 2021, 63, 102457. [Google Scholar]
- Osungunna, M.O.; Pitondo-Silva, A.; Silva, L.B.; Ré, A.C.S.; Marcato, P.D.; Massaro, T.N.C.; Polizello, A.C.M.; Aires, C.P. Effect of Chitosan-Coated Nanostructured Lipid Carrier on Escherichia coli Biofilms. BioNanoScience 2021, 11, 762–769. [Google Scholar] [CrossRef]
- Gabrielyan, L.; Hakobyan, L.; Hovhannisyan, A.; Trchounian, A. Effects of iron oxide (Fe3O4) nanoparticles on Escherichia coli antibiotic-resistant strains. J. Appl. Microbiol. 2019, 126, 1108–1116. [Google Scholar] [CrossRef]
- Li, D.; Chen, S.; Zhang, K.; Gao, N.; Zhang, M.; Albasher, G.; Shi, J.; Wang, C. The interaction of Ag2O nanoparticles with Escherichia coli: Inhibition–sterilization process. Sci. Rep. 2021, 11, 1703. [Google Scholar] [CrossRef]
- Vu, X.H.; Duong, T.T.T.; Pham, T.T.H.; Trinh, D.K.; Nguyen, X.H.; Dang, V.-S. Synthesis and study of silver nanoparticles for antibacterial activity against Escherichia coli and Staphylococcus aureus. Adv. Nat. Sci. Nanosci. Nanotechnol. 2018, 9, 025019. [Google Scholar] [CrossRef]
- Davarpanah, A.M.; Rahdar, A.; Dastnae, M.A.; Zeybek, O.; Beyzaei, H. (1-x) BaFe12O19/xCoFe2O4 hard/soft magnetic nanocomposites: Synthesis, physical characterization, and antibacterial activities study. J. Mol. Struct. 2019, 1175, 445–449. [Google Scholar] [CrossRef]
- Akbarzadeh, I.; Keramati, M.; Azadi, A.; Afzali, E.; Shahbazi, R.; Norouzian, D.; Bakhshandeh, H. Optimization, physicochemical characterization, and antimicrobial activity of a novel simvastatin nano-niosomal gel against E. coli and S. aureus. Chem. Phys. Lipids 2021, 234, 105019. [Google Scholar] [CrossRef]
- Zahi, M.R.; El Hattab, M.; Liang, H.; Yuan, Q. Enhancing the antimicrobial activity of d-limonene nanoemulsion with the inclusion of ε-polylysine. Food Chem. 2017, 221, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhong, Q. Physical and antimicrobial properties of neutral nanoemulsions self-assembled from alkaline thyme oil and sodium caseinate mixtures. Int. J. Biol. Macromol. 2020, 148, 1046–1052. [Google Scholar] [CrossRef]
- Sowińska, M.; Laskowska, A.; Guśpiel, A.; Solecka, J.; Bochynska-Czyż, M.; Lipkowski, A.W.; Trzeciak, K.; Urbanczyk-Lipkowska, Z. Bioinspired Amphiphilic Peptide Dendrimers as Specific and Effective Compounds against Drug Resistant Clinical Isolates of E. coli. Bioconjugate Chem. 2018, 29, 3571–3585. [Google Scholar] [CrossRef]
- Niaz, T.; Shabbir, S.; Noor, T.; Imran, M. Antimicrobial and antibiofilm potential of bacteriocin loaded nano-vesicles functionalized with rhamnolipids against foodborne pathogens. LWT 2019, 116, 108583. [Google Scholar] [CrossRef]
- Canciu, A.; Tertis, M.; Hosu, O.; Cernat, A.; Cristea, C.; Graur, F. Modern analytical techniques for detection of bacteria in surface and wastewaters. Sustainability 2021, 13, 7229. [Google Scholar] [CrossRef]
- Reali, S.; Najib, E.Y.; Balázs, K.E.T.; Tan, A.C.H.; Váradi, L.; Hibbs, D.E.; Groundwater, P.W. Novel diagnostics for point-of-care bacterial detection and identification. RSC Adv. 2019, 9, 21486–21497. [Google Scholar] [CrossRef]
- Wei, X.; Zhou, W.; Sanjay, S.T.; Zhang, J.; Jin, Q.; Xu, F.; Dominguez, D.C.; Li, X. Multiplexed instrument-free bar-chart spinchip integrated with nanoparticle-mediated magnetic aptasensors for visual quantitative detection of multiple pathogens. Anal. Chem. 2018, 90, 9888–9896. [Google Scholar] [CrossRef] [PubMed]
- Draz, M.S.; Lakshminaraasimulu, N.K.; Krishnakumar, S.; Battalapalli, D.; Vasan, A.; Kanakasabapathy, M.K.; Sreeram, A.; Kallakuri, S.; Thirumalaraju, P.; Li, Y. Motion-based immunological detection of Zika virus using Pt-nanomotors and a cellphone. ACS Nano 2018, 12, 5709–5718. [Google Scholar] [CrossRef]
- Wang, X.-F.; Gao, P.; Liu, Y.-F.; Li, H.-F.; Lu, F. Predicting thermophilic proteins by machine learning. Curr. Bioinform. 2020, 15, 493–502. [Google Scholar] [CrossRef]
- Niu, M.; Lin, Y.; Zou, Q. sgRNACNN: Identifying sgRNA on-target activity in four crops using ensembles of convolutional neural networks. Plant Mol. Biol. 2021, 105, 483–495. [Google Scholar] [CrossRef]
- Sun, S.; Xu, L.; Zou, Q.; Wang, G. BP4RNAseq: A babysitter package for retrospective and newly generated RNA-seq data analyses using both alignment-based and alignment-free quantification method. Bioinformatics 2021, 37, 1319–1321. [Google Scholar] [CrossRef]
- Bowman, D.M.; Ludlow, K. Assessing the impact of a for government review on the nanotechnology regulatory landscape. Monash UL Rev. 2012, 38, 168. [Google Scholar]
- Allan, J.; Belz, S.; Hoeveler, A.; Hugas, M.; Okuda, H.; Patri, A.; Rauscher, H.; Silva, P.; Slikker, W.; Sokull-Kluettgen, B. Regulatory landscape of nanotechnology and nanoplastics from a global perspective. Regul. Toxicol. Pharmacol. 2021, 122, 104885. [Google Scholar] [CrossRef]
- Mirza, M.A.; Iqbal, Z.; Mishra, H. FDC in nanotechnology: Regulatory landscape. In Nanocarriers for the Delivery of Combination Drugs; Elsevier: Amsterdam, The Netherlands, 2021; pp. 473–496. [Google Scholar]
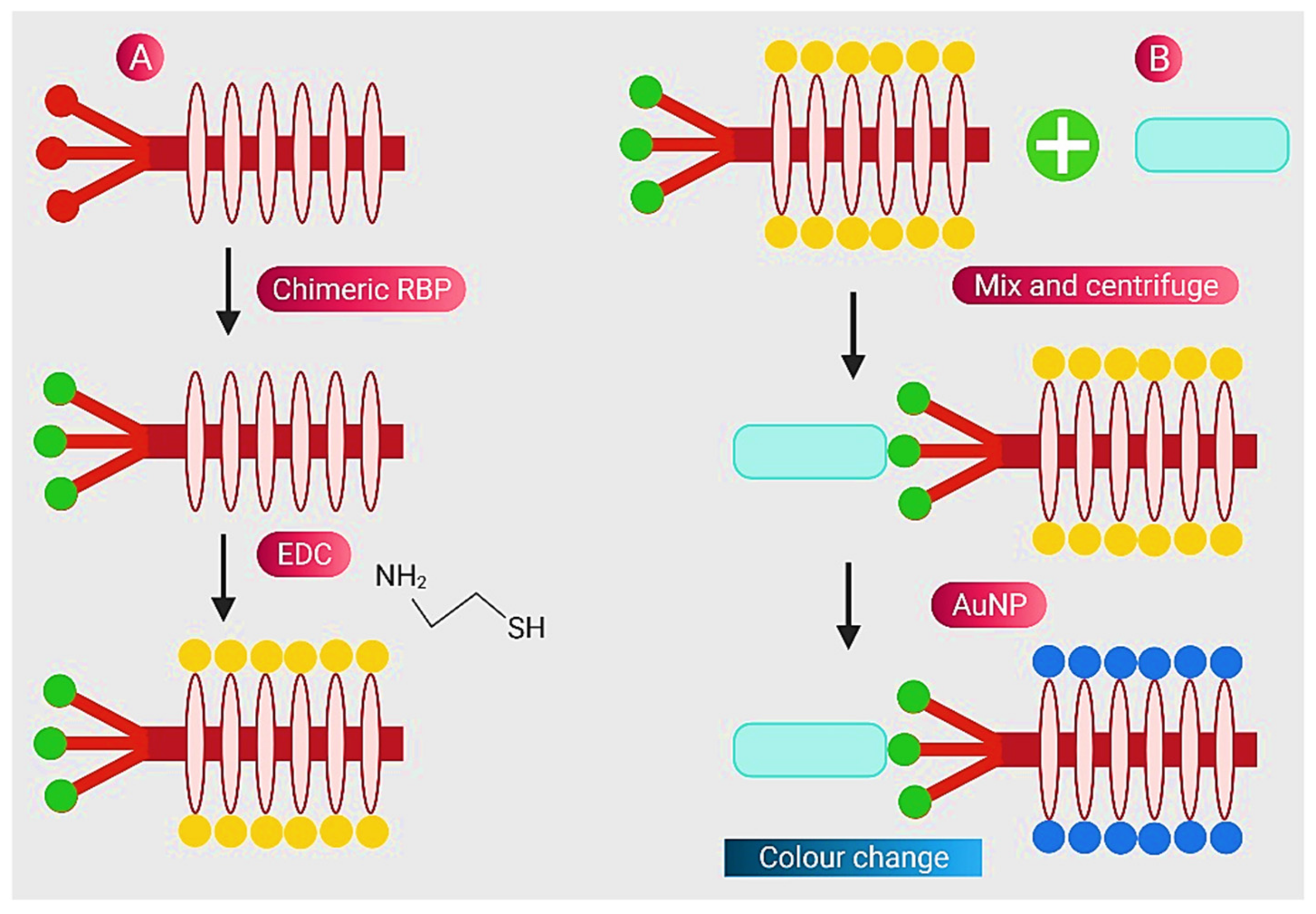
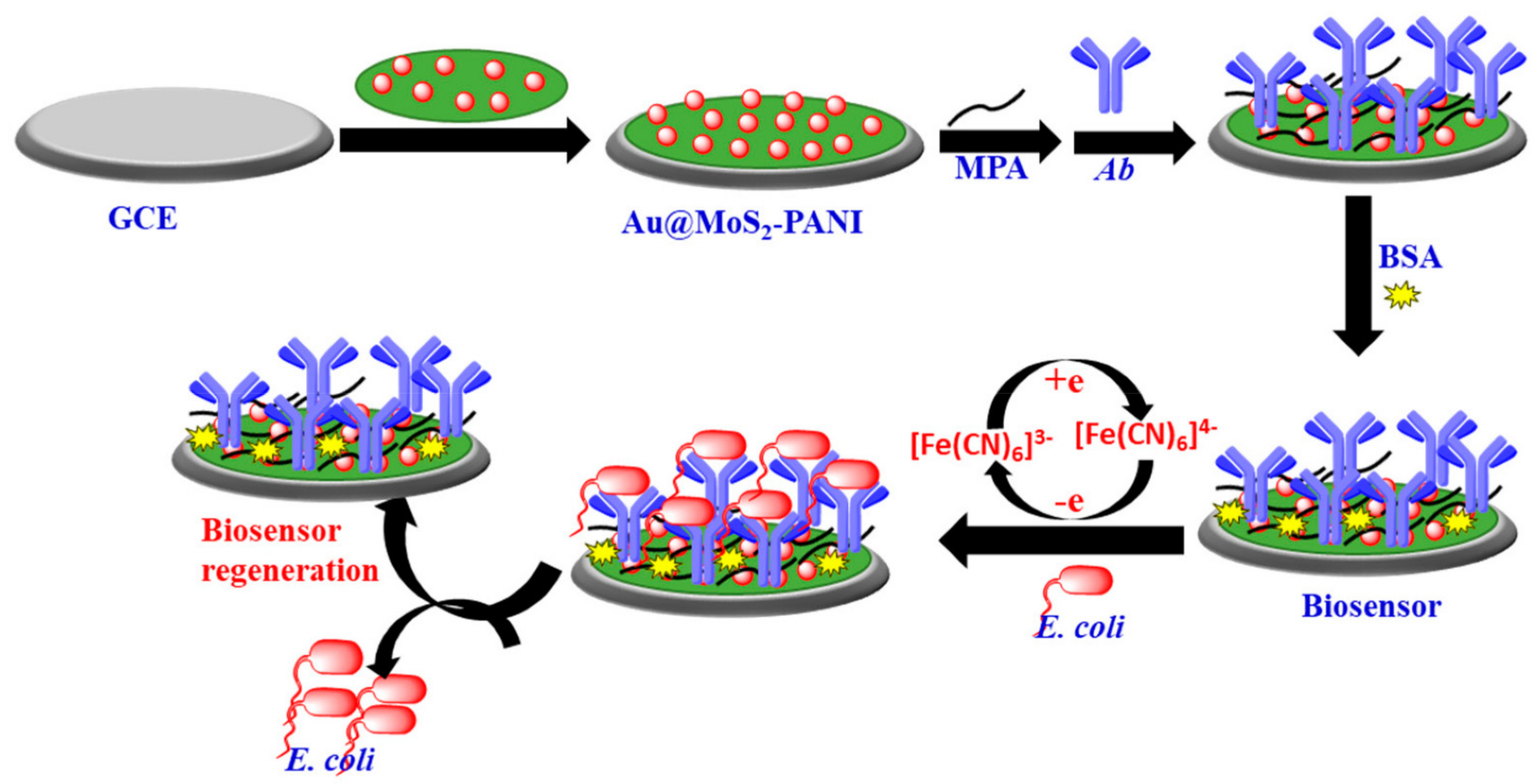
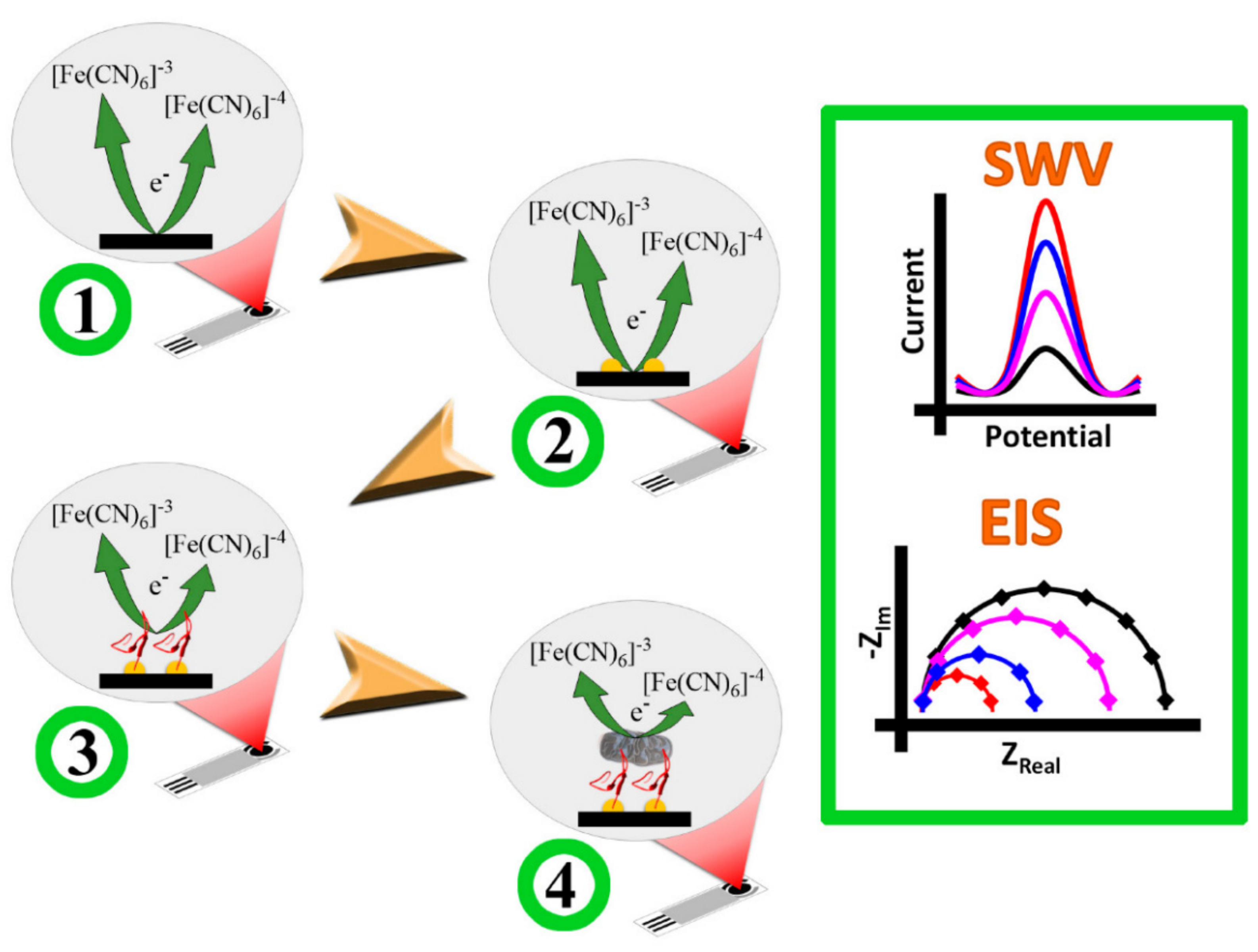
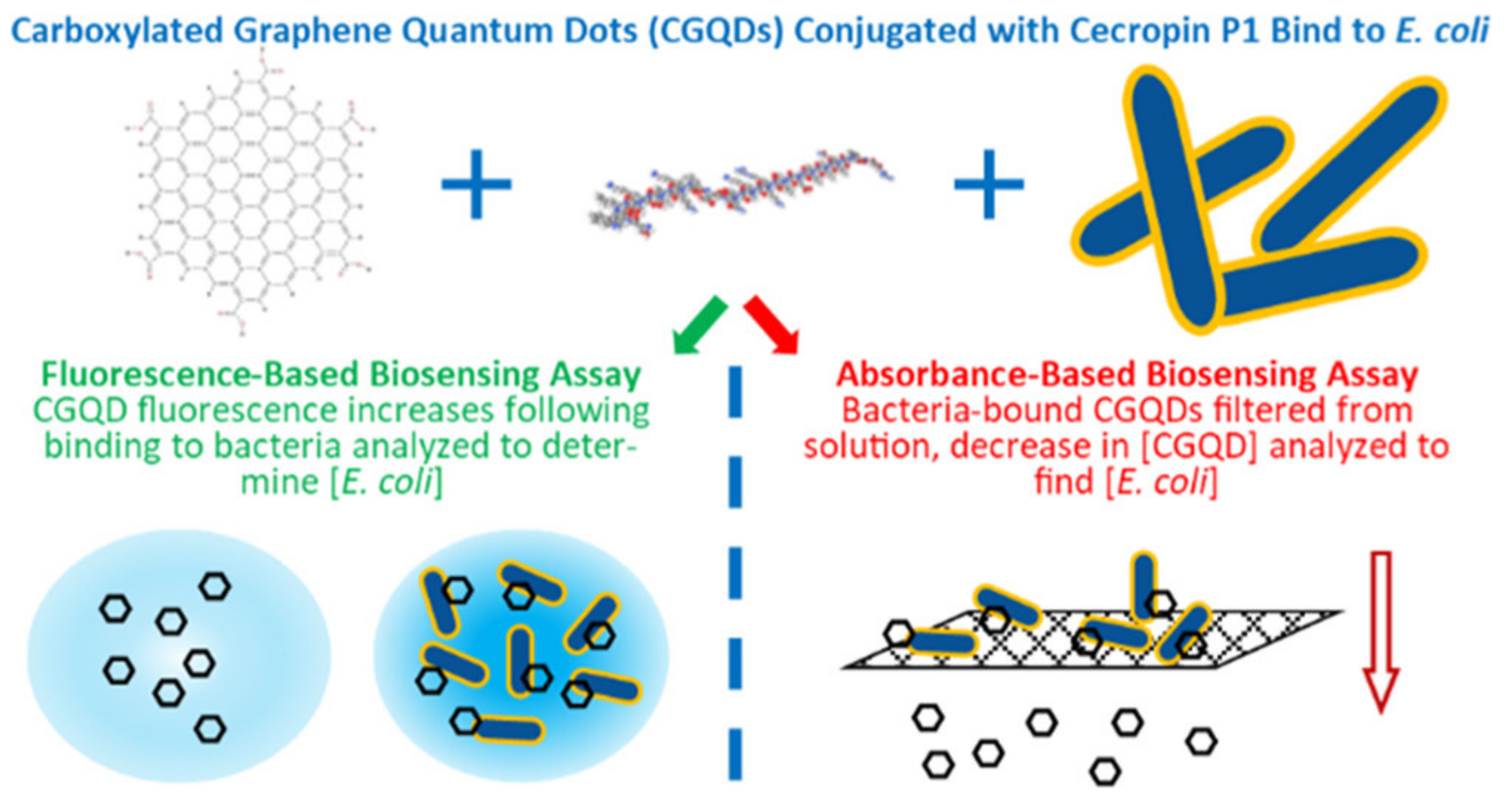

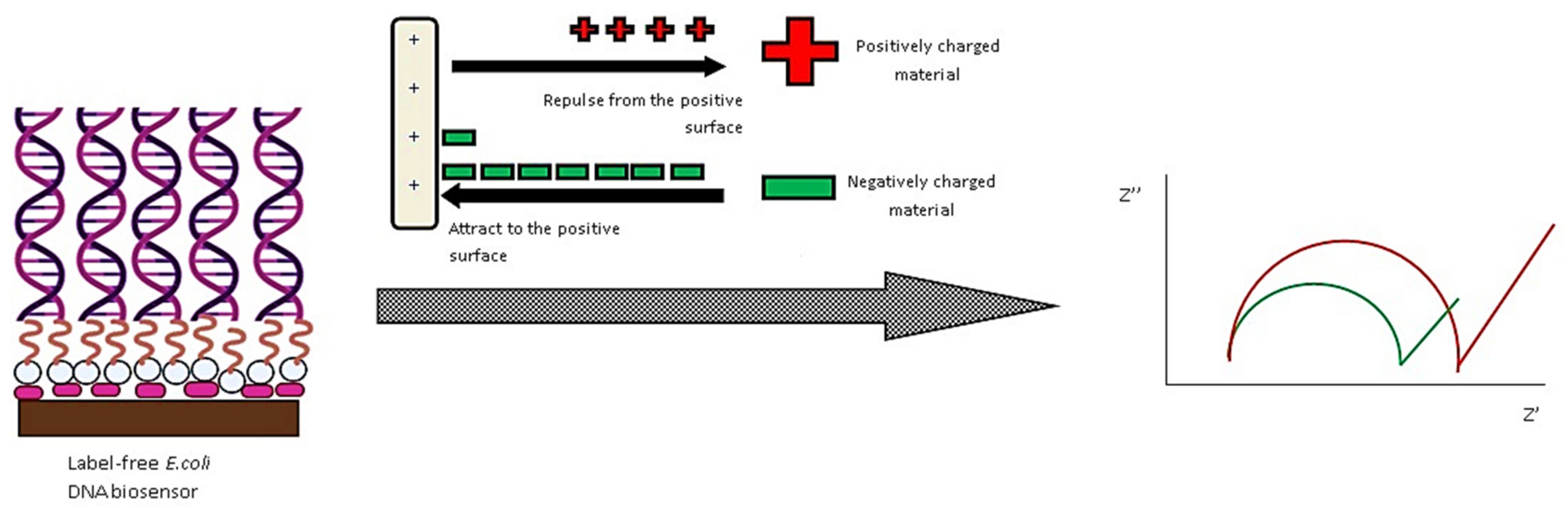


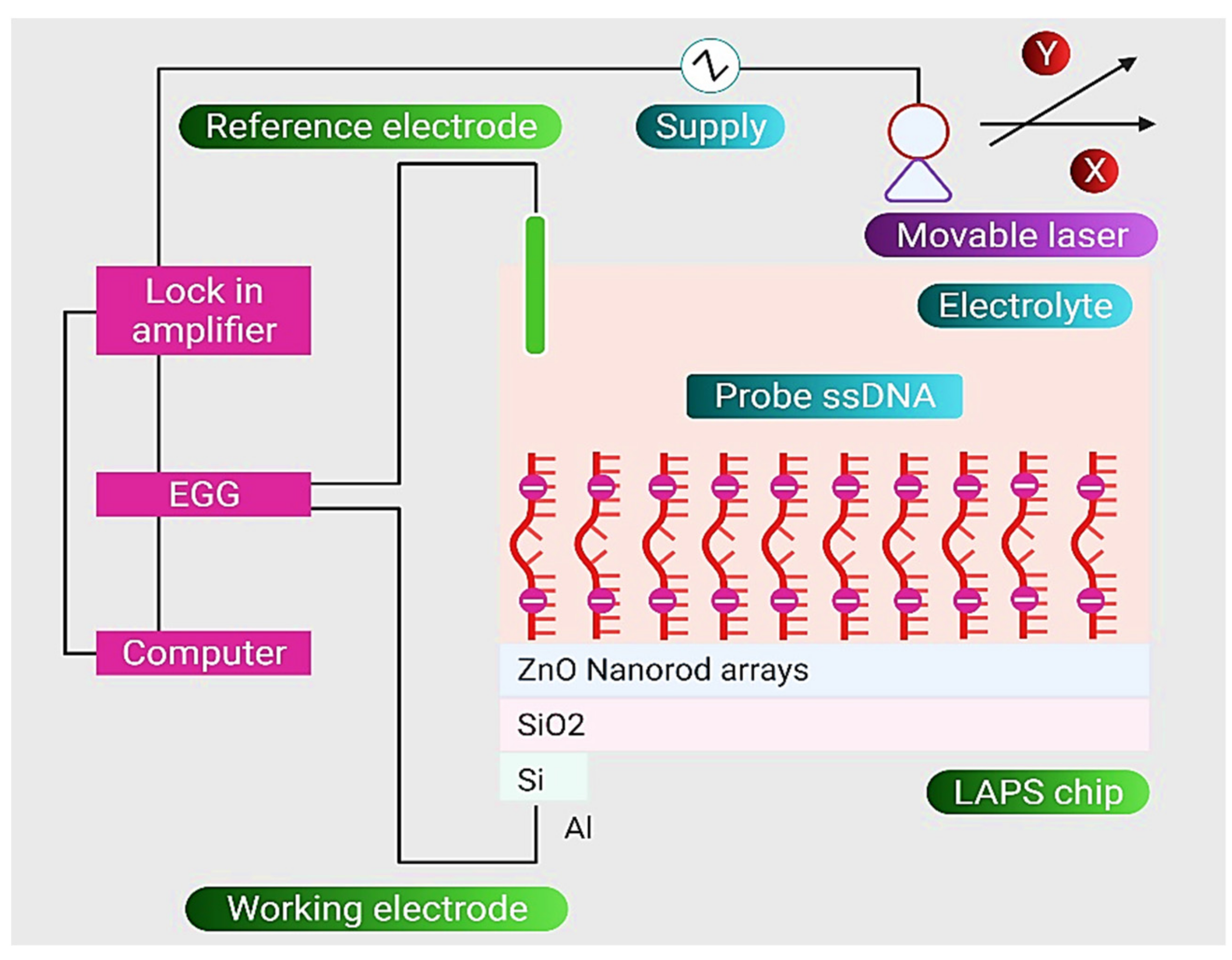


| Nanostructure | Type | Key Feature | Ref. |
|---|---|---|---|
| Au NPs | Labeled gold NPs | A perfect Detect of E. coli 500 CFU mL−1 in 1 mL of sample | [75] |
| DNA-gold NPs | Detection of bacteria with low concentration of 2 × 103 CFU mL−1 | [76] | |
| Protein-gold NPs | No cross-reactivity for Gram-negative pathogens | [77] | |
| gold@MoS₂–PANI nanocomposite | 10 CFU mL−1 for LOD in just 30 min | [78] | |
| Peptide-gold NPs | LOD and LOQ for E. coli measurement was 2 and 6 CFU mL−1, respectively | [79] | |
| Protein-gold NPs | A perfect LOD of 4 × 103 CFU mL−1, reusable potential, and wide-range analysis of 2 × 104–2 × 107 CFU mL−1 for E. coli detection | [80] | |
| Ag NPs | Ag-gold alloy nanohole arrays | Label-free detection, wide-range analysis of 103–108 CFU mL−1 and LOD of 59 CFU mL−1 | [81] |
| Polymer-Ag NPs | Wide range analysis of 0.001–100 ng mL−1 and 10–107 CFU mL−1 | [82] | |
| QDs | Mannose-ZnTe QDs | Good selectivity and a perfect linearity range of 1.0 × 105–1.0 × 108 CFU mL−1 toward E. coli | [83] |
| CdS QDs@MOF | Suitable linear range of 10–108 CFU mL−1, LOD of 3 CFU mL−1 and S/N of 3 | [84] | |
| carboxylated graphene QD | Detection of pathogen in drinking water in low concentrations of E. coli (103–106 CFU mL−1) | [85] | |
| Carbon nanomaterials | Carbon dot/ZnO/PANI | Perfect selectivity and good LOD of 1.3 × 10−18 M E. coli in water | [86] |
| SWCNTs | Detect the presence of specific bacteria based on metabolic fingerprint and differentiate among other pathogens | [87] | |
| Aptamer-BC-Ni NPs | Selective detection of E. coli with a LOD of 10 CFU mL−1 and wide range detection of 100–105 CFU mL−1 in juice, water, and fecal | [88] | |
| POE-SWCNTs | Multiplexed detection and a LOD of 102 CFU mL−1 within 2 min | [84] | |
| MOF | Tb-BTC | Wide Detection range of 1.3 × 102–1.3 × 108 CFU mL−1 and LOD of 3 CFU mL−1 | [89] |
| Cu3(BTC)2-PANI | High sensitivity, LOD of 2 CFU mL−1, and short answer time of 2 min | [90] | |
| Silica NPs | SNP-RB | Wide detection range of 10–105 CFU mL−1 and LOD of 8 CFU mL−1 | [91] |
| DNA-HSMs | Wide detection range of 1 × 10−10–1 × 10−5 µM with R2 of 0.982 and LOD of 1.95 × 10−15 µM | [92] | |
| Magnetic NPs | Antibody-MNBs | No need for pre-enrichment, LOD of 104.45 CFU mL−1 that equal to 1400 bacterial 25 μL and response time less than 60 min | [93] |
| Antibody-Fe3O4 | An LOD of 7.4 × 104 and 8.0 × 105 CFU ml−1 in pure culture and ground beef samples | [94] | |
| Fe3O4@ gold | No need for pre-enrichment and LOD of <1 log CFU mL−1 | [95] | |
| ZnO NPs | Antibody-piezoelectric ZnO | Wide linear detection of 103–107 CFU mL−1 and LOD of 103 CFU mL−1 | [96] |
| DNA-ZnO Nanorod | An LOD of 1.0 × 102 CFU mL−1 for target ssDNA of E. coli | [97] |
| No. | Nanocarriers | Drug | Size (nm) | Action | Ref. |
|---|---|---|---|---|---|
| A. | Polymeric nanocarriers | ||||
| 1. Chitosan nanocarrier | Nettle oil | 208.3–369.4 | Inhibit E. coli (zone of inhibition: 4.11–3.95 cm) | [107] | |
| 2. Chitosan nanocomposites | Cranberry proanthocyanin | 122.8 to 618.7 | Agglutination and inhibition of E. coli to invade epithelial cells | [108] | |
| 3. Mannosylated chitosan | ------ | 180 ± 5 | MIC 17.91 μg/mL against E. coli, treatment of acute cystitis in mice | [109] | |
| 4. Surface charge conversion nanocarrier | Chlorin e6 (Ce6) | 80.9 to 181.8 | Inhibit resistant E. coli strain and other bacteria, antibiofilm | [110] | |
| 5. Multifunctional PLGA NPs | Caffeic acid phenethyl and juglone | 151 to 196 | Synergistic effect in eliminating E. coli, S. aureus, and leishmania | [111] | |
| 6. Cationic acrylate copolyvidone NPs | Iodine | ~200 | Inhibit E. coli (99% efficiency) and S. aureus | [112] | |
| 7. PCL NPs | Chlorhexidine | 152 ± 37 | Prevent E. coli growth and proliferation | [113] | |
| B. | Lipidic nanocarriers | ||||
| 1. NLCs | Ceftriaxone | 86 | Inhibit E. coli | [114] | |
| 2. SLNs | Carvacrol | 14.9–25.3 | Inhibit E. coli and S. aureus | [115] | |
| 3. DPPC liposomes | BSP16Lys endolysin | 303 | Reduced E. coli CFU/mL | [116] | |
| 4. DODAB lipidic vesicle/disk | Gramicidin | 61–247 | Kill E. coli | [117] | |
| 5. Rhamnosomes nanovesicles | Nisin | 209 ± 4 | Activity against E. coli, Listeria monocytogenes, S. aureus, and P. aeruginosa biofilms | [128] | |
| C. | Metallic NPs | ||||
| 1. Iron oxide NPs | ------ | 10.64 ± 4.73 | Retard growth of E. coli antibiotic-resistant strains | [120] | |
| 2. Magnetite NPs | ------ | 19 | MIC 2.8 µg/mL against E. coli, biocompatible to organs | [57] | |
| 3. ZnO NPs | Plant extracts | 20–47 | Activity against E. coli and S. aureus | [52] | |
| 4. Ag NPs | ------ | 30 | E. coli inhibition and sterilization | [121] | |
| 5. Ag NPs | ------ | 8 ± 4 | E. coli inhibition | [122] | |
| D. | Magnetic nanoparticles | ||||
| BaFe12O19/xCoFe2O4 | ------ | 71–91 | Suitable saturation magnetization and magnetic coercivity. Effective against bacterial (E. coli) and fungal strains | [123] | |
| E. | Hybrid nanoparticles | ||||
| 1. Polymer-lipid (chitosan-glycerol monostearate) NPs | Ceftriaxone | 188–720 | Higher mortality rate of E. coli | [118] | |
| 2. Chitosan coated nanostructured lipidic carriers (NLCs) | ------ | 292.9 ± 2.5 | Inhibit E. coli biofilm formation on catheter | [119] | |
| F. | Niosomal gel | Simvastatin | 168 | Inhibit E. coli and S. aureus | [124] |
| G. | Nanoemulsion | ||||
| 1. ε-polylysine nanoemulsion | D-limonene | 12.21–15.65 | Strong synergistic action against E. coli, Bacillus subtilis, S. aureus etc. | [125] | |
| 2. Sodium caseinate based self-assembled nanoemulsion | Thyme oil | 90–200 | Antibacterial activity against E. coli and S. aureus | [126] | |
| H. | Bioinspired peptide-based dendrimers | ------ | ------ | Activity against clinical isolates of antibiotic-resistant ESBL-producing and MDR isolates of E. coli | [127] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barani, M.; Zeeshan, M.; Kalantar-Neyestanaki, D.; Farooq, M.A.; Rahdar, A.; Jha, N.K.; Sargazi, S.; Gupta, P.K.; Thakur, V.K. Nanomaterials in the Management of Gram-Negative Bacterial Infections. Nanomaterials 2021, 11, 2535. https://doi.org/10.3390/nano11102535
Barani M, Zeeshan M, Kalantar-Neyestanaki D, Farooq MA, Rahdar A, Jha NK, Sargazi S, Gupta PK, Thakur VK. Nanomaterials in the Management of Gram-Negative Bacterial Infections. Nanomaterials. 2021; 11(10):2535. https://doi.org/10.3390/nano11102535
Chicago/Turabian StyleBarani, Mahmood, Mahira Zeeshan, Davood Kalantar-Neyestanaki, Muhammad Asim Farooq, Abbas Rahdar, Niraj Kumar Jha, Saman Sargazi, Piyush Kumar Gupta, and Vijay Kumar Thakur. 2021. "Nanomaterials in the Management of Gram-Negative Bacterial Infections" Nanomaterials 11, no. 10: 2535. https://doi.org/10.3390/nano11102535
APA StyleBarani, M., Zeeshan, M., Kalantar-Neyestanaki, D., Farooq, M. A., Rahdar, A., Jha, N. K., Sargazi, S., Gupta, P. K., & Thakur, V. K. (2021). Nanomaterials in the Management of Gram-Negative Bacterial Infections. Nanomaterials, 11(10), 2535. https://doi.org/10.3390/nano11102535












