Gold Nanoparticle-Assisted Virus Formation by Means of the Delivery of an Oncolytic Adenovirus Genome
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.1.1. Chemicals
2.1.2. Plasmids
2.1.3. Cell Lines
2.2. Synthesis of Nanoparticles and Complexes
2.2.1. PEI-AuNPs
2.2.2. PEI-AuNPs/DNA Complexes
2.3. Characterization of PEI-AuNPs and PEI-AuNPs/DNA Complexes
2.4. Transfection
2.4.1. Transfection Procedure
2.4.2. Evaluation of AuNPs-eGFP Transfection
2.5. Transfection of Nanoparticles Carrying Large DNA (pLR1 Plasmid)
2.5.1. Nanoparticles Characterization
2.5.2. Evaluation of AuNPs Efficacy for Large DNA Transfection
2.5.3. Transfection Efficacy of AuNPs/pLR1
2.6. Real Time PCR
2.7. Cell Viability Assay
3. Results
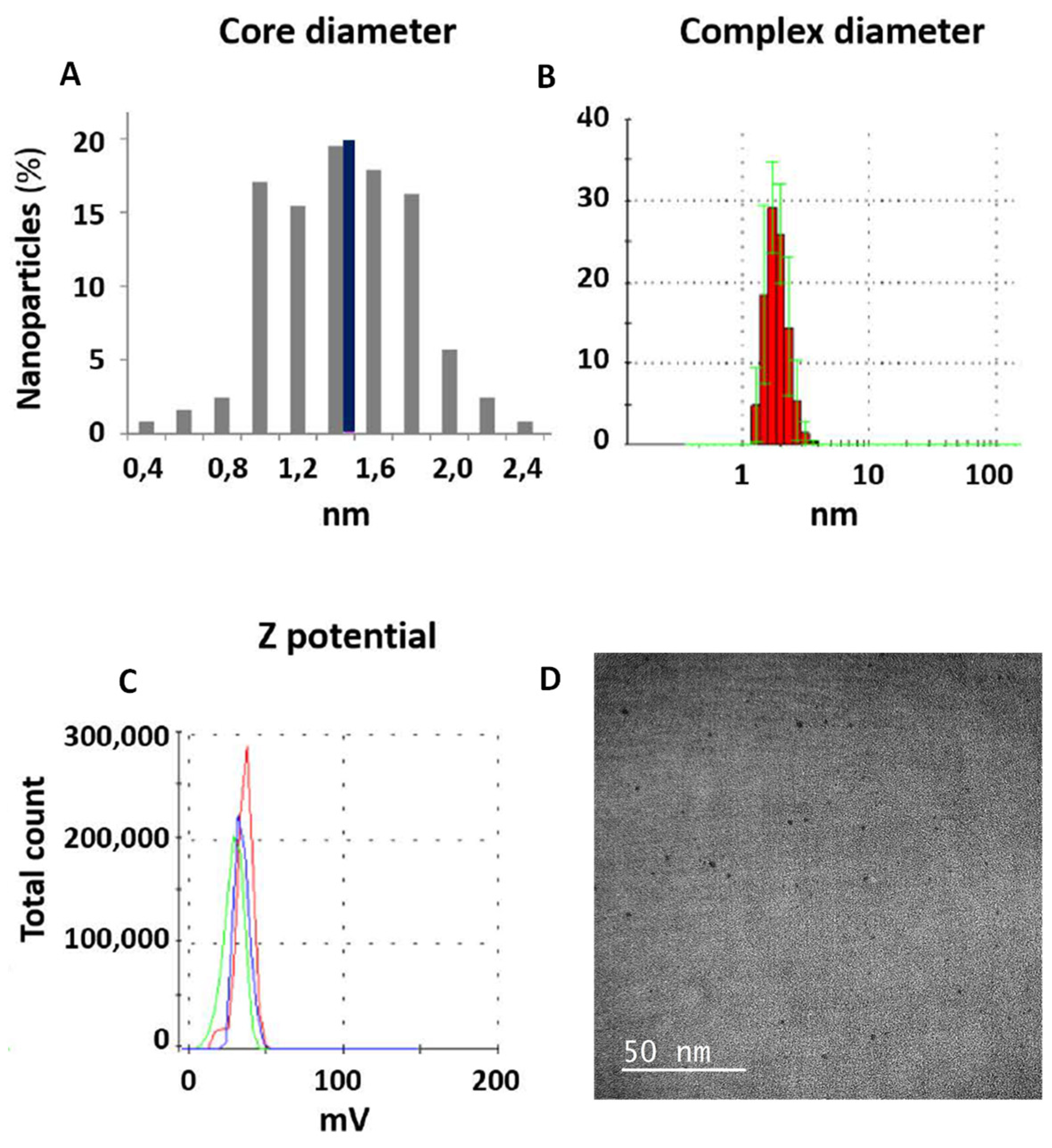
3.1. Characterization of PEI-AuNPs
3.2. Characterization of AuNPs/DNA COMPLEX USINg the p3CeGFP Small Plasmid
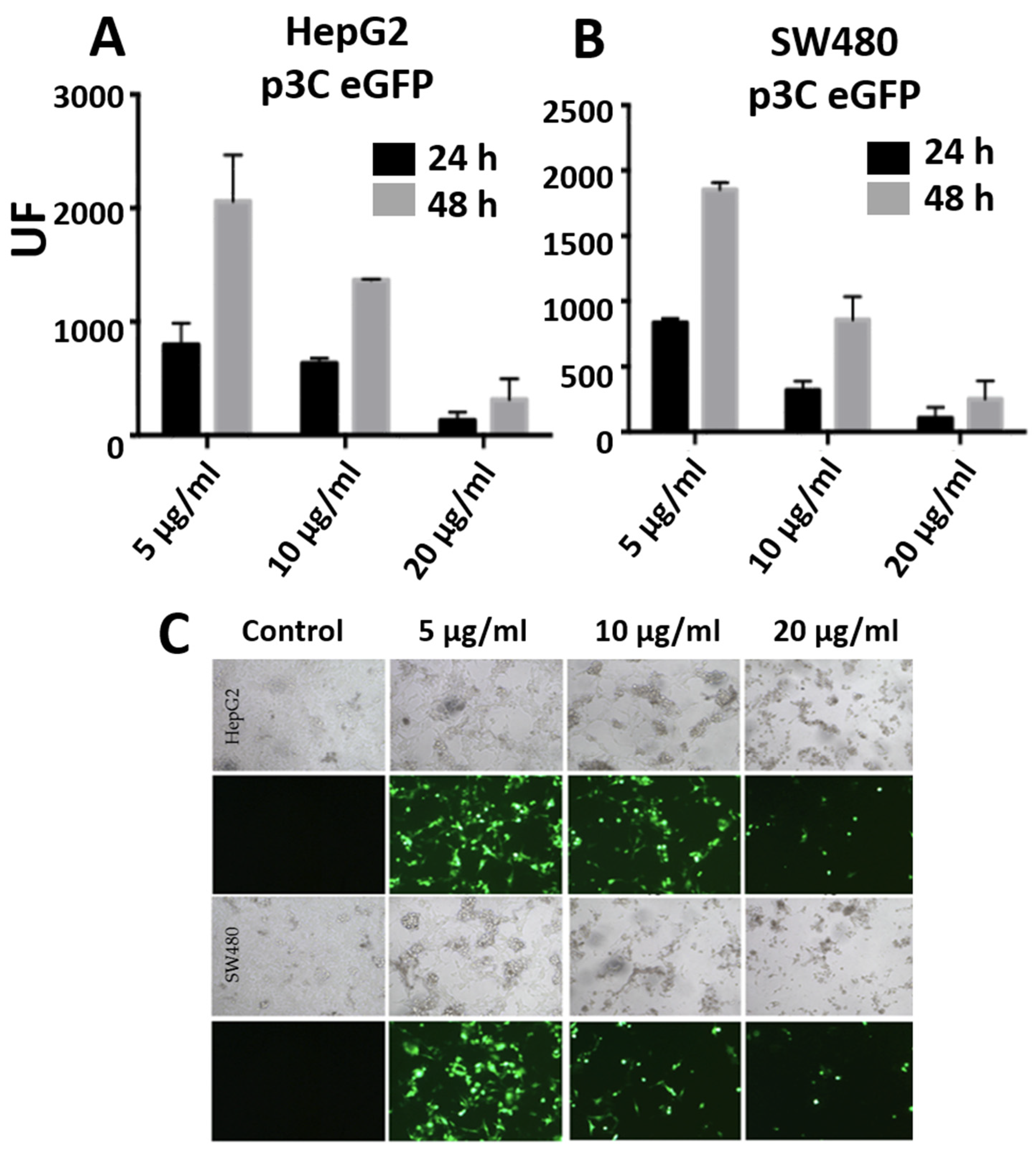
3.3. Transfections with PEI-AuNPs/p3C eGFP
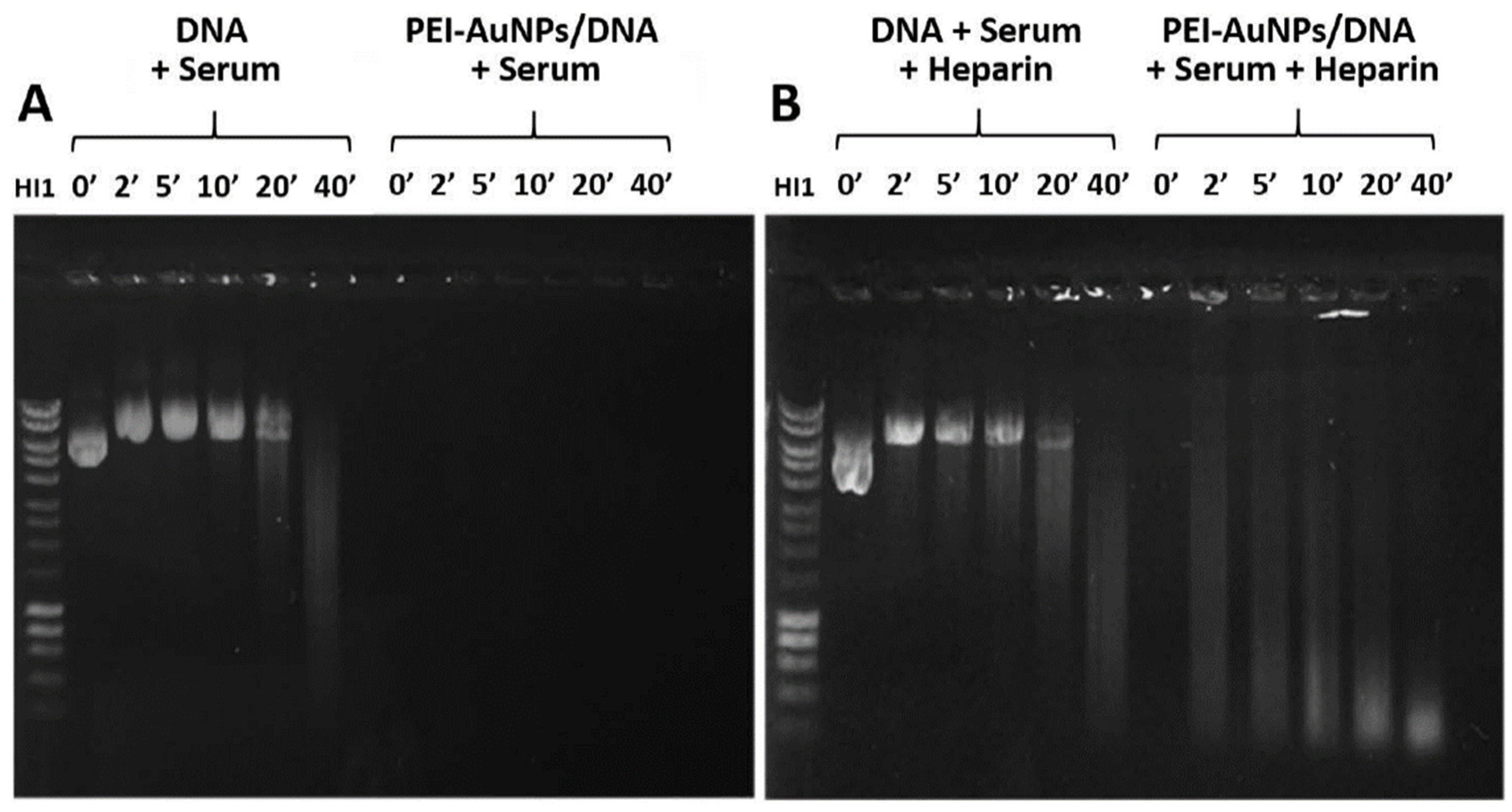
3.4. AuNPs Protection of DNA against Nucleases
3.5. Characterization of PEI-AuNPs/DNA Using an Oncolytic Adenovirus Genome Plasmid
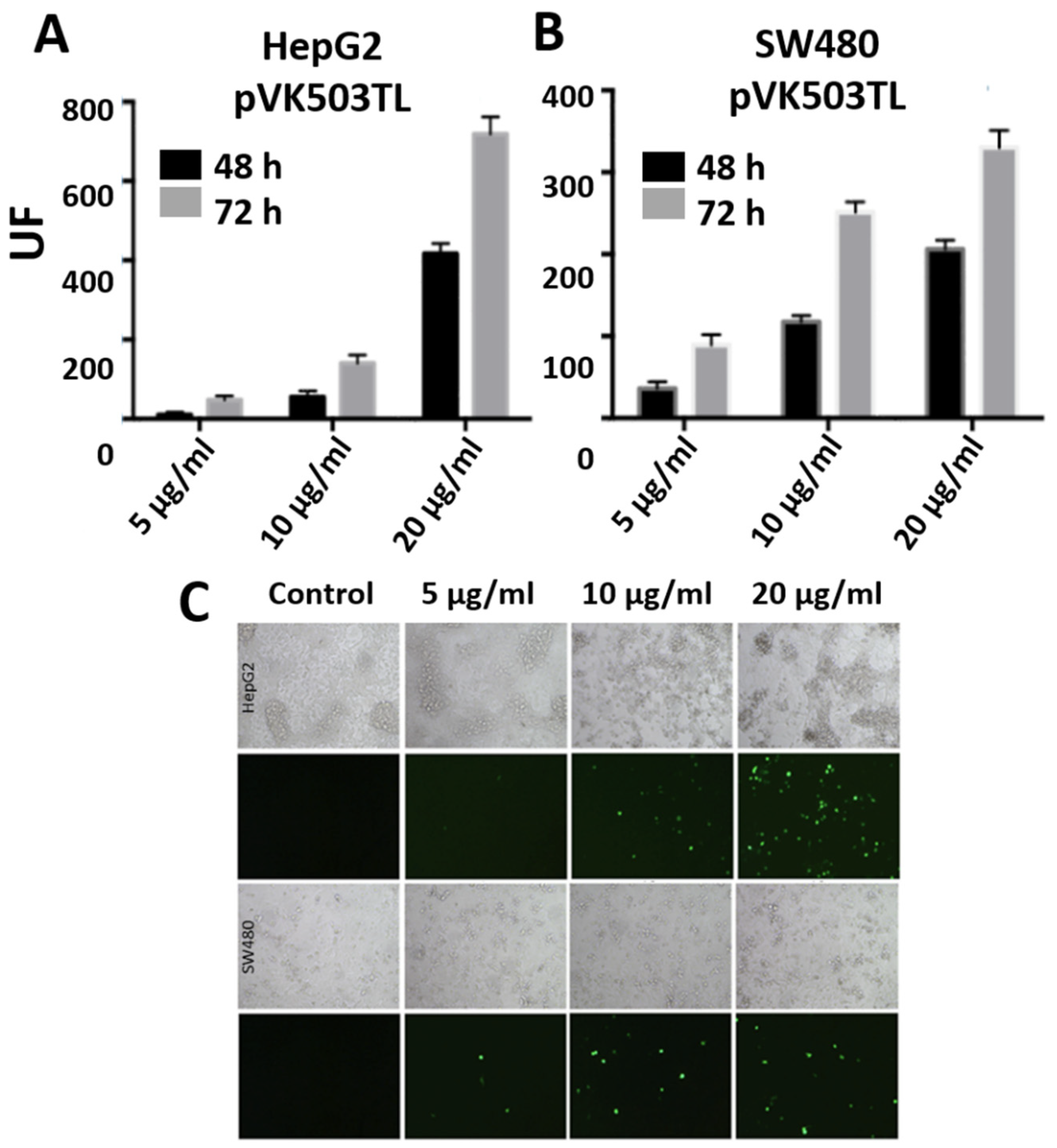
3.6. Transfections with Large Plasmids: PEI-AuNPs/pVK503TL and PEI-AuNPs/pLR1
3.7. Functional Effect of the Adenovirus Genome Expression after AuNPs-Mediated Transfection
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cebrián, V.; Martín-Saavedra, F.; Yagüe, C.; Arruebo, M.; Santamaría, J.; Vilaboa, N. Size-dependent transfection efficiency of PEI-coated gold nanoparticles. Acta Biomater. 2011, 7, 3645–3655. [Google Scholar] [CrossRef] [PubMed]
- Shukla, R.; Bansal, V.; Chaudhary, M.; Basu, A.; Bhonde, R.R.; Sastry, M. Biocompatibility of gold nanoparticles and their endocytotic fate inside the cellular compartment: A microscopic overview. Langmuir 2005, 21, 10644–10654. [Google Scholar] [CrossRef] [PubMed]
- Niidome, T.; Nakashima, K.; Takahashi, H.; Niidome, Y. Preparation of primary amine-modified gold nanoparticles and their transfection ability into cultivated cells. Chem. Commun. 2004, 17, 1978–1979. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, K.K.; McIntosh, C.M.; Simard, J.M.; Smith, S.W.; Rotello, V.M. Gold nanoparticle-mediated transfection of mammalian cells. Bioconjug Chem. 2002, 13, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.; Klibanov, A.M. Conjugation to gold nanoparticles enhances polyethylenimine’s transfer of plasmid DNA into mammalian cells. Proc. Natl. Acad. Sci. USA 2003, 100, 9138–9143. [Google Scholar] [CrossRef] [PubMed]
- Noh, S.M.; Kim, W.K.; Kim, S.J.; Kim, J.M.; Baek, K.H.; Oh, Y.K. Enhanced cellular delivery and transfection efficiency of plasmid DNA using positively charged biocompatible colloidal gold nanoparticles. Biochim. Biophys. Acta 2007, 1770, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.G.; Morse, S.V.; Copping, M.J.; Choi, J.J.; Vilar, R. Targeted Delivery of DNA-Au Nanoparticles across the Blood-Brain Barrier Using Focused Ultrasound. ChemMedChem 2018, 13, 1311–1314. [Google Scholar] [CrossRef]
- Mbatha, L.S.; Singh, M. Starburst Poly(amidoamine) Dendrimer Grafted Gold Nanoparticles as a Scaffold for Folic Acid-Targeted Plasmid DNA Delivery In Vitro. J. Nanosci. Nanotechnol. 2019, 19, 1959–1970. [Google Scholar] [CrossRef]
- Cobley, C.M.; Chen, J.; Cho, E.C.; Wang, L.V.; Xia, Y. Gold nanostructures: A class of multifunctional materials for biomedical applications. Chem. Soc. Rev. 2011, 40, 44–56. [Google Scholar] [CrossRef]
- Daniel, M.C.; Astruc, D. Gold nanoparticles: Assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology. Chem. Rev. 2004, 104, 293–346. [Google Scholar] [CrossRef]
- Cho, E.C.; Au, L.; Zhang, Q.; Xia, Y. The effects of size, shape, and surface functional group of gold nanostructures on their adsorption and internalization by cells. Small 2010, 6, 517–522. [Google Scholar] [CrossRef] [PubMed]
- Pissuwan, D.; Niidome, T.; Cortie, M.B. The forthcoming applications of gold nanoparticles in drug and gene delivery systems. J. Control. Release 2011, 149, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Rosi, N.L.; Giljohann, D.A.; Thaxton, C.S.; Lytton-Jean, A.K.R.; Han, M.S.; Mirkin, C.A. Oligonucleotidemodified gold nanoparticles for intracellular gene regulation. Science 2006, 312, 1027–1030. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.S.; Kim, C.-K.; Han, G.; Forbes, N.S.; Rotello, V.M. Efficient Gene Delivery Vectors by Tuning the Surface Charge Density of Amino Acid-Functionalized Gold Nanoparticles. Acs Nano 2008, 2, 2213–2218. [Google Scholar] [CrossRef] [PubMed]
- Massich, M.D.; Giljohann, D.A.; Seferos, D.S.; Ludlow, L.E.; Horvath, C.M.; Mirkin, C.A. Regulating Immune Response Using Polyvalent Nucleic Acid-Gold Nanoparticle Conjugates. Mol. Pharm. 2009, 6, 1934–1940. [Google Scholar] [CrossRef]
- Ryou, S.M.; Kim, S.; Jang, H.H.; Kim, J.H.; Yeom, J.H.; Eom, M.S.; Bae, J.; Han, M.S.; Lee, K. Delivery of shRNA using gold nanoparticle-DNA oligonucleotide conjugates as a universal carrier. Biochem. Biophys. Res. Commun. 2010, 398, 542–546. [Google Scholar] [CrossRef]
- Stobiecka, M.; Hepel, M. Double-shell gold nanoparticle-based DNA-carriers with poly-L-lysine binding surface. Biomaterials 2011, 32, 3312–3321. [Google Scholar] [CrossRef]
- Sharma, A.; Tandon, A.; Tovey, J.C.; Gupta, R.; Robertson, J.D.; Fortune, J.A.; Klibanov, A.M.; Cowden, J.W.; Rieger, F.G.; Mohan, R.R. Polyethylenimine-conjugated gold nanoparticles: Gene transfer potential and low toxicity in the cornea. Nanomed. Nanotechnol. Biol. Med. 2011, 7, 505–513. [Google Scholar] [CrossRef]
- Yan, X.; Blacklock, J.; Li, J.; Moehwald, H. One-Pot Synthesis of Polypeptide-Gold Nanoconjugates for in Vitro Gene Transfection. ACS Nano 2012, 6, 111–117. [Google Scholar] [CrossRef]
- Shan, Y.; Luo, T.; Peng, C.; Sheng, R.; Cao, A.; Cao, X.; Shen, M.; Guo, R.; Tomás, H.; Shi, X. Gene delivery using dendrimer-entrapped gold nanoparticles as nonviral vectors. Biomaterials 2012, 33, 3025–3035. [Google Scholar] [CrossRef]
- Trigueros, S.; Domènech, E.B.; Toulis, V.; Marfany, G. In Vitro Gene Delivery in Retinal Pigment Epithelium Cells by Plasmid DNA-Wrapped Gold Nanoparticles. Genes 2019, 10, 289. [Google Scholar] [CrossRef] [PubMed]
- Munsell, E.V.; Fang, B.; Sullivan, M.O. Histone-Mimetic Gold Nanoparticles as Versatile Scaffolds for Gene Transfer and Chromatin Analysis. Bioconjug Chem. 2018, 29, 3691–3704. [Google Scholar] [CrossRef]
- Dhanya, G.R.; Caroline, D.S.; Rekha, M.R.; Sreenivasan, K. Histidine and arginine conjugated starch-PEI and its corresponding gold nanoparticles for gene delivery. Int. J. Biol. Macromol. 2018, 120 Pt A, 999–1008. [Google Scholar] [CrossRef]
- Hersey, P.; Gallagher, S. Intralesional immunotherapy for melanoma. J. Surg. Oncol. 2014, 109, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Mastrangelo, M.J.; Maguire, H.C.; Eisenlohr, L.C.; Laughlin, C.E.; Monken, C.E.; McCue, P.A.; Kovatich, A.J.; Lattime, E.C. Intratumoral recombinant GM-CSF-encoding virus as gene therapy in patients with cutaneous melanoma. Cancer Gene Ther. 1999, 6, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Senzer, N.N.; Kaufman, H.L.; Amatruda, T.; Nemunaitis, M.; Reid, T.; Daniels, G.; Gonzalez, R.; Glaspy, J.; Whitman, E.; Harrington, K.; et al. Phase II clinical trial of a granulocyte-macrophage colony-stimulating factor-encoding, second-generation oncolytic herpesvirus in patients with unresectable metastatic melanoma. J. Clin. Oncol. 2009, 27, 5763–5771. [Google Scholar] [CrossRef]
- Goins, W.F.; Huang, S.; Cohen, J.B.; Glorioso, J.C. Engineering HSV-1 vectors for gene therapy. Methods Mol. Biol. 2014, 1144, 63–79. [Google Scholar]
- Dummer, R.; Rochlitz, C.; Velu, T.; Acres, B.; Limacher, J.M.; Bleuzen, P.; Lacoste, G.; Slos, P.; Romero, P.; Urosevic, M. Intralesional adenovirus-mediated interleukin-2 gene transfer for advanced solid cancers and melanoma. Mol. Ther. 2008, 16, 985–994. [Google Scholar] [CrossRef]
- Gupta, P.; Su, Z.Z.; Lebedeva, I.V.; Sarkar, D.; Sauane, M.; Emdad, L.; Bachelor, M.A.; Grant, S.; Curiel, D.T.; Dent, P.; et al. mda-7/IL-24, multifunctional cancer-specific apoptosis-inducing cytokine. Pharmacol. Ther. 2006, 111, 596–628. [Google Scholar] [CrossRef]
- Abbink, P.; Lemckert, A.A.; Ewald, B.A.; Lynch, D.M.; Denholtz, M.; Smits, S.; Holterman, L.; Damen, I.; Vogels, R.; Thorner, A.R.; et al. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J. Virol. 2007, 81, 4654–4663. [Google Scholar] [CrossRef]
- Mast, T.C.; Kierstead, L.; Gupta, S.B.; Nikas, A.A.; Kallas, E.G.; Novitsky, V.; Mbewe, B.; Pitisuttithum, P.; Schechter, M.; Vardas, E.; et al. International epidemiology of human pre-existing adenovirus (Ad) type-5, type-6, type-26 and type-36 neutralizing antibodies: Correlates of high Ad5 titers and implications for potential HIV vaccine trials. Vaccine 2010, 28, 950–957. [Google Scholar] [CrossRef] [PubMed]
- Barouch, D.H.; Kik, S.V.; Weverling, G.J.; Dilan, R.; King, S.L.; Maxfield, L.F.; Clark, S.; Brandariz, K.L.; Abbink, P.; Sinangil, F.; et al. International seroepidemiology of adenovirus serotypes 5, 26, 35, and 48 in pediatric and adult populations. Vaccine 2011, 29, 5203–5209. [Google Scholar] [CrossRef] [PubMed]
- Na, Y.; Nam, J.P.; Hong, J.; Oh, E.; Shin, H.C.; Kim, H.S.; Kim, S.W.; Yun, C.O. Systemic administration of human mesenchymal stromal cells infected with polymer-coated oncolytic adenovirus induces efficient pancreatic tumor homing and infiltration. J. Control. Release 2019, 305, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Kasala, D.; Yoon, A.R.; Hong, J.; Kim, S.W.; Yun, C.O. Evolving lessons on nanomaterial-coated viral vectors for local and systemic gene therapy. Nanomedicine 2016, 11, 1689–1713. [Google Scholar] [CrossRef]
- Kwon, O.J.; Kang, E.; Kim, S.; Yun, C.O. Viral genome DNA/lipoplexes elicit in situ oncolytic viral replication and potent antitumor efficacy via systemic delivery. J. Control. Release 2011, 155, 317–325. [Google Scholar] [CrossRef]
- Chieko, Y.; Katsuyuki, H.; Minako, K.; Koyama, Y. Oncolytic plasmid: A novel strategy for tumor immuno-gene therapy. Oncol. Lett. 2012, 3, 387–390. [Google Scholar]
- Rojas, J.J.; Guedan, S.; Searle, P.F.; Martinez-Quintanilla, J.; Gil-Hoyos, R.; Alcayaga-Miranda, F.; Cascallo, M.; Alemany, R. Minimal RB-responsive E1A promoter modification to attain potency, selectivity, and transgene-arming capacity in oncolytic adenoviruses. Mol. Ther. 2010, 18, 1960–1971. [Google Scholar] [CrossRef]
- Rincón, E.; Cejalvo, T.; Kanojia, D.; Alfranca, A.; Rodríguez-Milla, M.Á.; Hoyos, R.A.G.; Han, Y.; Zhang, L.; Alemany, R.; Lesniak, M.S.; et al. Mesenchymal stem cell carriers enhance antitumor efficacy of oncolytic adenoviruses in an immunocompetent mouse model. Oncotarget 2017, 8, 45415–45431. [Google Scholar] [CrossRef]
- Carette, J.E.; Graat, H.C.; Schagen, F.H.; Abou El Hassan, M.A.; Gerritsen, W.R.; van Beusechem, V.W. Replication-dependent transgene expression from a conditionally replicating adenovirus via alternative splicing to a heterologous splice-acceptor site. J. Gene Med. 2005, 7, 1053–1062. [Google Scholar] [CrossRef]
- Stanton, R.J.; McSharry, B.P.; Armstrong, M.; Tomasec, P.; Wilkinson, G.W. Re-engineering adenovirus vector systems to enable high-throughput analyses of gene function. Biotechniques 2008, 45, 659–662, 664–668. [Google Scholar] [CrossRef]
- Bayo-Puxan, N.; Cascallo, M.; Gros, A.; Huch, M.; Fillat, C.; Alemany, R. Role of the putative heparan sulfate glycosaminoglycan-binding site of the adenovirus type 5 fiber shaft on liver detargeting and knob-mediated retargeting. J. Gen Virol. 2006, 87 Pt 9, 2487–2495. [Google Scholar] [CrossRef]
- Brust, M.; Fink, J.; Bethell, D.; Schiffrin, D.J.; Kiely, C. Synthesis and reactions of functionalised gold nanoparticles. J. Chem. Soc. Chem. Commun. 1995, 16, 1655–1656. [Google Scholar] [CrossRef]
- Guillem, V.M.; Tormo, M.; Revert, F.; Benet, I.; García-Conde, J.; Crespo, A.; Aliño, S.F. Polyethyleneimine-based immunopolyplex for targeted gene transfer in human lymphoma cell lines. J. Gene Med. 2002, 4, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Stuchbury, T.; Shipton, M.I.C.H.A.E.L.; Norris, R.O.G.E.R.; Malthouse, J.P.G.; Brocklehurst, K.; Herbert, J.A.L.; Suschitzky, H. A reporter group delivery system with both absolute and selective specificity for thiol groups and an improved fluorescent probe containing the 7-nitrobenzo-2-oxa-1,3-diazole moiety. Biochem. J. 1975, 151, 417–432. [Google Scholar] [CrossRef] [PubMed]
- Moret, I.; Peris, J.E.; Guillem, V.M.; Benet, M.; Revert, F.; Dasi, F.; Crespo, A.; Aliño, S.F. Stability of PEI-DNA and DOTAP-DNA complexes: Effect of alkaline pH, heparin and serum. J. Control. Release 2001, 76, 169–181. [Google Scholar] [CrossRef]
- Lisitsyna, E.S.; Lygo, O.N.; Durandin, N.A.; Dement’eva, O.V.; Rudoi, V.M.; Kuzmin, V.A. Superquenching of SYBRGreen dye fluorescence in complex with DNA by gold nanoparticles. High Energy Chem. 2012, 46, 363–367. [Google Scholar] [CrossRef]
- Venkiteswaran, S.; Thomas, T.; Thomas, T.J. Selectivity of polyethyleneimines on DNA nanoparticle preparation and gene transport. Chem. Select. 2016, 1, 1144–1150. [Google Scholar] [CrossRef]
- Taranejoo, S.; Liu, J.; Verma, P.; Hourigan, K. A review of the developments of characteristics of PEI derivatives for gene delivery applications. J. Appl. Polym. Sci. 2015, 132, 42096. [Google Scholar] [CrossRef]
- Del Papa, J.; Parks, R.J. Adenoviral Vectors Armed with Cell Fusion-Inducing Proteins as Anti-Cancer Agents. Viruses 2017, 9, 13. [Google Scholar] [CrossRef]
- Oskuee, R.K.; Dabbaghi, M.; Gholami, L.; Taheri-Bojd, S.; Balali-Mood, M.; Mousavi, S.H.; Malaekeh-Nikouei, B. Investigating the influence of polyplex size on toxicity properties of polyethylenimine mediated gene delivery. Life Sci. 2018, 197, 101–108. [Google Scholar] [CrossRef]
- Thomas, T.J.; Tajmir-Riahi, H.A.; Pillai, C.K.S. Biodegradable Polymers for Gene Delivery. Molecules 2019, 24, 3744. [Google Scholar] [CrossRef] [PubMed]
- Miciak, J.J.; Hirshberg, J.; Bunz, F. Seamless assembly of recombinant adenoviral genomes from high-copy plasmids. PLoS ONE 2018, 13, e0199563. [Google Scholar] [CrossRef] [PubMed]



| Au-NPs | Initial Concentration PEI (mg/mL) | Core Diameter TEM (nm) | Hydrodynamic Diameter Zetasizer (nm) | Zeta Potential (mV) | Molecules PEI/Np |
|---|---|---|---|---|---|
| NP1 | 1 | 2.1 | 86.6 | 39 | 2.1 |
| NP2 | 2 | 1.8 | 6.5 | 35 | 3.8 |
| NP3 | 4 | 1.6 | 5.5 | 25 | 6.4 |
| NP4 | 8 | 1.5 | 2.1 | 34 | 9.4 |
| Au-NPs | PEI-AuNP/DNA Ratio (µg/µg) | Core Diameter TEM (nm) | Hydrodynamic Diameter Zetasizer (nm) | Zeta Potential (mV) |
|---|---|---|---|---|
| NP4 | 1:0 | 1.5 | 2.1 | 34 |
| 1:0.01 | 2.1 | 5.66 | 4.33 | |
| 1:0.07 | 2.1 | 6.97 | 11.67 | |
| 1:0.3 | 2.1 | 37.21 | 14.67 | |
| 1:0.6 | 2.1 | 49.06 | 25 | |
| 1:1.25 | 2.1 | 38.18 | 18.67 | |
| 1:2.5 | 2.1 | 66.78 | 27 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sendra, L.; Miguel, A.; Navarro-Plaza, M.C.; Herrero, M.J.; de la Higuera, J.; Cháfer-Pericás, C.; Aznar, E.; Marcos, M.D.; Martínez-Máñez, R.; Rojas, L.A.; et al. Gold Nanoparticle-Assisted Virus Formation by Means of the Delivery of an Oncolytic Adenovirus Genome. Nanomaterials 2020, 10, 1183. https://doi.org/10.3390/nano10061183
Sendra L, Miguel A, Navarro-Plaza MC, Herrero MJ, de la Higuera J, Cháfer-Pericás C, Aznar E, Marcos MD, Martínez-Máñez R, Rojas LA, et al. Gold Nanoparticle-Assisted Virus Formation by Means of the Delivery of an Oncolytic Adenovirus Genome. Nanomaterials. 2020; 10(6):1183. https://doi.org/10.3390/nano10061183
Chicago/Turabian StyleSendra, Luis, Antonio Miguel, M. Carmen Navarro-Plaza, María José Herrero, José de la Higuera, Consuelo Cháfer-Pericás, Elena Aznar, M. Dolores Marcos, Ramón Martínez-Máñez, Luis Alfonso Rojas, and et al. 2020. "Gold Nanoparticle-Assisted Virus Formation by Means of the Delivery of an Oncolytic Adenovirus Genome" Nanomaterials 10, no. 6: 1183. https://doi.org/10.3390/nano10061183
APA StyleSendra, L., Miguel, A., Navarro-Plaza, M. C., Herrero, M. J., de la Higuera, J., Cháfer-Pericás, C., Aznar, E., Marcos, M. D., Martínez-Máñez, R., Rojas, L. A., Alemany, R., & Aliño, S. F. (2020). Gold Nanoparticle-Assisted Virus Formation by Means of the Delivery of an Oncolytic Adenovirus Genome. Nanomaterials, 10(6), 1183. https://doi.org/10.3390/nano10061183






