Physico-Chemically Distinct Nanomaterials Synthesized from Derivates of a Poly(Anhydride) Diversify the Spectrum of Loadable Antibiotics
Abstract
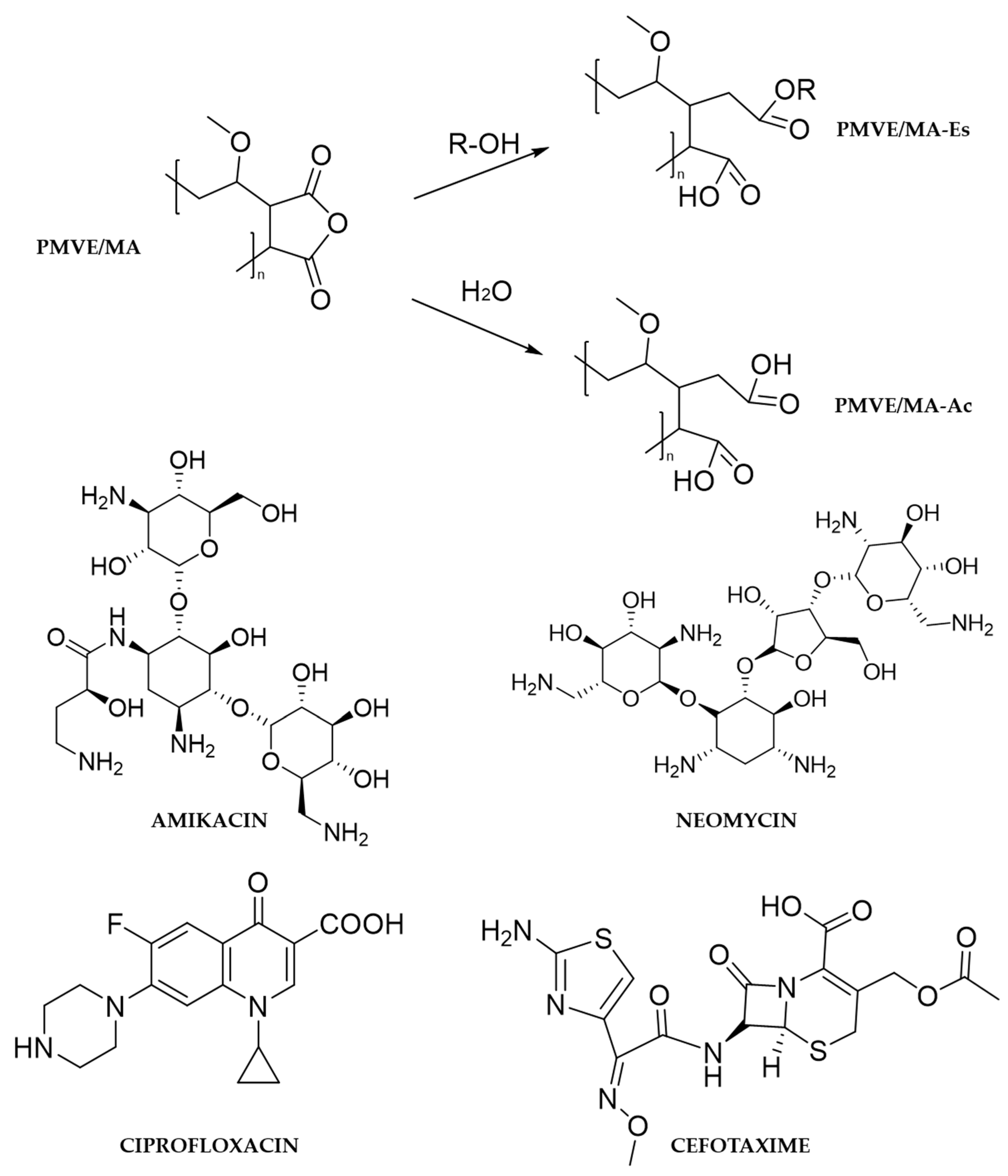
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Nanofibers by Electrospinning
2.3. Preparation of Nanoparticles by Solvent Displacement
2.4. Average Size and Zeta Potential Determination of Nanoparticles
2.5. Microscopy
2.6. HPLC Analysis
2.7. Antibacterial Assays
2.8. Data Analysis and Graphics
3. Results
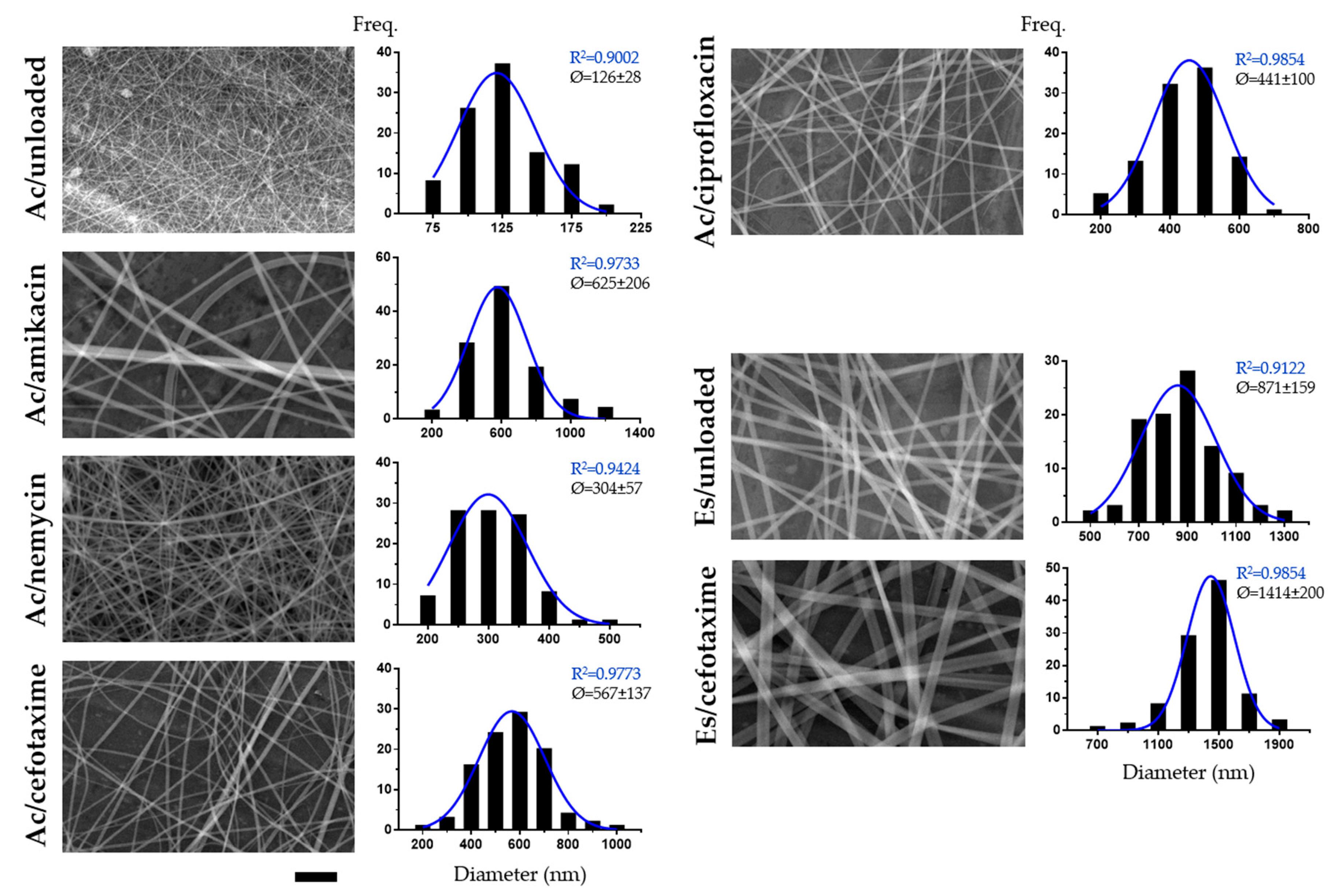
3.1. Electrospinnability of Formulations and Characterization of Obtained Nanofibers
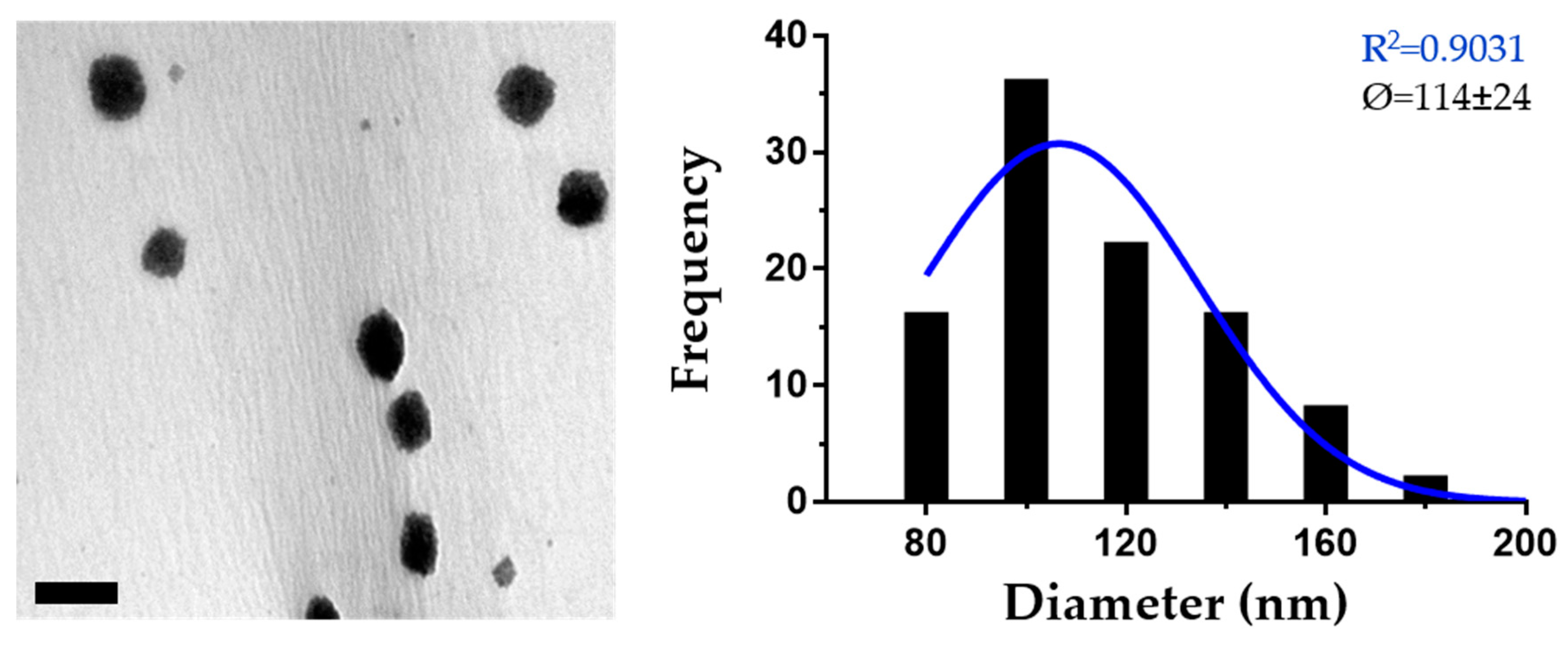
3.2. Optimization of the Preparation of PMVE/MA-Es Nanoparticles and Their Characterization
3.3. Characterization of PMVE/MA-Es Nanoparticles Loaded with Antibiotics
3.4. Analysis of the Antibacterial Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Coates, A.R.; Halls, G.; Hu, Y. Novel classes of antibiotics or more of the same? Br. J. Pharmacol. 2011, 163, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Merelli, M.; Temperoni, C.; Astilean, A. New antibiotics for bad bugs: Where are we? Ann. Clin. Microbiol. Antimicrob. 2013, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Leucuta, S.E. Nanotechnology for delivery of drugs and biomedical applications. Curr. Clin. Pharmacol. 2010, 5, 257–280. [Google Scholar] [CrossRef] [PubMed]
- Mahapatro, A.; Singh, D.K. Biodegradable nanoparticles are excellent vehicle for site directed in-vivo delivery of drugs and vaccines. J. Nanobiotechnol. 2011, 9, 55. [Google Scholar] [CrossRef]
- Gunn, J.; Zhang, M. Polyblend nanofibers for biomedical applications: Perspectives and challenges. Trends Biotechnol. 2010, 28, 189–197. [Google Scholar] [CrossRef]
- Vasita, R.; Katti, D.S. Nanofibers and their applications in tissue engineering. Int. J. Nanomed. 2006, 1, 15–30. [Google Scholar] [CrossRef]
- Guo, G.; Fu, S.; Zhou, L.; Liang, H.; Fan, M.; Luo, F.; Qian, Z.; Wei, Y. Preparation of curcumin loaded poly(epsilon-caprolactone)-poly(ethylene glycol)-poly(epsilon-caprolactone) nanofibers and their in vitro antitumor activity against glioma 9l cells. Nanoscale 2011, 3, 3825–3832. [Google Scholar] [CrossRef]
- Yoo, J.J.; Kim, C.; Chung, C.W.; Jeong, Y.I.; Kang, D.H. 5-aminolevulinic acid-incorporated poly(vinyl alcohol) nanofiber-coated metal stent for application in photodynamic therapy. Int. J. Nanomed. 2012, 7, 1997–2005. [Google Scholar]
- Lucio, D.; Martinez-Oharriz, M.C.; Gonzalez-Navarro, C.J.; Navarro-Herrera, D.; Gonzalez-Gaitano, G.; Radulescu, A.; Irache, J.M. Coencapsulation of cyclodextrins into poly(anhydride) nanoparticles to improve the oral administration of glibenclamide. A screening on c. Elegans. Colloids Surf. B Biointerfaces 2018, 163, 64–72. [Google Scholar] [CrossRef]
- Martinez-Ortega, L.; Mira, A.; Fernandez-Carvajal, A.; Mateo, C.R.; Mallavia, R.; Falco, A. Development of a new delivery system based on drug-loadable electrospun nanofibers for psoriasis treatment. Pharmaceutics 2019, 11, 14. [Google Scholar] [CrossRef]
- Mira, A.; Mateo, C.R.; Mallavia, R.; Falco, A. Poly (methyl vinyl ether-alt-maleic acid) and ethyl monoester as building polymers for drug-loadable electrospun nanofibers. Sci. Rep. 2017, 7, 17205. [Google Scholar] [CrossRef] [PubMed]
- Iglesias, T.; de Cerain, A.L.; Irache, J.; Martín-Arbella, N.; Wilcox, M.; Pearson, J.; Azqueta, A. Evaluation of the cytotoxicity, genotoxicity and mucus permeation capacity of several surface modified poly (anhydride) nanoparticles designed for oral drug delivery. Int. J. Pharm. 2017, 517, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Gaton, L.; Espuelas, S.; Larraneta, E.; Reviakine, I.; Yate, L.A.; Irache, J.M. Pegylated poly(anhydride) nanoparticles for oral delivery of docetaxel. Eur. J. Pharm. Sci. 2018, 118, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Guillo, R.; Martinez-Tome, M.J.; Kahveci, Z.; Torres, I.; Falco, A.; Mallavia, R.; Mateo, C.R. Synthesis and characterization of a novel green cationic polyfluorene and its potential use as a fluorescent membrane probe. Polymers 2018, 10, 938. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Guilló, R.; Falco, A.; Martínez-Tomé, M.J.; Mateo, C.R.; Herrero, M.A.; Vázquez, E.; Mallavia, R. Advantageous microwave-assisted suzuki polycondensation for the synthesis of aniline-fluorene alternate copolymers as molecular model with solvent sensing properties. Polymers 2018, 10, 215. [Google Scholar] [CrossRef] [PubMed]
- Kahveci, Z.; Vazquez-Guillo, R.; Martinez-Tome, M.J.; Mallavia, R.; Mateo, C.R. New red-emitting conjugated polyelectrolyte: Stabilization by interaction with biomolecules and potential use as drug carriers and bioimaging probes. ACS Appl Mater. Interfaces 2016, 8, 1958–1969. [Google Scholar] [CrossRef]
- Kumari, A.; Yadav, S.K.; Yadav, S.C. Biodegradable polymeric nanoparticles based drug delivery systems. Colloids Surf. B Biointerfaces 2010, 75, 1–18. [Google Scholar] [CrossRef]
- Sharma, J.; Lizu, M.; Stewart, M.; Zygula, K.; Lu, Y.; Chauhan, R.; Yan, X.; Guo, Z.; Wujcik, E.K.; Wei, S. Multifunctional nanofibers towards active biomedical therapeutics. Polymers 2015, 7, 186–219. [Google Scholar] [CrossRef]
- Arbos, P.; Wirth, M.; Arangoa, M.A.; Gabor, F.; Irache, J.M. Gantrez an as a new polymer for the preparation of ligand-nanoparticle conjugates. J. Control. Release 2002, 83, 321–330. [Google Scholar] [CrossRef]
- Cockerill, F.; Wikler, M.; Alder, J.; Dudley, M.; Eliopoulos, G.; Ferraro, M.; Hardy, D.; Hecht, D.; Hindler, J.; Patel, J. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: Approved Standard; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012. [Google Scholar]
- Goke, K.; Lorenz, T.; Repanas, A.; Schneider, F.; Steiner, D.; Baumann, K.; Bunjes, H.; Dietzel, A.; Finke, J.H.; Glasmacher, B.; et al. Novel strategies for the formulation and processing of poorly water-soluble drugs. Eur. J. Pharm. Biopharm. 2018, 126, 40–56. [Google Scholar] [CrossRef]
- Yu, D.G.; Li, J.J.; Williams, G.R.; Zhao, M. Electrospun amorphous solid dispersions of poorly water-soluble drugs: A review. J. Control. Release 2018, 292, 91–110. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Rath, G.; Singh, R.; Goyal, A.K. Nanofibers: An effective tool for controlled and sustained drug delivery. Curr. Drug. Deliv. 2018, 15, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Son, Y.J.; Kim, W.J.; Yoo, H.S. Therapeutic applications of electrospun nanofibers for drug delivery systems. Arch. Pharm. Res. 2014, 37, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Yu, D.; Zhu, L.; Branford-White, C.; White, K.; Chatterton, N.P. Electrospun diclofenac sodium loaded eudragit(r) l 100-55 nanofibers for colon-targeted drug delivery. Int. J. Pharm. 2011, 408, 200–207. [Google Scholar] [CrossRef]
- Samprasit, W.; Akkaramongkolporn, P.; Ngawhirunpat, T.; Rojanarata, T.; Kaomongkolgit, R.; Opanasopit, P. Fast releasing oral electrospun pvp/cd nanofiber mats of taste-masked meloxicam. Int. J. Pharm. 2015, 487, 213–222. [Google Scholar] [CrossRef]
- Nezarati, R.M.; Eifert, M.B.; Cosgriff-Hernandez, E. Effects of humidity and solution viscosity on electrospun fiber morphology. Tissue Eng. Part C Methods 2013, 19, 810–819. [Google Scholar] [CrossRef]
- Reda, R.I.; Wen, M.M.; El-Kamel, A.H. Ketoprofen-loaded eudragit electrospun nanofibers for the treatment of oral mucositis. Int. J. Nanomed. 2017, 12, 2335. [Google Scholar] [CrossRef]
- Canbolat, M.F.; Celebioglu, A.; Uyar, T. Drug delivery system based on cyclodextrin-naproxen inclusion complex incorporated in electrospun polycaprolactone nanofibers. Colloids Surf. B Biointerfaces 2014, 115, 15–21. [Google Scholar] [CrossRef]
- Irache, J.M.; Huici, M.; Konecny, M.; Espuelas, S.; Campanero, M.A.; Arbos, P. Bioadhesive properties of gantrez nanoparticles. Molecules 2005, 10, 126–145. [Google Scholar] [CrossRef]
- Salman, H.H.; Azcarate, I.G. Nanoparticles Comprising Esters of Poly (Methyl Vinyl Ether-Co-Maleic Anhydride) and Uses Thereof. U.S. Patent No. 9,351,940, 31 May 2016. [Google Scholar]
- Kaur, I.P.; Bhandari, R.; Bhandari, S.; Kakkar, V. Potential of solid lipid nanoparticles in brain targeting. J. Control. Release 2008, 127, 97–109. [Google Scholar] [CrossRef]
- Hunter, R.J. Zeta Potential in Colloid Science: Principles and Applications; Academic Press: Cambridge, MA, USA, 2013; Volume 2. [Google Scholar]
- Kovacevic, A.; Savic, S.; Vuleta, G.; Muller, R.H.; Keck, C.M. Polyhydroxy surfactants for the formulation of lipid nanoparticles (sln and nlc): Effects on size, physical stability and particle matrix structure. Int. J. Pharm. 2011, 406, 163–172. [Google Scholar] [CrossRef]
- Agüeros, M.; Zabaleta, V.; Espuelas, S.; Campanero, M.; Irache, J. Increased oral bioavailability of paclitaxel by its encapsulation through complex formation with cyclodextrins in poly (anhydride) nanoparticles. J. Control. Release 2010, 145, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Araujo, R.S.; Garcia, G.M.; Vilela, J.M.C.; Andrade, M.S.; Oliveira, L.A.M.; Kano, E.K.; Lange, C.C.; Brito, M.; Brandao, H.M.; Mosqueira, V.C.F. Cloxacillin benzathine-loaded polymeric nanocapsules: Physicochemical characterization, cell uptake, and intramammary antimicrobial effect. Mater. Sci. Eng. C Mater. Biol. Appl. 2019, 104, 110006. [Google Scholar] [CrossRef] [PubMed]
- Brandhonneur, N.; Hatahet, T.; Amela-Cortes, M.; Molard, Y.; Cordier, S.; Dollo, G. Molybdenum cluster loaded plga nanoparticles: An innovative theranostic approach for the treatment of ovarian cancer. Eur. J. Pharm. Biopharm. 2018, 125, 95–105. [Google Scholar] [CrossRef]
- Lopalco, A.; Ali, H.; Denora, N.; Rytting, E. Oxcarbazepine-loaded polymeric nanoparticles: Development and permeability studies across in vitro models of the blood-brain barrier and human placental trophoblast. Int. J. Nanomed. 2015, 10, 1985–1996. [Google Scholar]
- Calleja, P.; Espuelas, S.; Vauthier, C.; Ponchel, G.; Irache, J.M. Controlled release, intestinal transport, and oral bioavailablity of paclitaxel can be considerably increased using suitably tailored pegylated poly(anhydride) nanoparticles. J. Pharm. Sci. 2015, 104, 2877–2886. [Google Scholar] [CrossRef]
- Sabaeifard, P.; Abdi-Ali, A.; Soudi, M.R.; Gamazo, C.; Irache, J.M. Amikacin loaded plga nanoparticles against pseudomonas aeruginosa. Eur. J. Pharm. Sci. 2016, 93, 392–398. [Google Scholar] [CrossRef]
- Zaki, N.M.; Hafez, M.M. Enhanced antibacterial effect of ceftriaxone sodium-loaded chitosan nanoparticles against intracellular salmonella typhimurium. AAPS PharmSciTech 2012, 13, 411–421. [Google Scholar] [CrossRef]
- Mushtaq, S.; Khan, J.A.; Rabbani, F.; Latif, U.; Arfan, M.; Yameen, M.A. Biocompatible biodegradable polymeric antibacterial nanoparticles for enhancing the effects of a third-generation cephalosporin against resistant bacteria. J. Med. Microbiol. 2017, 66, 318–327. [Google Scholar] [CrossRef]
- Sonam; Chaudhary, H.; Kumar, V. Taguchi design for optimization and development of antibacterial drug-loaded plga nanoparticles. Int. J. Biol. Macromol. 2014, 64, 99–105. [Google Scholar] [CrossRef]
- Natan, M.; Banin, E. From nano to micro: Using nanotechnology to combat microorganisms and their multidrug resistance. FEMS Microbiol. Rev. 2017, 41, 302–322. [Google Scholar] [CrossRef] [PubMed]
| Conc. (mg/g) | HDD (nm) | PDI (a.u.) | ZP (mV) |
|---|---|---|---|
| 1.25 | 84 ± 5 | 0.11 ± 0.04 | −26 ± 10 |
| 2.5 | 139 ± 26 | 0.19 ± 0.03 | −5 ± 4 |
| 5 | 106 ± 4 | 0.10 ± 0.04 | −5 ± 6 |
| 10 | 202 ± 11 | 0.20 ± 0.06 | −35 ± 9 |
| 20 | 230 ± 1 | 0.06 ± 0.01 | −32 ± 1 |
| Antibiotic | HDD (nm) | PDI (a.u.) | ZP (mV) | EE (%) |
|---|---|---|---|---|
| None | 195 ± 7 | 0.15 ± 0.04 | −38 ± 13 | - |
| Amikacin | 253 ± 5 | 0.17 ± 0.07 | −39 ± 2 | 14 ± 4 |
| Neomycin | 209 ± 4 | 0.20 ± 0.07 | −29 ± 7 | 40 ± 3 |
| Cefotaxime | 156 ± 6 | 0.29 ± 0.03 | −31 ± 8 | 69 ± 7 |
| Ciprofloxacin | 256 ± 8 | 0.17 ± 0.01 | −16 ± 3 | 59 ± 8 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mira, A.; Sainz-Urruela, C.; Codina, H.; Jenkins, S.I.; Rodriguez-Diaz, J.C.; Mallavia, R.; Falco, A. Physico-Chemically Distinct Nanomaterials Synthesized from Derivates of a Poly(Anhydride) Diversify the Spectrum of Loadable Antibiotics. Nanomaterials 2020, 10, 486. https://doi.org/10.3390/nano10030486
Mira A, Sainz-Urruela C, Codina H, Jenkins SI, Rodriguez-Diaz JC, Mallavia R, Falco A. Physico-Chemically Distinct Nanomaterials Synthesized from Derivates of a Poly(Anhydride) Diversify the Spectrum of Loadable Antibiotics. Nanomaterials. 2020; 10(3):486. https://doi.org/10.3390/nano10030486
Chicago/Turabian StyleMira, Amalia, Carlos Sainz-Urruela, Helena Codina, Stuart I. Jenkins, Juan Carlos Rodriguez-Diaz, Ricardo Mallavia, and Alberto Falco. 2020. "Physico-Chemically Distinct Nanomaterials Synthesized from Derivates of a Poly(Anhydride) Diversify the Spectrum of Loadable Antibiotics" Nanomaterials 10, no. 3: 486. https://doi.org/10.3390/nano10030486
APA StyleMira, A., Sainz-Urruela, C., Codina, H., Jenkins, S. I., Rodriguez-Diaz, J. C., Mallavia, R., & Falco, A. (2020). Physico-Chemically Distinct Nanomaterials Synthesized from Derivates of a Poly(Anhydride) Diversify the Spectrum of Loadable Antibiotics. Nanomaterials, 10(3), 486. https://doi.org/10.3390/nano10030486







