Effects of Nanopillar Size and Spacing on Mechanical Perturbation and Bactericidal Killing Efficiency
Abstract
:1. Introduction
2. Materials and Methods
2.1. Geometry and Properties of a Nanopillar
2.2. Geometry and Properties of a Gram-Negative Bacteria
2.3. Boundary Conditions of the Bacteria-Nanopattern System
2.4. Energy Considerations
2.5. Envelope Stress State at Equilibrium
2.6. Discontinuities
2.7. Comparison to Previous Experimental Results
2.8. Range of the Parametric Study
2.9. Solution and Plotting
3. Results
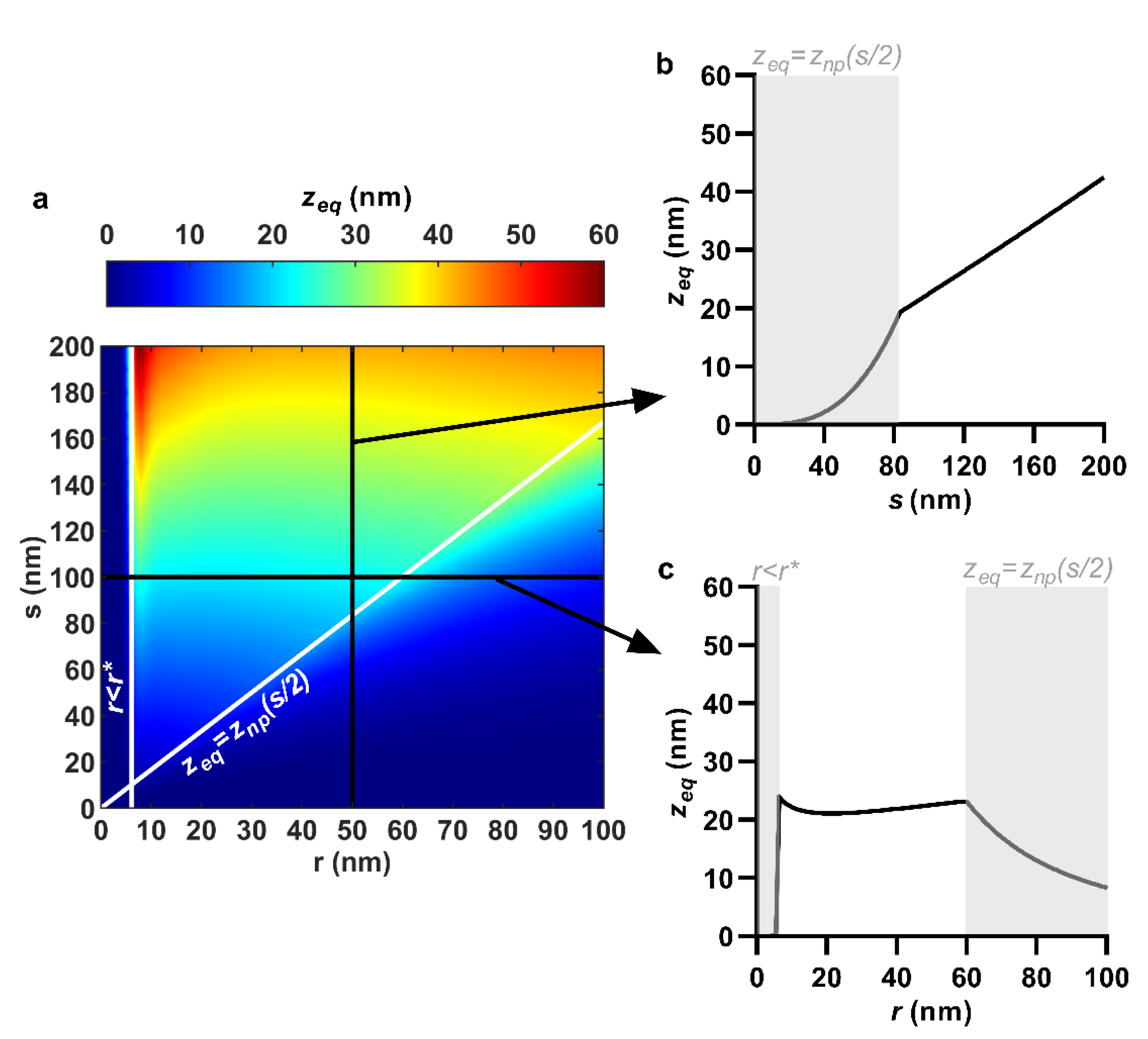
3.1. Equilibrium Sinking Depth
3.2. Areal Stress
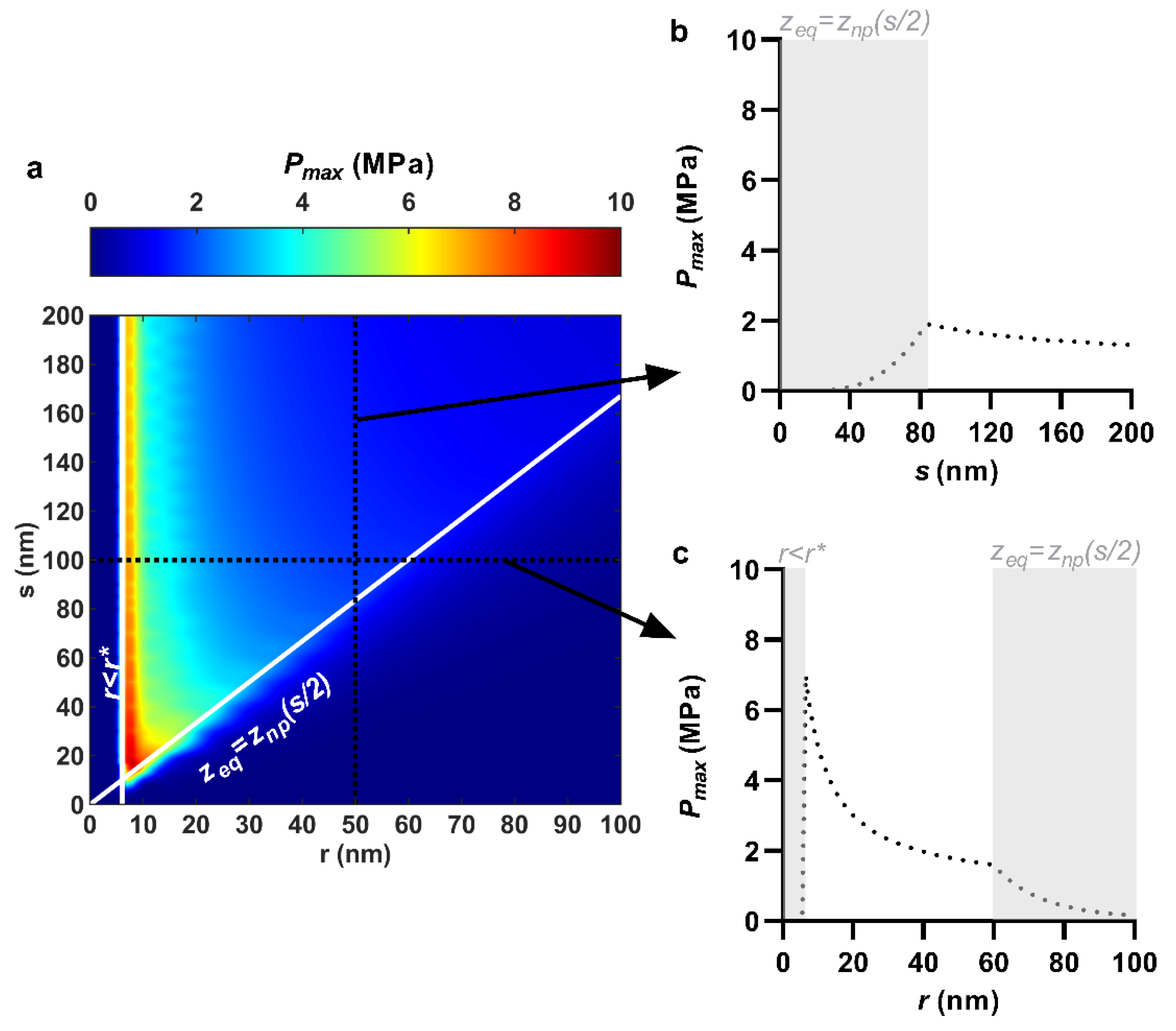
3.3. Maximum Contact Pressure
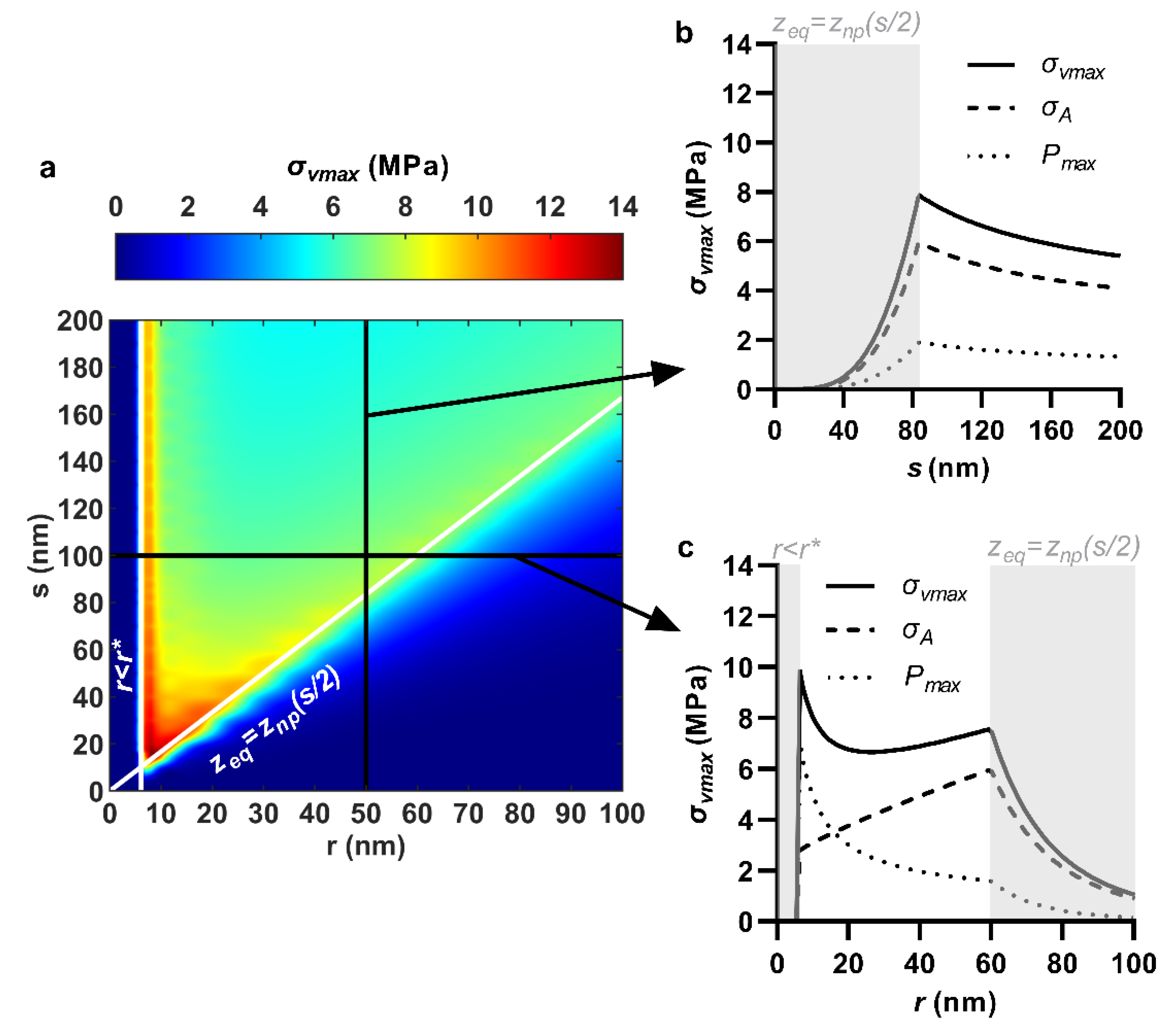
3.4. Maximum Von Mises Stress
3.5. Effects of Nanopillar Height and Pattern Ordering
3.6. Comparison to Previous Experimental Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Linklater, D.P.; Baulin, V.A.; Juodkazis, S.; Crawford, R.J.; Stoodley, P.; Ivanova, E.P. Mechano-bactericidal actions of nanostructured surfaces. Nat. Rev. Genet. 2021, 19, 8–22. [Google Scholar] [CrossRef]
- Modaresifar, K.; Azizian, S.; Ganjian, M.; Fratila-Apachitei, L.E.; Zadpoor, A.A. Bactericidal effects of nanopatterns: A systematic review. Acta Biomater. 2019, 83, 29–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanova, E.P.; Hasan, J.; Webb, H.; Truong, V.K.; Watson, G.; Watson, J.; Baulin, V.; Pogodin, S.; Wang, J.; Tobin, M.; et al. Natural Bactericidal Surfaces: Mechanical Rupture of Pseudomonas aeruginosa Cells by Cicada Wings. Small 2012, 8, 2489–2494. [Google Scholar] [CrossRef] [PubMed]
- Arias, S.L.; DeVorkin, J.; Spear, J.C.; Civantos, A.; Allain, J.P. Bacterial Envelope Damage Inflicted by Bioinspired Nanostructures Grown in a Hydrogel. ACS Appl. Bio Mater. 2020, 3, 7974–7988. [Google Scholar] [CrossRef]
- Wu, S.; Zuber, F.; Maniura-Weber, K.; Brugger, J.; Ren, Q. Nanostructured surface topographies have an effect on bactericidal activity. J. Nanobiotechnol. 2018, 16, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhadra, C.M.; Truong, V.K.; Pham, V.T.H.; Al Kobaisi, M.; Seniutinas, G.; Wang, J.; Juodkazis, S.; Crawford, R.; Ivanova, E.P. Antibacterial titanium nano-patterned arrays inspired by dragonfly wings. Sci. Rep. 2015, 5, 16817. [Google Scholar] [CrossRef] [Green Version]
- Hasan, J.; Jain, S.; Chatterjee, K. Nanoscale Topography on Black Titanium Imparts Multi-biofunctional Properties for Orthopedic Applications. Sci. Rep. 2017, 7, 41118. [Google Scholar] [CrossRef] [Green Version]
- Wandiyanto, J.V.; Cheeseman, S.; Truong, V.K.; Al Kobaisi, M.; Bizet, C.; Juodkazis, S.; Thissen, H.; Crawford, R.J.; Ivanova, E.P. Outsmarting superbugs: Bactericidal activity of nanostructured titanium surfaces against methicillin- and gentamicin-resistantStaphylococcus aureusATCC 33592. J. Mater. Chem. B 2019, 7, 4424–4443. [Google Scholar] [CrossRef]
- Hasan, J.; Jain, S.; Padmarajan, R.; Purighalla, S.; Sambandamurthy, V.K.; Chatterjee, K. Multi-scale surface topography to minimize adherence and viability of nosocomial drug-resistant bacteria. Mater. Des. 2018, 140, 332–344. [Google Scholar] [CrossRef] [PubMed]
- Bhadra, C.M.; Werner, M.; Baulin, V.A.; Truong, V.K.; Al Kobaisi, M.; Nguyen, S.H.; Balcytis, A.; Juodkazis, S.; Wang, J.Y.; Mainwaring, D.E.; et al. Subtle Variations in Surface Properties of Black Silicon Surfaces Influence the Degree of Bactericidal Efficiency. Nano-Micro Lett. 2018, 10, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Linklater, D.P.; Nguyen, H.K.D.; Bhadra, C.M.; Juodkazis, S.; Ivanova, E.P. Influence of nanoscale topology on bactericidal efficiency of black silicon surfaces. Nanotechnology 2017, 28, 245301. [Google Scholar] [CrossRef] [PubMed]
- Velic, A.; Hasan, J.; Li, Z.; Yarlagadda, P.K.D.V. Mechanics of Bacterial Interaction and Death on Nanopatterned Surfaces. Biophys. J. 2021, 120, 217–231. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, J.; Mantell, J.; Neal, C.; Gholinia, A.; Verkade, P.; Nobbs, A.H.; Su, B. Antibacterial effects of nanopillar surfaces are mediated by cell impedance, penetration and induction of oxidative stress. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Michalska, M.; Gambacorta, F.; Divan, R.; Aranson, I.S.; Sokolov, A.; Noirot, P.; Laible, P.D. Tuning antimicrobial properties of biomimetic nanopatterned surfaces. Nanoscale 2018, 10, 6639–6650. [Google Scholar] [CrossRef]
- Watson, G.S.; Green, D.W.; Watson, J.A.; Zhou, Z.; Li, X.; Cheung, G.S.P.; Gellender, M. A Simple Model for Binding and Rupture of Bacterial Cells on Nanopillar Surfaces. Adv. Mater. Interfaces 2019, 6, 1801646. [Google Scholar] [CrossRef]
- Modaresifar, K.; Kunkels, L.B.; Ganjian, M.; Tümer, N.; Hagen, C.W.; Otten, L.G.; Hagedoorn, P.-L.; Angeloni, L.; Ghatkesar, M.K.; Fratila-Apachitei, L.E.; et al. Deciphering the Roles of Interspace and Controlled Disorder in the Bactericidal Properties of Nanopatterns against Staphylococcus aureus. Nanomaterials 2020, 10, 347. [Google Scholar] [CrossRef] [Green Version]
- Pogodin, S.; Hasan, J.; Baulin, V.A.; Webb, H.; Truong, V.K.; Nguyen, T.H.P.; Boshkovikj, V.; Fluke, C.; Watson, G.; Watson, J.; et al. Biophysical Model of Bacterial Cell Interactions with Nanopatterned Cicada Wing Surfaces. Biophys. J. 2013, 104, 835–840. [Google Scholar] [CrossRef] [Green Version]
- Genova, L.A.; Roberts, M.F.; Wong, Y.-C.; Harper, C.E.; Santiago, A.G.; Fu, B.; Srivastava, A.; Jung, W.; Wang, L.M.; Łukasz, K.; et al. Mechanical stress compromises multicomponent efflux complexes in bacteria. Proc. Natl. Acad. Sci. USA 2019, 116, 25462–25467. [Google Scholar] [CrossRef]
- Nowlin, K.; Boseman, A.; Covell, A.; LaJeunesse, D. Adhesion-dependent rupturing of Saccharomyces cerevisiae on biological antimicrobial nanostructured surfaces. J. R. Soc. Interface 2015, 12, 20140999. [Google Scholar] [CrossRef] [Green Version]
- Dickson, M.N.; Liang, E.I.; Rodriguez, L.A.; Vollereaux, N.; Yee, A.F. Nanopatterned polymer surfaces with bactericidal properties. Biointerphases 2015, 10, 021010. [Google Scholar] [CrossRef] [Green Version]
- Kelleher, S.M.; Habimana, O.; Lawler, J.; Reilly, B.O.; Daniels, S.; Casey, E.; Cowley, A. Cicada Wing Surface Topography: An Investigation into the Bactericidal Properties of Nanostructural Features. ACS Appl. Mater. Interfaces 2015, 8, 14966–14974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazell, G.; Fisher, L.E.; Murray, W.A.; Nobbs, A.H.; Su, B. Bioinspired bactericidal surfaces with polymer nanocone arrays. J. Colloid Interface Sci. 2018, 528, 389–399. [Google Scholar] [CrossRef] [Green Version]
- Shahali, H.; Hasan, J.; Mathews, A.; Wang, H.; Yan, C.; Tesfamichael, T.; Yarlagadda, P.K.D.V. Multi-biofunctional properties of three species of cicada wings and biomimetic fabrication of nanopatterned titanium pillars. J. Mater. Chem. B 2019, 7, 1300–1310. [Google Scholar] [CrossRef] [PubMed]
- Li, X. Bactericidal mechanism of nanopatterned surfaces. Phys. Chem. Chem. Phys. 2016, 18, 1311–1316. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, T. Enhancement and suppression effects of a nanopatterned surface on bacterial adhesion. Phys. Rev. E 2016, 93, 052419. [Google Scholar] [CrossRef]
- Xiao, K.; Cao, X.; Chen, X.; Hu, H.; Wu, C. Bactericidal efficacy of nanopatterned surface tuned by topography. J. Appl. Phys. 2020, 128, 064701. [Google Scholar] [CrossRef]
- Xue, F.; Liu, J.; Guo, L.; Zhang, L.; Li, Q. Theoretical study on the bactericidal nature of nanopatterned surfaces. J. Theor. Biol. 2015, 385, 1–7. [Google Scholar] [CrossRef]
- Mirzaali, M.J.; Van Dongen, I.C.P.; Tumer, N.; Weinans, H.; Yavari, S.A.; Zadpoor, A.A.; Van Dongen, I. In-silico quest for bactericidal but non-cytotoxic nanopatterns. Nanotechnology 2018, 29, 43LT02. [Google Scholar] [CrossRef] [Green Version]
- Maleki, E.; Mirzaali, M.J.; Guagliano, M.; Bagherifard, S. Analyzing the mechano-bactericidal effect of nano-patterned surfaces on different bacteria species. Surf. Coat. Technol. 2021, 408, 126782. [Google Scholar] [CrossRef]
- Carniello, V.; Peterson, B.W.; van der Mei, H.C.; Busscher, H.J. Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth. Adv. Colloid Interface Sci. 2018, 261, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, E.P.; Linklater, D.P.; Werner, M.; Baulin, V.A.; Xu, X.; Vrancken, N.; Rubanov, S.; Hanssen, E.; Wandiyanto, J.; Truong, V.K.; et al. The multi-faceted mechano-bactericidal mechanism of nanostructured surfaces. Proc. Natl. Acad. Sci. USA 2020, 117, 12598–12605. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Jericho, M.; Pink, D.; Beveridge, T. Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. J. Bacteriol. 1999, 181, 6865–6875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picas, L.; Rico, F.; Scheuring, S. Direct Measurement of the Mechanical Properties of Lipid Phases in Supported Bilayers. Biophys. J. 2012, 102, L01–L03. [Google Scholar] [CrossRef] [Green Version]
- Song, F.; Lee, K.L.; Soh, A.K.; Zhu, F.; Bai, Y.L. Experimental studies of the material properties of the forewing of cicada(Homóptera, Cicàdidae). J. Exp.Biol. 2004, 207, 3035–3042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pianigiani, M.; Kirchner, R.; Sovernigo, E.; Pozzato, A.; Tormen, M.; Schift, H. Effect of nanoimprint on the elastic modulus of PMMA: Comparison between standard and ultrafast thermal NIL. Microelectron. Eng. 2016, 155, 85–91. [Google Scholar] [CrossRef]
- Kim, J.-H.; Mun, C.; Ma, J.; Park, S.-G.; Lee, S.; Kim, C.S. Simple Fabrication of Transparent, Colorless, and Self-Disinfecting Polyethylene Terephthalate Film via Cold Plasma Treatment. Nanomaterials 2020, 10, 949. [Google Scholar] [CrossRef]
- Hasan, J.; Webb, H.; Truong, V.K.; Pogodin, S.; Baulin, V.; Watson, G.S.; Watson, J.; Crawford, R.; Ivanova, E.P. Selective bactericidal activity of nanopatterned superhydrophobic cicada Psaltoda claripennis wing surfaces. Appl. Microbiol. Biotechnol. 2012, 97, 9257–9262. [Google Scholar] [CrossRef]
- Hwang, H.; Paracini, N.; Parks, J.; Lakey, J.; Gumbart, J.C. Distribution of mechanical stress in the Escherichia coli cell envelope. Biochim. Biophys. Acta (BBA)-Biomembr. 2018, 1860, 2566–2575. [Google Scholar] [CrossRef]
- Rojas, E.R.; Billings, G.; Odermatt, P.D.; Auer, G.K.; Zhu, L.; Miguel, A.; Chang, F.; Weibel, D.B.; Theriot, J.A.; Huang, K.C. The outer membrane is an essential load-bearing element in Gram-negative bacteria. Nat. Cell Biol. 2018, 559, 617–621. [Google Scholar] [CrossRef]
- Samsudin, F.; Boags, A.; Piggot, T.J.; Khalid, S. Braun’s Lipoprotein Facilitates OmpA Interaction with the Escherichia coli Cell Wall. Biophys. J. 2017, 113, 1496–1504. [Google Scholar] [CrossRef] [Green Version]
- Erickson, H.P. How bacterial cell division might cheat turgor pressure—A unified mechanism of septal division in Gram-positive and Gram-negative bacteria. BioEssays 2017, 39, 1700045. [Google Scholar] [CrossRef]
- Agudo-Canalejo, J.; Discher, D.E. Biomembrane Adhesion to Substrates Topographically Patterned with Nanopits. Biophys. J. 2018, 115, 1292–1306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deserno, M. Fluid lipid membranes: From differential geometry to curvature stresses. Chem. Phys. Lipids 2015, 185, 11–45. [Google Scholar] [CrossRef]
- Wong, F.; Amir, A. Mechanics and Dynamics of Bacterial Cell Lysis. Biophys. J. 2019, 116, 2378–2389. [Google Scholar] [CrossRef]
- Van Oss, C.J. Long-range and short-range mechanisms of hydrophobic attraction and hydrophilic repulsion in specific and aspecific interactions. J. Mol. Recognit. 2003, 16, 177–190. [Google Scholar] [CrossRef]
- Ciavarella, M.; Joe, J.; Papangelo, A.; Barber, J.R. The role of adhesion in contact mechanics. J. R. Soc. Interface 2019, 16, 20180738. [Google Scholar] [CrossRef] [Green Version]
- Norouzi, D.; Müller, M.M.; Deserno, M. How to determine local elastic properties of lipid bilayer membranes from atomic-force-microscope measurements: A theoretical analysis. Phys. Rev. E 2006, 74, 061914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Zhang, X.; Sun, J.; Dang, Z.; Li, J.; Li, X.; Chen, T. The effects of surface topography of nanostructure arrays on cell adhesion. Phys. Chem. Chem. Phys. 2018, 20, 22946–22951. [Google Scholar] [CrossRef] [PubMed]
- Canham, P. The minimum energy of bending as a possible explanation of the biconcave shape of the human red blood cell. J. Theor. Biol. 1970, 26, 61–81. [Google Scholar] [CrossRef]
- Helfrich, W. Elastic properties of lipid bilayers: Theory and possible experiments. Z. Naturforsch. C 1973, 28, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Shahane, G.; Ding, W.; Palaiokostas, M.; Orsi, M. Physical properties of model biological lipid bilayers: Insights from all-atom molecular dynamics simulations. J. Mol. Model. 2019, 25, 76. [Google Scholar] [CrossRef] [Green Version]
- Volokh, K. Modeling failure of soft anisotropic materials with application to arteries. J. Mech. Behav. Biomed. Mater. 2011, 4, 1582–1594. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, J.D.; Holzapfel, G.A. Mechanics, mechanobiology, and modeling of human abdominal aorta and aneurysms. J. Biomech. 2012, 45, 805–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valiei, A.; Lin, N.; Bryche, J.-F.; McKay, G.; Canva, M.; Charette, P.G.; Nguyen, D.; Moraes, C.; Tufenkji, N. Hydrophilic Mechano-Bactericidal Nanopillars Require External Forces to Rapidly Kill Bacteria. Nano Lett. 2020, 20, 5720–5727. [Google Scholar] [CrossRef]
- Mainwaring, D.E.; Nguyen, S.H.; Webb, H.; Jakubov, T.; Tobin, M.; Lamb, R.N.; Wu, A.; Marchant, R.; Crawford, R.; Ivanova, E.P. The nature of inherent bactericidal activity: Insights from the nanotopology of three species of dragonfly. Nanoscale 2016, 8, 6527–6534. [Google Scholar] [CrossRef] [PubMed]
- Hazell, G.; May, P.W.; Taylor, P.; Nobbs, A.; Welch, C.C.; Su, B. Studies of black silicon and black diamond as materials for antibacterial surfaces. Biomater. Sci. 2018, 6, 1424–1432. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhang, Q.; Yan, T.; Jiang, Z.; Zhang, X.; Zuo, Y.Y. Quantitatively Predicting Bacterial Adhesion Using Surface Free Energy Determined with a Spectrophotometric Method. Environ. Sci. Technol. 2015, 49, 6164–6171. [Google Scholar] [CrossRef] [Green Version]
- Park, B.-J.; Abu-Lail, N.I. The role of the pH conditions of growth on the bioadhesion of individual and lawns of pathogenic Listeria monocytogenes cells. J. Colloid Interface Sci. 2011, 358, 611–620. [Google Scholar] [CrossRef] [Green Version]
- Fialho, J.F.; Naves, E.A.; Bernardes, P.C.; Ferreira, D.C.; Dos Anjos, L.D.; Gelamo, R.V.; De Sá, J.P.; De Andrade, N.J. Stainless steel and polyethylene surfaces functionalized with silver nanoparticles. Food Sci. Technol. Int. 2018, 24, 87–94. [Google Scholar] [CrossRef]
- D’Errico, J. Surface Fitting Using Gridfit. Available online: https://www.mathworks.com/matlabcentral/fileexchange/8998-surface-fitting-using-gridfit (accessed on 10 December 2020).
- Kim, S.J.; Chang, J.; Singh, M. Peptidoglycan architecture of Gram-positive bacteria by solid-state NMR. Biochim. Biophys. Acta (BBA)-Biomembr. 2015, 1848, 350–362. [Google Scholar] [CrossRef] [Green Version]
- Loskill, P.; Pereira, P.; Jung, P.; Bischoff, M.; Herrmann, M.; Pinho, M.G.; Jacobs, K. Reduction of the Peptidoglycan Crosslinking Causes a Decrease in Stiffness of the Staphylococcus aureus Cell Envelope. Biophys. J. 2014, 107, 1082–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amir, A.; Babaeipour, F.; McIntosh, D.B.; Nelson, D.R.; Jun, S. Bending forces plastically deform growing bacterial cell walls. Proc. Natl. Acad. Sci. USA 2014, 111, 5778–5783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.E.; Zhang, Z.; Thomas, C.R.; Moxham, K.E.; Middelberg, A.P.J. The mechanical properties of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 9871–9874. [Google Scholar] [CrossRef] [Green Version]
- Thwaites, J.J.; Surana, U.C.; Jones, A.M. Mechanical properties of Bacillus subtilis cell walls: Effects of ions and lysozyme. J. Bacteriol. 1991, 173, 204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stocks, S.M.; Thomas, C.R. Viability, strength, and fragmentation of Saccharopolyspora erythraea in submerged fermentation. Biotechnol. Bioeng. 2001, 75, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Evans, E.; Heinrich, V.; Ludwig, F.; Rawicz, W. Dynamic Tension Spectroscopy and Strength of Biomembranes. Biophys. J. 2003, 85, 2342–2350. [Google Scholar] [CrossRef] [Green Version]
- Tolpekina, T.V.; Otter, W.K.D.; Briels, W.J. Simulations of stable pores in membranes: System size dependence and line tension. J. Chem. Phys. 2004, 121, 8014–8020. [Google Scholar] [CrossRef] [PubMed]
- Bavi, N.; Nakayama, Y.; Bavi, O.; Cox, C.D.; Qin, Q.; Martinac, B. Biophysical implications of lipid bilayer rheometry for mechanosensitive channels. Proc. Natl. Acad. Sci. USA 2014, 111, 13864–13869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stocks, S.M.; Thomas, C.R. Strength of mid-logarithmic and stationary phaseSaccharopolyspora erythraea hyphae during a batch fermentation in defined nitrate-limited medium. Biotechnol. Bioeng. 2001, 73, 370–378. [Google Scholar] [CrossRef]
- Harimawan, A.; Rajasekar, A.; Ting, Y.-P. Bacteria attachment to surfaces–AFM force spectroscopy and physicochemical analyses. J. Colloid Interface Sci. 2011, 364, 213–218. [Google Scholar] [CrossRef]
- Román-Kustas, J.; Hoffman, J.B.; Reed, J.H.; Gonsalves, A.E.; Oh, J.; Li, L.; Hong, S.; Jo, K.D.; Dana, C.; Miljkovic, N.; et al. Molecular and Topographical Organization: Influence on Cicada Wing Wettability and Bactericidal Properties. Adv. Mater. Interfaces 2020, 7, 2000112. [Google Scholar] [CrossRef] [Green Version]
- Diu, T.; Faruqui, N.; Sjöström, T.; Lamarre, B.; Jenkinson, H.; Su, B.; Ryadnov, M.G. Cicada-inspired cell-instructive nanopatterned arrays. Sci. Rep. 2015, 4, 7122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Cheung, G.S.; Watson, G.S.; Watson, J.A.; Lin, S.; Schwarzkopf, L.; Green, D.W. The nanotipped hairs of gecko skin and biotemplated replicas impair and/or kill pathogenic bacteria with high efficiency. Nanoscale 2016, 8, 18860–18869. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Velic, A.; Jaggessar, A.; Tesfamichael, T.; Li, Z.; Yarlagadda, P.K.D.V. Effects of Nanopillar Size and Spacing on Mechanical Perturbation and Bactericidal Killing Efficiency. Nanomaterials 2021, 11, 2472. https://doi.org/10.3390/nano11102472
Velic A, Jaggessar A, Tesfamichael T, Li Z, Yarlagadda PKDV. Effects of Nanopillar Size and Spacing on Mechanical Perturbation and Bactericidal Killing Efficiency. Nanomaterials. 2021; 11(10):2472. https://doi.org/10.3390/nano11102472
Chicago/Turabian StyleVelic, Amar, Alka Jaggessar, Tuquabo Tesfamichael, Zhiyong Li, and Prasad K. D. V. Yarlagadda. 2021. "Effects of Nanopillar Size and Spacing on Mechanical Perturbation and Bactericidal Killing Efficiency" Nanomaterials 11, no. 10: 2472. https://doi.org/10.3390/nano11102472
APA StyleVelic, A., Jaggessar, A., Tesfamichael, T., Li, Z., & Yarlagadda, P. K. D. V. (2021). Effects of Nanopillar Size and Spacing on Mechanical Perturbation and Bactericidal Killing Efficiency. Nanomaterials, 11(10), 2472. https://doi.org/10.3390/nano11102472







