In Vitro and In Vivo Models for Evaluating the Oral Toxicity of Nanomedicines
Abstract
1. Introduction
2. Mechanisms of Oral Nanotoxicity
2.1. Oxidative Stress, Immune Responses, and Inflammation
2.2. Nanotoxicity of Intracellular Organelles
2.3. Genotoxicity
2.4. Nanotoxicity of Gut Barrier
2.5. Nanotoxicity and Luminal Environment
2.6. Nanotoxicity Involves Microbiota
3. In Vitro Toxicity Models for Orally Delivered NFs
3.1. Static and Dynamic Culture Models
3.2. 3D Cell Culture Models
3.3. Models that Involve the Gut Microbiome
3.4. Supplementary Test for Determining NP Induced Mutagenicity
4. In Vivo Models
4.1. Invertebrates
4.2. Zebrafish (Danio rerio)
4.3. Rodent Models
4.4. Nonhuman Primate Models
5. Toxicological Endpoints of Observing
5.1. In Vitro Cell Viability, Cytotoxicity, and Cell Proliferation
5.2. Detecting the Oxidative Stress and Inflammation
5.2.1. ROS/RNS Detection
5.2.2. Detecting the Oxidative Stress-Induced Damages
5.2.3. The Oxidative Response System
5.2.4. Detections with the Multi-Omics Platform
5.3. Cell Monolayer Permeability and Tight Junction Assays
5.4. Clonogenic Assay
5.5. Genotoxicity
5.5.1. Chromosome Aberration
5.5.2. Single-Cell Gel Electrophoresis (Comet Assay)
5.5.3. Histochemical Approach
5.5.4. Immunohistochemical Approach
5.5.5. Measurements of the Gene Expression Changes
5.5.6. Measuring Oxidized Guanosine
5.6. Method Validation
6. Strategies for Making Nontoxic Oral NFs
7. Conclusions and Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shahiwala, A. Formulation approaches in enhancement of patient compliance to oral drug therapy. Expert Opin. Drug Deliv. 2011, 8, 1521–1529. [Google Scholar] [CrossRef] [PubMed]
- Schultz, H.B.; Meola, T.R.; Thomas, N.; Prestidge, C.A. Oral formulation strategies to improve the bioavailability and mitigate the food effect of abiraterone acetate. Int. J. Pharm. 2020, 577, 119069. [Google Scholar] [CrossRef]
- Raz, N.R.; Akbarzadeh, T.M.; Tafaghodi, M. Bioinspired Nanonetworks for Targeted Cancer Drug Delivery. IEEE Trans. Nanobiosci. 2015, 14, 894–906. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.; Zhang, X.Q.; Tang, M.X.; Liu, Q.; Zhou, G. Anti-PD-L1-modified and ATRA-loaded nanoparticles for immuno-treatment of oral dysplasia and oral squamous cell carcinoma. Nanomedicine 2020, 15, 951–968. [Google Scholar] [CrossRef] [PubMed]
- Sladek, S.; McCartney, F.; Eskander, M.; Dunne, D.J.; Santos-Martinez, M.J.; Benetti, F.; Tajber, L.; Brayden, D.J. An Enteric-Coated Polyelectrolyte Nanocomplex Delivers Insulin in Rat Intestinal Instillations when Combined with a Permeation Enhancer. Pharmaceutics 2020, 12, 259. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, Y.; Cheng, J.; Guo, J.; Zhang, Q.; Zhang, X.; Ren, J.; Wang, F.; Huang, J.; Hu, H.; et al. A Proresolving Peptide Nanotherapy for Site-Specific Treatment of Inflammatory Bowel Disease by Regulating Proinflammatory Microenvironment and Gut Microbiota. Adv. Sci. 2019, 6, 1900610. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Zhang, M.; Lama, S.; Wang, L.; Merlin, D. Natural-lipid nanoparticle-based therapeutic approach to deliver 6-shogaol and its metabolites M2 and M13 to the colon to treat ulcerative colitis. J. Control. Release 2020, 323, 293–310. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, F.; Li, S.; Liu, R.; Jin, T.; Dou, Y.; Zhou, Z.; Zhang, J. A Multifunctional Nanotherapy for Targeted Treatment of Colon Cancer by Simultaneously Regulating Tumor Microenvironment. Theranostics 2019, 9, 3732–3753. [Google Scholar] [CrossRef] [PubMed]
- Khare, T.; Palakurthi, S.S.; Shah, B.M.; Palakurthi, S.; Khare, S. Natural Product-Based Nanomedicine in Treatment of Inflammatory Bowel Disease. Int. J. Mol. Sci. 2020, 21, 3956. [Google Scholar] [CrossRef]
- Elkateb, H.; Tatham, L.M.; Cauldbeck, H.; Niezabitowska, E.; Owen, A.; Rannard, S.; McDonald, T. Optimization of the synthetic parameters of lipid polymer hybrid nanoparticles dual loaded with darunavir and ritonavir for the treatment of HIV. Int. J. Pharm. 2020, 588, 119794. [Google Scholar] [CrossRef]
- Jain, S.; Harde, H.; Indulkar, A.; Agrawal, A.K. Improved stability and immunological potential of tetanus toxoid containing surface engineered bilosomes following oral administration. Nanomedicine 2014, 10, 431–440. [Google Scholar] [CrossRef] [PubMed]
- Truzzi, E.; Bongio, C.; Sacchetti, F.; Maretti, E.; Montanari, M.; Iannuccelli, V.; Vismara, E.; Leo, E. Self-Assembled Lipid Nanoparticles for Oral Delivery of Heparin-Coated Iron Oxide Nanoparticles for Theranostic Purposes. Molecules 2017, 22, 963. [Google Scholar] [CrossRef] [PubMed]
- Pocock, K.; Delon, L.C.; Khatri, A.; Prestidge, C.; Gibson, R.; Barbe, C.; Thierry, B. Uptake of silica particulate drug carriers in an intestine-on-a-chip: Towards a better in vitro model of nanoparticulate carrier and mucus interactions. Biomater. Sci. 2019, 7, 2410–2420. [Google Scholar] [CrossRef]
- Farjadian, F.; Ghasemi, A.; Gohari, O.; Roointan, A.; Karimi, M.; Hamblin, M.R. Nanopharmaceuticals and nanomedicines currently on the market: Challenges and opportunities. Nanomedicine 2019, 14, 93–126. [Google Scholar] [CrossRef] [PubMed]
- Ammendolia, M.G.; Iosi, F.; Maranghi, F.; Tassinari, R.; Cubadda, F.; Aureli, F.; Raggi, A.; Superti, F.; Mantovani, A.; De Berardis, B. Short-term oral exposure to low doses of nano-sized TiO2 and potential modulatory effects on intestinal cells. Food Chem. Toxicol. 2017, 102, 63–75. [Google Scholar] [CrossRef]
- Li, W.; Li, Y.; Liu, Z.; Kerdsakundee, N.; Zhang, M.; Zhang, F.; Liu, X.; Bauleth-Ramos, T.; Lian, W.; Makila, E.; et al. Hierarchical structured and programmed vehicles deliver drugs locally to inflamed sites of intestine. Biomaterials 2018, 185, 322–332. [Google Scholar] [CrossRef]
- Yang, P.; Hong, W.; Zhou, P.; Chen, B.; Xu, H. Nano and bulk ZnO trigger diverse Zn-transport-related gene transcription in distinct regions of the small intestine in mice after oral exposure. Biochem. Biophys. Res. Commun. 2017, 493, 1364–1369. [Google Scholar] [CrossRef]
- Zou, P.; Chen, H.; Paholak, H.J.; Sun, D. Noninvasive fluorescence resonance energy transfer imaging of in vivo premature drug release from polymeric nanoparticles. Mol. Pharm. 2013, 10, 4185–4194. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, M.; Merlin, D. Advances in Plant-derived Edible Nanoparticle-based lipid Nano-drug Delivery Systems as Therapeutic Nanomedicines. J. Mater. Chem. B 2018, 6, 1312–1321. [Google Scholar] [CrossRef]
- Clift, M.J.; Rothen-Rutishauser, B.; Brown, D.M.; Duffin, R.; Donaldson, K.; Proudfoot, L.; Guy, K.; Stone, V. The impact of different nanoparticle surface chemistry and size on uptake and toxicity in a murine macrophage cell line. Toxicol. Appl. Pharm. 2008, 232, 418–427. [Google Scholar] [CrossRef]
- Gorth, D.J.; Rand, D.M.; Webster, T.J. Silver nanoparticle toxicity in Drosophila: Size does matter. Int. J. Nanomed. 2011, 6, 343–350. [Google Scholar] [CrossRef]
- Cui, X.; Bao, L.; Wang, X.; Chen, C. The Nano–Intestine Interaction: Understanding the Location-Oriented Effects of Engineered Nanomaterials in the Intestine. Small 2020, 16, 1907665. [Google Scholar] [CrossRef]
- Osborne, O.J.; Lin, S.; Chang, C.H.; Ji, Z.; Yu, X.; Wang, X.; Lin, S.; Xia, T.; Nel, A.E. Organ-Specific and Size-Dependent Ag Nanoparticle Toxicity in Gills and Intestines of Adult Zebrafish. ACS Nano 2015, 9, 9573–9584. [Google Scholar] [CrossRef]
- Sergent, J.A.; Paget, V.; Chevillard, S. Toxicity and genotoxicity of nano-SiO2 on human epithelial intestinal HT-29 cell line. Ann. Occup. Hyg. 2012, 56, 622–630. [Google Scholar] [CrossRef][Green Version]
- Friedman, A.L.; Panaitescu, E.; Richter, C.; Menon, L. High-aspect ratio nano-noodles of alumina and titania. J. Nanosci. Nanotechnol. 2008, 8, 5864–5868. [Google Scholar] [CrossRef] [PubMed]
- Koch, K.; Schulte, A.J.; Fischer, A.; Gorb, S.N.; Barthlott, W. A fast, precise and low-cost replication technique for nano- and high-aspect-ratio structures of biological and artificial surfaces. Bioinspir. Biomim. 2008, 3, 046002. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Santos, A.; Kaur, G.; Evdokiou, A.; Losic, D. Structurally engineered anodic alumina nanotubes as nano-carriers for delivery of anticancer therapeutics. Biomaterials 2014, 35, 5517–5526. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kaur, G.; Zysk, A.; Liapis, V.; Hay, S.; Santos, A.; Losic, D.; Evdokiou, A. Systematic in vitro nanotoxicity study on anodic alumina nanotubes with engineered aspect ratio: Understanding nanotoxicity by a nanomaterial model. Biomaterials 2015, 46, 117–130. [Google Scholar] [CrossRef]
- Libralato, G.; Galdiero, E.; Falanga, A.; Carotenuto, R.; de Alteriis, E.; Guida, M. Toxicity Effects of Functionalized Quantum Dots, Gold and Polystyrene Nanoparticles on Target Aquatic Biological Models: A Review. Molecules 2017, 22, 1439. [Google Scholar] [CrossRef]
- Kikkeri, R.; Padler-Karavani, V.; Diaz, S.; Verhagen, A.; Yu, H.; Cao, H.; Langereis, M.A.; De Groot, R.J.; Chen, X.; Varki, A. Quantum dot nanometal surface energy transfer based biosensing of sialic acid compositions and linkages in biological samples. Anal. Chem. 2013, 85, 3864–3870. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, M. Review of in vitro toxicological research of quantum dot and potentially involved mechanisms. Sci. Total Environ. 2018, 625, 940–962. [Google Scholar] [CrossRef] [PubMed]
- Ju, L.; Zhang, G.; Zhang, C.; Sun, L.; Jiang, Y.; Yan, C.; Duerksen-Hughes, P.J.; Zhang, X.; Zhu, X.; Chen, F.F.; et al. Quantum dot-related genotoxicity perturbation can be attenuated by PEG encapsulation. Mutat. Res. 2013, 753, 54–64. [Google Scholar] [CrossRef]
- Li, Y.; Hou, X.; Yang, C.; Pang, Y.; Li, X.; Jiang, G.; Liu, Y. Photoprotection of Cerium Oxide Nanoparticles against UVA radiation-induced Senescence of Human Skin Fibroblasts due to their Antioxidant Properties. Sci. Rep. 2019, 9, 2595. [Google Scholar] [CrossRef] [PubMed]
- Rubio, L.; Annangi, B.; Vila, L.; Hernandez, A.; Marcos, R. Antioxidant and anti-genotoxic properties of cerium oxide nanoparticles in a pulmonary-like cell system. Arch. Toxicol. 2016, 90, 269–278. [Google Scholar] [CrossRef]
- Xia, T.; Kovochich, M.; Liong, M.; Madler, L.; Gilbert, B.; Shi, H.; Yeh, J.I.; Zink, J.I.; Nel, A.E. Comparison of the mechanism of toxicity of zinc oxide and cerium oxide nanoparticles based on dissolution and oxidative stress properties. ACS Nano 2008, 2, 2121–2134. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Li, J.; Iwai, H.; Mei, Q.; Fujita, D.; Su, H.; Chen, H.; Hanagata, N. Formation of nano-bio-complex as nanomaterials dispersed in a biological solution for understanding nanobiological interactions. Sci. Rep. 2012, 2, 406. [Google Scholar] [CrossRef] [PubMed]
- Muller, K.H.; Kulkarni, J.; Motskin, M.; Goode, A.; Winship, P.; Skepper, J.N.; Ryan, M.P.; Porter, A.E. pH-dependent toxicity of high aspect ratio ZnO nanowires in macrophages due to intracellular dissolution. ACS Nano 2010, 4, 6767–6779. [Google Scholar] [CrossRef]
- Chen, Z.; Meng, H.; Xing, G.; Chen, C.; Zhao, Y.; Jia, G.; Wang, T.; Yuan, H.; Ye, C.; Zhao, F.; et al. Acute toxicological effects of copper nanoparticles in vivo. Toxicol. Lett. 2006, 163, 109–120. [Google Scholar] [CrossRef]
- Falinski, M.M.; Garland, M.A.; Hashmi, S.M.; Tanguay, R.L.; Zimmerman, J.B. Establishing structure-property-hazard relationships for multi-walled carbon nanotubes: The role of aggregation, surface charge, and oxidative stress on embryonic zebrafish mortality. Carbon N. Y. 2019, 155, 587–600. [Google Scholar] [CrossRef]
- Yang, C.; Zhang, M.; Sung, J.; Wang, L.; Jung, Y.; Merlin, D. Autologous Exosome Transfer: A New Personalised Treatment Concept to Prevent Colitis in a Murine Model. J. Crohns Colitis 2020, 14, 841–855. [Google Scholar] [CrossRef]
- Wang, D.; Sun, M.; Zhang, Y.; Chen, Z.; Zang, S.; Li, G.; Li, G.; Clark, A.R.; Huang, J.; Si, L. Enhanced therapeutic efficacy of a novel colon-specific nanosystem loading emodin on DSS-induced experimental colitis. Phytomedicine 2020, 78, 153293. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Merlin, D. Nanoparticle-Mediated Drug Delivery Systems For The Treatment Of IBD: Current Perspectives. Int. J. Nanomed. 2019, 14, 8875–8889. [Google Scholar] [CrossRef]
- Ramesh, N.; Mandal, A.K.A. Pharmacokinetic, toxicokinetic, and bioavailability studies of epigallocatechin-3-gallate loaded solid lipid nanoparticle in rat model. Drug Dev. Ind. Pharm. 2019, 45, 1506–1514. [Google Scholar] [CrossRef]
- Joshi, J.C.; Bhardwaj, A.; Roy, I.; Gulati, K.; Ray, A. Experimental Studies on the Systemic Toxicity and Biodistribution of Synthesized Calcium Phosphate Nanoparticles After Oral Administration in Rats. Pharm. Nanotechnol. 2016, 4, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Gundogdu, E.; Yurdasiper, A. Drug transport mechanism of oral antidiabetic nanomedicines. Int. J. Endocrinol. Metab. 2014, 12, e8984. [Google Scholar] [CrossRef] [PubMed]
- Hirano, A.; Wada, M.; Tanaka, T.; Kataura, H. Oxidative Stress of Carbon Nanotubes on Proteins Is Mediated by Metals Originating from the Catalyst Remains. ACS Nano 2019, 13, 1805–1816. [Google Scholar] [CrossRef]
- Lanone, S.; Andujar, P.; Kermanizadeh, A.; Boczkowski, J. Determinants of carbon nanotube toxicity. Adv. Drug Deliv. Rev. 2013, 65, 2063–2069. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Guo, L.; Morris, D.; Kane, A.B.; Hurt, R.H. Targeted Removal of Bioavailable Metal as a Detoxification Strategy for Carbon Nanotubes. Carbon N. Y. 2008, 46, 489–500. [Google Scholar] [CrossRef]
- Yan, J.; Wang, D.; Li, K.; Chen, Q.; Lai, W.; Tian, L.; Lin, B.; Tan, Y.; Liu, X.; Xi, Z. Toxic effects of the food additives titanium dioxide and silica on the murine intestinal tract: Mechanisms related to intestinal barrier dysfunction involved by gut microbiota. Environ. Toxicol. Pharm. 2020, 103485. [Google Scholar] [CrossRef]
- Cornu, R.; Chretien, C.; Pellequer, Y.; Martin, H.; Beduneau, A. Small silica nanoparticles transiently modulate the intestinal permeability by actin cytoskeleton disruption in both Caco-2 and Caco-2/HT29-MTX models. Arch. Toxicol. 2020, 94, 1191–1202. [Google Scholar] [CrossRef]
- Mercier-Bonin, M.; Despax, B.; Raynaud, P.; Houdeau, E.; Thomas, M. Mucus and microbiota as emerging players in gut nanotoxicology: The example of dietary silver and titanium dioxide nanoparticles. Crit. Rev. Food Sci. Nutr. 2018, 58, 1023–1032. [Google Scholar] [CrossRef] [PubMed]
- Brun, E.; Barreau, F.; Veronesi, G.; Fayard, B.; Sorieul, S.; Chaneac, C.; Carapito, C.; Rabilloud, T.; Mabondzo, A.; Herlin-Boime, N.; et al. Titanium dioxide nanoparticle impact and translocation through ex vivo, in vivo and in vitro gut epithelia. Part. Fibre Toxicol. 2014, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Tournebize, J.; Sapin-Minet, A.; Bartosz, G.; Leroy, P.; Boudier, A. Pitfalls of assays devoted to evaluation of oxidative stress induced by inorganic nanoparticles. Talanta 2013, 116, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Santos, A.; Evdokiou, A.; Losic, D. An overview of nanotoxicity and nanomedicine research: Principles, progress and implications for cancer therapy. J. Mater. Chem. B 2015, 3, 7153–7172. [Google Scholar] [CrossRef]
- Moldogazieva, N.T.; Mokhosoev, I.M.; Feldman, N.B.; Lutsenko, S.V. ROS and RNS signalling: Adaptive redox switches through oxidative/nitrosative protein modifications. Free Radic. Res. 2018, 52, 507–543. [Google Scholar] [CrossRef]
- Moldogazieva, N.T.; Mokhosoev, I.M.; Mel’nikova, T.I.; Zavadskiy, S.P.; Kuz’menko, A.N.; Terentiev, A.A. Dual Character of Reactive Oxygen, Nitrogen, and Halogen Species: Endogenous Sources, Interconversions and Neutralization. Biochemistry 2020, 85, S56–S78. [Google Scholar] [CrossRef]
- Van der Paal, J.; Hong, S.H.; Yusupov, M.; Gaur, N.; Oh, J.S.; Short, R.D.; Szili, E.J.; Bogaerts, A. How membrane lipids influence plasma delivery of reactive oxygen species into cells and subsequent DNA damage: An experimental and computational study. Phys. Chem. Chem. Phys. 2019, 21, 19327–19341. [Google Scholar] [CrossRef]
- Ahmadinejad, F.; Geir Moller, S.; Hashemzadeh-Chaleshtori, M.; Bidkhori, G.; Jami, M.S. Molecular Mechanisms behind Free Radical Scavengers Function against Oxidative Stress. Antioxidants 2017, 6, 51. [Google Scholar] [CrossRef]
- Indo, H.P.; Yen, H.C.; Nakanishi, I.; Matsumoto, K.; Tamura, M.; Nagano, Y.; Matsui, H.; Gusev, O.; Cornette, R.; Okuda, T.; et al. A mitochondrial superoxide theory for oxidative stress diseases and aging. J. Clin. Biochem. Nutr. 2015, 56, 1–7. [Google Scholar] [CrossRef]
- Gao, L.; Zhuang, J.; Nie, L.; Zhang, J.; Zhang, Y.; Gu, N.; Wang, T.; Feng, J.; Yang, D.; Perrett, S.; et al. Intrinsic peroxidase-like activity of ferromagnetic nanoparticles. Nat. Nanotechnol. 2007, 2, 577–583. [Google Scholar] [CrossRef]
- Husain, N.; Mahmood, R. Copper(II) generates ROS and RNS, impairs antioxidant system and damages membrane and DNA in human blood cells. Environ. Sci. Pollut. Res. Int. 2019, 26, 20654–20668. [Google Scholar] [CrossRef] [PubMed]
- Cummins, T.D.; Higdon, A.N.; Kramer, P.A.; Chacko, B.K.; Riggs, D.W.; Salabei, J.K.; Dell’Italia, L.J.; Zhang, J.; Darley-Usmar, V.M.; Hill, B.G. Utilization of fluorescent probes for the quantification and identification of subcellular proteomes and biological processes regulated by lipid peroxidation products. Free Radic. Biol. Med. 2013, 59, 56–68. [Google Scholar] [CrossRef] [PubMed]
- Popova, M.P.; Popov, C.S. Damage to subcellular structures evoked by lipid peroxidation. Z. Nat. C J. Biosci. 2002, 57, 361–365. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Funnell, W.R.; Maysinger, D. Three-dimensional reconstruction of cell nuclei, internalized quantum dots and sites of lipid peroxidation. J. Nanobiotechnol. 2006, 4, 10. [Google Scholar] [CrossRef][Green Version]
- Majewski, M.; Lis, B.; Olas, B.; Ognik, K.; Juskiewicz, J. Dietary supplementation with copper nanoparticles influences the markers of oxidative stress and modulates vasodilation of thoracic arteries in young Wistar rats. PLoS ONE 2020, 15, e0229282. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.; Goswami, M.; Yadav, K.; Chaudhary, D. Oxidative Stress and Nano-Toxicity Induced by TiO2 and ZnO on WAG Cell Line. PLoS ONE 2015, 10, e0127493. [Google Scholar] [CrossRef]
- Suzuki, Y.J.; Carini, M.; Butterfield, D.A. Protein Carbonylation. Antioxid. Redox Signal. 2010, 12, 3. [Google Scholar] [CrossRef]
- Wang, C.C.; Wang, S.; Xia, Q.; He, W.; Yin, J.J.; Fu, P.P.; Li, J.H. Phototoxicity of zinc oxide nanoparticles in HaCaT keratinocytes-generation of oxidative DNA damage during UVA and visible light irradiation. J. Nanosci. Nanotechnol. 2013, 13, 3880–3888. [Google Scholar] [CrossRef]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef]
- Arus, B.A.; Souza, D.G.; Bellaver, B.; Souza, D.O.; Goncalves, C.A.; Quincozes-Santos, A.; Bobermin, L.D. Resveratrol modulates GSH system in C6 astroglial cells through heme oxygenase 1 pathway. Mol. Cell Biochem. 2017, 428, 67–77. [Google Scholar] [CrossRef]
- Haddad, J.J.; Harb, H.L. L-gamma-Glutamyl-L-cysteinyl-glycine (glutathione; GSH) and GSH-related enzymes in the regulation of pro- and anti-inflammatory cytokines: A signaling transcriptional scenario for redox(y) immunologic sensor(s)? Mol. Immunol. 2005, 42, 987–1014. [Google Scholar] [CrossRef]
- Abudayyak, M.; Guzel, E.; Ozhan, G. Cupric Oxide Nanoparticles Induce Cellular Toxicity in Liver and Intestine Cell Lines. Adv. Pharm. Bull. 2020, 10, 213–220. [Google Scholar] [CrossRef]
- Shakeel, M.; Jabeen, F.; Shabbir, S.; Asghar, M.S.; Khan, M.S.; Chaudhry, A.S. Toxicity of Nano-Titanium Dioxide (TiO2-NP) Through Various Routes of Exposure: A Review. Biol. Trace Elem. Res. 2016, 172, 1–36. [Google Scholar] [CrossRef]
- Shukla, R.K.; Kumar, A.; Vallabani, N.V.; Pandey, A.K.; Dhawan, A. Titanium dioxide nanoparticle-induced oxidative stress triggers DNA damage and hepatic injury in mice. Nanomedicine 2014, 9, 1423–1434. [Google Scholar] [CrossRef]
- Shin, D.; Moon, H.W.; Oh, Y.; Kim, K.; Kim, D.D.; Lim, C.J. Defensive Properties of Ginsenoside Re against UV-B-Induced Oxidative Stress through Up-Regulating Glutathione and Superoxide Dismutase in HaCaT Keratinocytes. Iran. J. Pharm. Res. 2018, 17, 249–260. [Google Scholar]
- Abbasalipourkabir, R.; Moradi, H.; Zarei, S.; Asadi, S.; Salehzadeh, A.; Ghafourikhosroshahi, A.; Mortazavi, M.; Ziamajidi, N. Toxicity of zinc oxide nanoparticles on adult male Wistar rats. Food Chem. Toxicol. 2015, 84, 154–160. [Google Scholar] [CrossRef]
- Lucarelli, M.; Gatti, A.M.; Savarino, G.; Quattroni, P.; Martinelli, L.; Monari, E.; Boraschi, D. Innate defence functions of macrophages can be biased by nano-sized ceramic and metallic particles. Eur. Cytokine Netw. 2004, 15, 339–346. [Google Scholar]
- Li, Q.; Wang, H.; Peng, H.; Huyan, T.; Cacalano, N.A. Exosomes: Versatile Nano Mediators of Immune Regulation. Cancers 2019, 11, 1557. [Google Scholar] [CrossRef]
- Poon, W.L.; Alenius, H.; Ndika, J.; Fortino, V.; Kolhinen, V.; Mesceriakovas, A.; Wang, M.; Greco, D.; Lahde, A.; Jokiniemi, J.; et al. Nano-sized zinc oxide and silver, but not titanium dioxide, induce innate and adaptive immunity and antiviral response in differentiated THP-1 cells. Nanotoxicology 2017, 11, 936–951. [Google Scholar] [CrossRef]
- Sun, Z.Y.; Chen, P.G.; Liu, Y.F.; Shi, L.; Zhang, B.D.; Wu, J.J.; Zhao, Y.F.; Chen, Y.X.; Li, Y.M. Self-Assembled Nano-Immunostimulant for Synergistic Immune Activation. ChemBioChem 2017, 18, 1721–1729. [Google Scholar] [CrossRef]
- Elsabahy, M.; Wooley, K.L. Cytokines as biomarkers of nanoparticle immunotoxicity. Chem. Soc. Rev. 2013, 42, 5552–5576. [Google Scholar] [CrossRef] [PubMed]
- Schulz, M.; Olubummo, A.; Binder, W.H. Beyond the lipid-bilayer: Interaction of polymers and nanoparticles with membranes. Soft Matter 2012, 8, 4849. [Google Scholar] [CrossRef]
- Patel, S.; Ashwanikumar, N.; Robinson, E.; DuRoss, A.; Sun, C.; Murphy-Benenato, K.E.; Mihai, C.; Almarsson, O.; Sahay, G. Boosting Intracellular Delivery of Lipid Nanoparticle-Encapsulated mRNA. Nano Lett. 2017, 17, 5711–5718. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Nie, W.; He, Z.; Yang, J.; Shao, B.; Ma, X.; Zhang, X.; Bi, Z.; Sun, L.; Liang, X.; et al. Carbon black nanoparticles induce cell necrosis through lysosomal membrane permeabilization and cause subsequent inflammatory response. Theranostics 2020, 10, 4589–4605. [Google Scholar] [CrossRef]
- Domenech, M.; Marrero-Berrios, I.; Torres-Lugo, M.; Rinaldi, C. Lysosomal membrane permeabilization by targeted magnetic nanoparticles in alternating magnetic fields. ACS Nano 2013, 7, 5091–5101. [Google Scholar] [CrossRef] [PubMed]
- Gorojod, R.M.; Alaimo, A.; Porte Alcon, S.; Pomilio, C.; Saravia, F.; Kotler, M.L. The autophagic- lysosomal pathway determines the fate of glial cells under manganese- induced oxidative stress conditions. Free Radic. Biol. Med. 2015, 87, 237–251. [Google Scholar] [CrossRef]
- Gao, W.; Cao, W.; Zhang, H.; Li, P.; Xu, K.; Tang, B. Targeting lysosomal membrane permeabilization to induce and image apoptosis in cancer cells by multifunctional Au-ZnO hybrid nanoparticles. Chem. Commun. 2014, 50, 8117–8120. [Google Scholar] [CrossRef]
- Van de Vyver, T.; Bogaert, B.; De Backer, L.; Joris, F.; Guagliardo, R.; Van Hoeck, J.; Merckx, P.; Van Calenbergh, S.; Ramishetti, S.; Peer, D.; et al. Cationic Amphiphilic Drugs Boost the Lysosomal Escape of Small Nucleic Acid Therapeutics in a Nanocarrier-Dependent Manner. ACS Nano 2020, 14, 4774–4791. [Google Scholar] [CrossRef]
- Liu, H.; Lai, W.; Liu, X.; Yang, H.; Fang, Y.; Tian, L.; Li, K.; Nie, H.; Zhang, W.; Shi, Y.; et al. Exposure to copper oxide nanoparticles triggers oxidative stress and endoplasmic reticulum (ER)-stress induced toxicology and apoptosis in male rat liver and BRL-3A cell. J. Hazard. Mater. 2020, 401, 123349. [Google Scholar] [CrossRef]
- Zhou, Y.; Hong, F.; Tian, Y.; Zhao, X.; Hong, J.; Ze, Y.; Wang, L. Nanoparticulate titanium dioxide-inhibited dendritic development is involved in apoptosis and autophagy of hippocampal neurons in offspring mice. Toxicol. Res. 2017, 6, 889–901. [Google Scholar] [CrossRef]
- Di Meo, S.; Reed, T.T.; Venditti, P.; Victor, V.M. Role of ROS and RNS Sources in Physiological and Pathological Conditions. Oxid. Med. Cell. Longev. 2016, 2016, 1245049. [Google Scholar] [CrossRef]
- Long, J.; Ma, W.; Yu, Z.; Liu, H.; Cao, Y. Multi-walled carbon nanotubes (MWCNTs) promoted lipid accumulation in THP-1 macrophages through modulation of endoplasmic reticulum (ER) stress. Nanotoxicology 2019, 13, 938–951. [Google Scholar] [CrossRef] [PubMed]
- Adams, C.J.; Kopp, M.C.; Larburu, N.; Nowak, P.R.; Ali, M.M.U. Structure and Molecular Mechanism of ER Stress Signaling by the Unfolded Protein Response Signal Activator IRE1. Front. Mol. Biosci. 2019, 6, 11. [Google Scholar] [CrossRef]
- Chen, R.J.; Chen, Y.Y.; Liao, M.Y.; Lee, Y.H.; Chen, Z.Y.; Yan, S.J.; Yeh, Y.L.; Yang, L.X.; Lee, Y.L.; Wu, Y.H.; et al. The Current Understanding of Autophagy in Nanomaterial Toxicity and Its Implementation in Safety Assessment-Related Alternative Testing Strategies. Int. J. Mol. Sci. 2020, 21, 2387. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.W.; Xia, T.; Lee, Y.H.; Chen, C.W.; Tsai, J.C.; Wang, Y.J. Cationic polystyrene nanospheres induce autophagic cell death through the induction of endoplasmic reticulum stress. Nanoscale 2015, 7, 736–746. [Google Scholar] [CrossRef] [PubMed]
- Landsiedel, R.; Kapp, M.D.; Schulz, M.; Wiench, K.; Oesch, F. Genotoxicity investigations on nanomaterials: Methods, preparation and characterization of test material, potential artifacts and limitations--many questions, some answers. Mutat. Res. 2009, 681, 241–258. [Google Scholar] [CrossRef] [PubMed]
- Ling, C.; An, H.; Li, L.; Wang, J.; Lu, T.; Wang, H.; Hu, Y.; Song, G.; Liu, S. Genotoxicity Evaluation of Titanium Dioxide Nanoparticles In Vitro: A Systematic Review of the Literature and Meta-analysis. Biol. Trace Elem. Res. 2020. [Google Scholar] [CrossRef]
- Shan, W.; Cui, Y.; Liu, M.; Wu, L.; Xiang, Y.; Guo, Q.; Zhang, Z.; Huang, Y. Systematic evaluation of the toxicity and biodistribution of virus mimicking mucus-penetrating DLPC-NPs as oral drug delivery system. Int. J. Pharm. 2017, 530, 89–98. [Google Scholar] [CrossRef]
- Proquin, H.; Rodriguez-Ibarra, C.; Moonen, C.G.; Urrutia Ortega, I.M.; Briede, J.J.; de Kok, T.M.; van Loveren, H.; Chirino, Y.I. Titanium dioxide food additive (E171) induces ROS formation and genotoxicity: Contribution of micro and nano-sized fractions. Mutagenesis 2017, 32, 139–149. [Google Scholar] [CrossRef]
- Kawanishi, M.; Ogo, S.; Ikemoto, M.; Totsuka, Y.; Ishino, K.; Wakabayashi, K.; Yagi, T. Genotoxicity and reactive oxygen species production induced by magnetite nanoparticles in mammalian cells. J. Toxicol. Sci. 2013, 38, 503–511. [Google Scholar] [CrossRef]
- Priyadarsini, S.; Sahoo, S.K.; Sahu, S.; Mukherjee, S.; Hota, G.; Mishra, M. Oral administration of graphene oxide nano-sheets induces oxidative stress, genotoxicity, and behavioral teratogenicity in Drosophila melanogaster. Environ. Sci. Pollut. Res. Int. 2019, 26, 19560–19574. [Google Scholar] [CrossRef] [PubMed]
- Mahaye, N.; Thwala, M.; Cowan, D.A.; Musee, N. Genotoxicity of metal based engineered nanoparticles in aquatic organisms: A review. Mutat. Res. 2017, 773, 134–160. [Google Scholar] [CrossRef]
- Magdolenova, Z.; Collins, A.; Kumar, A.; Dhawan, A.; Stone, V.; Dusinska, M. Mechanisms of genotoxicity. A review of in vitro and in vivo studies with engineered nanoparticles. Nanotoxicology 2014, 8, 233–278. [Google Scholar] [CrossRef]
- Barabadi, H.; Najafi, M.; Samadian, H.; Azarnezhad, A.; Vahidi, H.; Mahjoub, M.A.; Koohiyan, M.; Ahmadi, A. A Systematic Review of the Genotoxicity and Antigenotoxicity of Biologically Synthesized Metallic Nanomaterials: Are Green Nanoparticles Safe Enough for Clinical Marketing? Medicina 2019, 55, 439. [Google Scholar] [CrossRef] [PubMed]
- Galeone, A.; Vecchio, G.; Malvindi, M.A.; Brunetti, V.; Cingolani, R.; Pompa, P.P. In vivo assessment of CdSe-ZnS quantum dots: Coating dependent bioaccumulation and genotoxicity. Nanoscale 2012, 4, 6401–6407. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhou, P.; Yang, Y.; Liu, J.; Widjaya, A.S.; Jiang, Y. Development of arsenic trioxide sustained-release pellets for reducing toxicity and improving compliance. Drug Dev. Ind. Pharm. 2020, 1–30. [Google Scholar] [CrossRef]
- Yang, G.F.; Li, X.H.; Zhao, Z.; Wang, W.B. Preparation, characterization, in vivo and in vitro studies of arsenic trioxide Mg-Fe ferrite magnetic nanoparticles. Acta Pharm. Sin. 2009, 30, 1688–1693. [Google Scholar] [CrossRef]
- Kang, T.; Guan, R.; Chen, X.; Song, Y.; Jiang, H.; Zhao, J. In vitro toxicity of different-sized ZnO nanoparticles in Caco-2 cells. Nanoscale Res. Lett. 2013, 8, 496. [Google Scholar] [CrossRef]
- Hong, T.K.; Tripathy, N.; Son, H.J.; Ha, K.T.; Jeong, H.S.; Hahn, Y.B. A comprehensive in vitro and in vivo study of ZnO nanoparticles toxicity. J. Mater. Chem. B 2013, 1, 2985–2992. [Google Scholar] [CrossRef]
- Abbasi-Oshaghi, E.; Mirzaei, F.; Mirzaei, A. Effects of ZnO nanoparticles on intestinal function and structure in normal/high fat diet-fed rats and Caco-2 cells. Nanomedicine 2018, 13, 2791–2816. [Google Scholar] [CrossRef]
- Choi, J.S.; Kim, R.O.; Yoon, S.; Kim, W.K. Developmental Toxicity of Zinc Oxide Nanoparticles to Zebrafish (Danio rerio): A Transcriptomic Analysis. PLoS ONE 2016, 11, e0160763. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kong, M.; Zhou, Z.; Yan, D.; Yu, X.; Cheng, X.; Feng, C.; Liu, Y.; Chen, X. Mechanism of surface charge triggered intestinal epithelial tight junction opening upon chitosan nanoparticles for insulin oral delivery. Carbohydr. Polym. 2017, 157, 596–602. [Google Scholar] [CrossRef]
- Hsu, L.W.; Lee, P.L.; Chen, C.T.; Mi, F.L.; Juang, J.H.; Hwang, S.M.; Ho, Y.C.; Sung, H.W. Elucidating the signaling mechanism of an epithelial tight-junction opening induced by chitosan. Biomaterials 2012, 33, 6254–6263. [Google Scholar] [CrossRef] [PubMed]
- Vllasaliu, D.; Exposito-Harris, R.; Heras, A.; Casettari, L.; Garnett, M.; Illum, L.; Stolnik, S. Tight junction modulation by chitosan nanoparticles: Comparison with chitosan solution. Int. J. Pharm. 2010, 400, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, B.; Kolli, A.R.; Esch, M.B.; Abaci, H.E.; Shuler, M.L.; Hickman, J.J. TEER measurement techniques for in vitro barrier model systems. J. Lab. Autom. 2015, 20, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Einspanier, R.; Schoen, J. Transepithelial electrical resistance (TEER): A functional parameter to monitor the quality of oviduct epithelial cells cultured on filter supports. Histochem. Cell Biol. 2015, 144, 509–515. [Google Scholar] [CrossRef]
- Ude, V.C.; Brown, D.M.; Viale, L.; Kanase, N.; Stone, V.; Johnston, H.J. Impact of copper oxide nanomaterials on differentiated and undifferentiated Caco-2 intestinal epithelial cells; assessment of cytotoxicity, barrier integrity, cytokine production and nanomaterial penetration. Part. Fibre Toxicol. 2017, 14, 31. [Google Scholar] [CrossRef]
- Ferruzza, S.; Scacchi, M.; Scarino, M.L.; Sambuy, Y. Iron and copper alter tight junction permeability in human intestinal Caco-2 cells by distinct mechanisms. Toxicol. In Vitro 2002, 16, 399–404. [Google Scholar] [CrossRef]
- Wang, B.; Feng, W.Y.; Wang, T.C.; Jia, G.; Wang, M.; Shi, J.W.; Zhang, F.; Zhao, Y.L.; Chai, Z.F. Acute toxicity of nano- and micro-scale zinc powder in healthy adult mice. Toxicol. Lett. 2006, 161, 115–123. [Google Scholar] [CrossRef]
- Piccapietra, F.; Sigg, L.; Behra, R. Colloidal stability of carbonate-coated silver nanoparticles in synthetic and natural freshwater. Environ. Sci. Technol. 2012, 46, 818–825. [Google Scholar] [CrossRef]
- van der Zande, M.; Jemec Kokalj, A.; Spurgeon, D.J.; Loureiro, S.; Silva, P.V.; Khodaparast, Z.; Drobne, D.; Clark, N.J.; van den Brink, N.W.; Baccaro, M.; et al. The gut barrier and the fate of engineered nanomaterials: A view from comparative physiology. Environ. Sci. Nano 2020, 7, 1874–1898. [Google Scholar] [CrossRef]
- Nikazar, S.; Sivasankarapillai, V.S.; Rahdar, A.; Gasmi, S.; Anumol, P.S.; Shanavas, M.S. Revisiting the cytotoxicity of quantum dots: An in-depth overview. Biophys. Rev. 2020, 12, 703–718. [Google Scholar] [CrossRef]
- Ishida, N.; Hosokawa, Y.; Imaeda, T.; Hatanaka, T. Reduction of the Cytotoxicity of Copper (II) Oxide Nanoparticles by Coating with a Surface-Binding Peptide. Appl. Biochem. Biotechnol. 2020, 190, 645–659. [Google Scholar] [CrossRef]
- Pranantyo, D.; Liu, P.; Zhong, W.; Kang, E.T.; Chan-Park, M.B. Antimicrobial Peptide-Reduced Gold Nanoclusters with Charge-Reversal Moieties for Bacterial Targeting and Imaging. Biomacromolecules 2019, 20, 2922–2933. [Google Scholar] [CrossRef] [PubMed]
- Gitrowski, C.; Al-Jubory, A.R.; Handy, R.D. Uptake of different crystal structures of TiO(2) nanoparticles by Caco-2 intestinal cells. Toxicol. Lett. 2014, 226, 264–276. [Google Scholar] [CrossRef]
- Axson, J.L.; Stark, D.I.; Bondy, A.L.; Capracotta, S.S.; Maynard, A.D.; Philbert, M.A.; Bergin, I.L.; Ault, A.P. Rapid Kinetics of Size and pH-Dependent Dissolution and Aggregation of Silver Nanoparticles in Simulated Gastric Fluid. J. Phys. Chem. C Nanomater. Interfaces 2015, 119, 20632–20641. [Google Scholar] [CrossRef]
- Zhu, L.; Pelaz, B.; Chakraborty, I.; Parak, W.J. Investigating Possible Enzymatic Degradation on Polymer Shells around Inorganic Nanoparticles. Int. J. Mol. Sci. 2019, 20, 935. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81. [Google Scholar] [CrossRef]
- Gulati, M.; Singh, S.K.; Corrie, L.; Kaur, I.P.; Chandwani, L. Delivery routes for faecal microbiota transplants: Available, anticipated and aspired. Pharm. Res. 2020, 159, 104954. [Google Scholar] [CrossRef]
- Mohajeri, M.H.; Brummer, R.J.M.; Rastall, R.A.; Weersma, R.K.; Harmsen, H.J.M.; Faas, M.; Eggersdorfer, M. The role of the microbiome for human health: From basic science to clinical applications. Eur. J. Nutr. 2018, 57, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Ng, S.C. The Gut Microbiota in the Pathogenesis and Therapeutics of Inflammatory Bowel Disease. Front. Microbiol. 2018, 9, 2247. [Google Scholar] [CrossRef]
- Festi, D.; Schiumerini, R.; Eusebi, L.H.; Marasco, G.; Taddia, M.; Colecchia, A. Gut microbiota and metabolic syndrome. World J. Gastroenterol. 2014, 20, 16079–16094. [Google Scholar] [CrossRef]
- Xie, Y.; Wu, B.; Zhang, X.X.; Yin, J.; Mao, L.; Hu, M. Influences of graphene on microbial community and antibiotic resistance genes in mouse gut as determined by high-throughput sequencing. Chemosphere 2016, 144, 1306–1312. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Lu, J.; Lin, G.; Su, H.; Sun, J.; Luan, T. Dysbiosis of gut microbiota by dietary exposure of three graphene-family materials in zebrafish (Danio rerio). Environ. Pollut. 2019, 254, 112969. [Google Scholar] [CrossRef]
- Lamas, B.; Martins Breyner, N.; Houdeau, E. Impacts of foodborne inorganic nanoparticles on the gut microbiota-immune axis: Potential consequences for host health. Part. Fibre Toxicol. 2020, 17, 19. [Google Scholar] [CrossRef]
- Agans, R.T.; Gordon, A.; Hussain, S.; Paliy, O. Titanium Dioxide Nanoparticles Elicit Lower Direct Inhibitory Effect on Human Gut Microbiota Than Silver Nanoparticles. Toxicol. Sci. 2019, 172, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Fondevila, M.; Herrer, R.; Casallas, M.C.; Abecia, L.; Ducha, J.J. Silver nanoparticles as a potential antimicrobial additive for weaned pigs. Anim. Feed Sci. Technol. 2009, 150, 259–269. [Google Scholar] [CrossRef]
- Frohlich, E.E.; Frohlich, E. Cytotoxicity of Nanoparticles Contained in Food on Intestinal Cells and the Gut Microbiota. Int. J. Mol. Sci. 2016, 17, 509. [Google Scholar] [CrossRef]
- Yang, C.; Merlin, D. Lipid-Based Drug Delivery Nanoplatforms for Colorectal Cancer Therapy. Nanomaterials 2020, 10, 1424. [Google Scholar] [CrossRef]
- Zhang, T.; Zhu, G.; Lu, B.; Peng, Q. Oral Nano-Delivery Systems for Colon Targeting Therapy. Pharm. Nanotechnol. 2017, 5, 83–94. [Google Scholar] [CrossRef]
- Mohammad-Beigi, H.; Hayashi, Y.; Zeuthen, C.M.; Eskandari, H.; Scavenius, C.; Juul-Madsen, K.; Vorup-Jensen, T.; Enghild, J.J.; Sutherland, D.S. Mapping and identification of soft corona proteins at nanoparticles and their impact on cellular association. Nat. Commun. 2020, 11, 4535. [Google Scholar] [CrossRef] [PubMed]
- Guerra, A.; Etienne-Mesmin, L.; Livrelli, V.; Denis, S.; Blanquet-Diot, S.; Alric, M. Relevance and challenges in modeling human gastric and small intestinal digestion. Trends Biotechnol. 2012, 30, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Uriot, O.; Galia, W.; Awussi, A.A.; Perrin, C.; Denis, S.; Chalancon, S.; Lorson, E.; Poirson, C.; Junjua, M.; Le Roux, Y.; et al. Use of the dynamic gastro-intestinal model TIM to explore the survival of the yogurt bacterium Streptococcus thermophilus and the metabolic activities induced in the simulated human gut. Food Microbiol. 2016, 53, 18–29. [Google Scholar] [CrossRef]
- Thumann, T.A.; Pferschy-Wenzig, E.M.; Aziz-Kalbhenn, H.; Ammar, R.M.; Rabini, S.; Moissl-Eichinger, C.; Bauer, R. Application of an in vitro digestion model to study the metabolic profile changes of an herbal extract combination by UHPLC-HRMS. Phytomedicine 2020, 71, 153221. [Google Scholar] [CrossRef]
- Santbergen, M.J.C.; van der Zande, M.; Gerssen, A.; Bouwmeester, H.; Nielen, M.W.F. Dynamic in vitro intestinal barrier model coupled to chip-based liquid chromatography mass spectrometry for oral bioavailability studies. Anal. Bioanal. Chem. 2020, 412, 1111–1122. [Google Scholar] [CrossRef]
- Mao, Y.; McClements, D.J. Influence of electrostatic heteroaggregation of lipid droplets on their stability and digestibility under simulated gastrointestinal conditions. Food Funct. 2012, 3, 1025–1034. [Google Scholar] [CrossRef] [PubMed]
- Antunes, F.; Andrade, F.; Araujo, F.; Ferreira, D.; Sarmento, B. Establishment of a triple co-culture in vitro cell models to study intestinal absorption of peptide drugs. Eur. J. Pharm. Biopharm. 2013, 83, 427–435. [Google Scholar] [CrossRef]
- Deng, C.; He, Y.; Feng, J.; Dong, Z.; Yao, Y.; Lu, F. Conditioned medium from 3D culture system of stromal vascular fraction cells accelerates wound healing in diabetic rats. Regen. Med. 2019, 14, 925–937. [Google Scholar] [CrossRef]
- Souza, R.F.; Schwartz, R.E.; Mashimo, H. Esophageal stem cells and 3D-cell culture models. Ann. N. Y. Acad. Sci. 2011, 1232, 316–322. [Google Scholar] [CrossRef]
- van der Hee, B.; Madsen, O.; Vervoort, J.; Smidt, H.; Wells, J.M. Congruence of Transcription Programs in Adult Stem Cell-Derived Jejunum Organoids and Original Tissue During Long-Term Culture. Front. Cell Dev. Biol. 2020, 8, 375. [Google Scholar] [CrossRef] [PubMed]
- Stieler Stewart, A.; Freund, J.M.; Blikslager, A.T.; Gonzalez, L.M. Intestinal Stem Cell Isolation and Culture in a Porcine Model of Segmental Small Intestinal Ischemia. J. Vis. Exp. 2018. [Google Scholar] [CrossRef] [PubMed]
- Pastula, A.; Middelhoff, M.; Brandtner, A.; Tobiasch, M.; Hohl, B.; Nuber, A.H.; Demir, I.E.; Neupert, S.; Kollmann, P.; Mazzuoli-Weber, G.; et al. Three-Dimensional Gastrointestinal Organoid Culture in Combination with Nerves or Fibroblasts: A Method to Characterize the Gastrointestinal Stem Cell Niche. Stem Cells Int. 2016, 2016, 3710836. [Google Scholar] [CrossRef]
- Grosheva, I.; Zheng, D.; Levy, M.; Polansky, O.; Lichtenstein, A.; Golani, O.; Dori-Bachash, M.; Moresi, C.; Shapiro, H.; Del Mare-Roumani, S.; et al. High-Throughput Screen Identifies Host and Microbiota Regulators of Intestinal Barrier Function. Gastroenterology 2020. [Google Scholar] [CrossRef] [PubMed]
- Samsa, L.A.; Williamson, I.A.; Magness, S.T. Quantitative Analysis of Intestinal Stem Cell Dynamics Using Microfabricated Cell Culture Arrays. Methods Mol. Biol. 2018, 1842, 139–166. [Google Scholar] [CrossRef]
- Verwei, M.; Minekus, M.; Zeijdner, E.; Schilderink, R.; Havenaar, R. Evaluation of two dynamic in vitro models simulating fasted and fed state conditions in the upper gastrointestinal tract (TIM-1 and tiny-TIM) for investigating the bioaccessibility of pharmaceutical compounds from oral dosage forms. Int. J. Pharm. 2016, 498, 178–186. [Google Scholar] [CrossRef]
- Diaz-Gimeno, P.; Cervello, I. Personalized medicine through three-dimensional cell-based culture systems in assisted reproductive technologies: How close are we? Fertil. Steril. 2020, 114, 520–521. [Google Scholar] [CrossRef]
- Shehzad, A.; Ravinayagam, V.; AlRumaih, H.; Aljafary, M.; Almohazey, D.; Almofty, S.; Al-Rashid, N.A.; Al-Suhaimi, E.A. Application of Three-dimensional (3D) Tumor Cell Culture Systems and Mechanism of Drug Resistance. Curr. Pharm. Des. 2019, 25, 3599–3607. [Google Scholar] [CrossRef]
- Jokinen, M.; Pittois, K.; van den Akker, S.; Gutschoven, I.; Assmuth, T.; Metz, T.; Lehtila, H.; Alanne, P. Multiphase matrix of silica, culture medium and air for 3D mammalian cell culture. Cytotechnology 2020, 72, 271–282. [Google Scholar] [CrossRef]
- Ong, L.J.Y.; Zhu, L.; Tan, G.J.S.; Toh, Y.C. Quantitative Image-Based Cell Viability (QuantICV) Assay for Microfluidic 3D Tissue Culture Applications. Micromachines (Basel) 2020, 11, 669. [Google Scholar] [CrossRef]
- van Duinen, V.; Trietsch, S.J.; Joore, J.; Vulto, P.; Hankemeier, T. Microfluidic 3D cell culture: From tools to tissue models. Curr. Opin. Biotechnol. 2015, 35, 118–126. [Google Scholar] [CrossRef]
- Gjorevski, N.; Sachs, N.; Manfrin, A.; Giger, S.; Bragina, M.E.; Ordonez-Moran, P.; Clevers, H.; Lutolf, M.P. Designer matrices for intestinal stem cell and organoid culture. Nature 2016, 539, 560–564. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Valadez, A.V.; Zuo, P.; Nie, Z. Microfluidic 3D cell culture: Potential application for tissue-based bioassays. Bioanalysis 2012, 4, 1509–1525. [Google Scholar] [CrossRef]
- Huh, D.; Hamilton, G.A.; Ingber, D.E. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011, 21, 745–754. [Google Scholar] [CrossRef]
- Kankala, R.K.; Wang, S.B.; Chen, A.Z. Microengineered Organ-on-a-chip Platforms towards Personalized Medicine. Curr. Pharm. Des. 2018, 24, 5354–5366. [Google Scholar] [CrossRef]
- Sun, W.; Luo, Z.; Lee, J.; Kim, H.J.; Lee, K.; Tebon, P.; Feng, Y.; Dokmeci, M.R.; Sengupta, S.; Khademhosseini, A. Organ-on-a-Chip for Cancer and Immune Organs Modeling. Adv. Healthc. Mater. 2019, 8, e1801363. [Google Scholar] [CrossRef]
- Ashammakhi, N.; Nasiri, R.; Barros, N.R.; Tebon, P.; Thakor, J.; Goudie, M.; Shamloo, A.; Martin, M.G.; Khademhosseini, A. Gut-on-a-chip: Current progress and future opportunities. Biomaterials 2020, 255, 120196. [Google Scholar] [CrossRef]
- Lee, J.; Choi, J.H.; Kim, H.J. Human gut-on-a-chip technology: Will this revolutionize our understanding of IBD and future treatments? Expert Rev. Gastroenterol. Hepatol. 2016, 10, 883–885. [Google Scholar] [CrossRef][Green Version]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef] [PubMed]
- Beaurivage, C.; Naumovska, E.; Chang, Y.X.; Elstak, E.D.; Nicolas, A.; Wouters, H.; van Moolenbroek, G.; Lanz, H.L.; Trietsch, S.J.; Joore, J.; et al. Development of a Gut-On-A-Chip Model for High Throughput Disease Modeling and Drug Discovery. Int. J. Mol. Sci. 2019, 20, 5661. [Google Scholar] [CrossRef]
- Mertz, L. Omics Tech, Gut-on-a-Chip, and Bacterial Engineering: New Approaches for Treating Inflammatory Bowel Diseases. IEEE Pulse 2016, 7, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Shin, W.; Hinojosa, C.D.; Ingber, D.E.; Kim, H.J. Human Intestinal Morphogenesis Controlled by Transepithelial Morphogen Gradient and Flow-Dependent Physical Cues in a Microengineered Gut-on-a-Chip. Iscience 2019, 15, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Li, H.; Collins, J.J.; Ingber, D.E. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc. Natl. Acad. Sci. USA 2016, 113, E7–E15. [Google Scholar] [CrossRef] [PubMed]
- Majeed, A.; Javed, F.; Akhtar, S.; Saleem, U.; Anwar, F.; Ahmad, B.; Nadhman, A.; Shahnaz, G.; Hussain, I.; Hussain, S.Z.; et al. Green synthesized selenium doped zinc oxide nano-antibiotic: Synthesis, characterization and evaluation of antimicrobial, nanotoxicity and teratogenicity potential. J. Mater. Chem. B 2020. [Google Scholar] [CrossRef]
- Bondue, P.; Lebrun, S.; Taminiau, B.; Everaert, N.; LaPointe, G.; Crevecoeur, S.; Daube, G.; Delcenserie, V. A toddler SHIME(R) model to study microbiota of young children. FEMS Microbiol. Lett. 2020, 367. [Google Scholar] [CrossRef]
- Van de Wiele, T.; Van den Abbeele, P.; Ossieur, W.; Possemiers, S.; Marzorati, M. The Simulator of the Human Intestinal Microbial Ecosystem (SHIME((R))). In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., Lopez-Exposito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer Nature Switzerland AG: Gewerbestrasse, Cham, Switzerland, 2015; pp. 305–317. [Google Scholar] [CrossRef]
- Venema, K. The TNO In Vitro Model of the Colon (TIM-2). In The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., Lopez-Exposito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer Nature Switzerland AG: Gewerbestrasse, Cham, Switzerland, 2015; pp. 293–304. [Google Scholar] [CrossRef]
- Alander, M.; De Smet, I.; Nollet, L.; Verstraete, W.; von Wright, A.; Mattila-Sandholm, T. The effect of probiotic strains on the microbiota of the Simulator of the Human Intestinal Microbial Ecosystem (SHIME). Int. J. Food Microbiol. 1999, 46, 71–79. [Google Scholar] [CrossRef]
- Giuliani, C.; Marzorati, M.; Innocenti, M.; Vilchez-Vargas, R.; Vital, M.; Pieper, D.H.; Van de Wiele, T.; Mulinacci, N. Dietary supplement based on stilbenes: A focus on gut microbial metabolism by the in vitro simulator M-SHIME(R). Food Funct. 2016, 7, 4564–4575. [Google Scholar] [CrossRef]
- Lambrecht, E.; Van Coillie, E.; Van Meervenne, E.; Boon, N.; Heyndrickx, M.; Van de Wiele, T. Commensal E. coli rapidly transfer antibiotic resistance genes to human intestinal microbiota in the Mucosal Simulator of the Human Intestinal Microbial Ecosystem (M-SHIME). Int. J. Food Microbiol. 2019, 311, 108357. [Google Scholar] [CrossRef]
- Marzorati, M.; Abbeele, P.V.D.; Bubeck, S.S.; Bayne, T.; Krishnan, K.; Young, A.; Mehta, D.; DeSouza, A. Bacillus subtilis HU58 and Bacillus coagulans SC208 Probiotics Reduced the Effects of Antibiotic-Induced Gut Microbiome Dysbiosis in An M-SHIME((R)) Model. Microorganisms 2020, 8, 1028. [Google Scholar] [CrossRef]
- Kortman, G.A.; Dutilh, B.E.; Maathuis, A.J.; Engelke, U.F.; Boekhorst, J.; Keegan, K.P.; Nielsen, F.G.; Betley, J.; Weir, J.C.; Kingsbury, Z.; et al. Microbial Metabolism Shifts Towards an Adverse Profile with Supplementary Iron in the TIM-2 In vitro Model of the Human Colon. Front. Microbiol. 2015, 6, 1481. [Google Scholar] [CrossRef]
- Sayago-Ayerdi, S.G.; Zamora-Gasga, V.M.; Venema, K. Prebiotic effect of predigested mango peel on gut microbiota assessed in a dynamic in vitro model of the human colon (TIM-2). Food Res. Int. 2019, 118, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Sayago-Ayerdi, S.G.; Zamora-Gasga, V.M.; Venema, K. Changes in gut microbiota in predigested Hibiscus sabdariffa L calyces and Agave (Agave tequilana weber) fructans assessed in a dynamic in vitro model (TIM-2) of the human colon. Food Res. Int. 2020, 132, 109036. [Google Scholar] [CrossRef] [PubMed]
- Yin, N.; Du, H.; Wang, P.; Cai, X.; Chen, P.; Sun, G.; Cui, Y. Interindividual variability of soil arsenic metabolism by human gut microbiota using SHIME model. Chemosphere 2017, 184, 460–466. [Google Scholar] [CrossRef]
- Garcia-Villalba, R.; Vissenaekens, H.; Pitart, J.; Romo-Vaquero, M.; Espin, J.C.; Grootaert, C.; Selma, M.V.; Raes, K.; Smagghe, G.; Possemiers, S.; et al. Gastrointestinal Simulation Model TWIN-SHIME Shows Differences between Human Urolithin-Metabotypes in Gut Microbiota Composition, Pomegranate Polyphenol Metabolism, and Transport along the Intestinal Tract. J. Agric. Food Chem. 2017, 65, 5480–5493. [Google Scholar] [CrossRef]
- Hill, C.; Paul Ross, R.; Stanton, C.; O’Toole, P.W. The Human Microbiome in Health and Disease. In Host-Pathogen Interaction; Unden, G., Thines, E., Schüffler, A., Eds.; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2016; pp. 57–76. [Google Scholar] [CrossRef]
- Becker, N.; Kunath, J.; Loh, G.; Blaut, M. Human intestinal microbiota: Characterization of a simplified and stable gnotobiotic rat model. Gut Microbes 2011, 2, 25–33. [Google Scholar] [CrossRef]
- Slezak, K.; Hanske, L.; Loh, G.; Blaut, M. Increased bacterial putrescine has no impact on gut morphology and physiology in gnotobiotic adolescent mice. Benef. Microbes 2013, 4, 253–266. [Google Scholar] [CrossRef]
- Krause, J.L.; Schaepe, S.S.; Fritz-Wallace, K.; Engelmann, B.; Rolle-Kampczyk, U.; Kleinsteuber, S.; Schattenberg, F.; Liu, Z.; Mueller, S.; Jehmlich, N.; et al. Following the community development of SIHUMIx—A new intestinal in vitro model for bioreactor use. Gut Microbes 2020, 11, 1116–1129. [Google Scholar] [CrossRef] [PubMed]
- Lengfelder, I.; Sava, I.G.; Hansen, J.J.; Kleigrewe, K.; Herzog, J.; Neuhaus, K.; Hofmann, T.; Sartor, R.B.; Haller, D. Complex Bacterial Consortia Reprogram the Colitogenic Activity of Enterococcus faecalis in a Gnotobiotic Mouse Model of Chronic, Immune-Mediated Colitis. Front. Immunol. 2019, 10, 1420. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Fritz, J.V.; Glaab, E.; Desai, M.S.; Greenhalgh, K.; Frachet, A.; Niegowska, M.; Estes, M.; Jager, C.; Seguin-Devaux, C.; et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat. Commun. 2016, 7, 11535. [Google Scholar] [CrossRef]
- von Martels, J.Z.H.; Sadaghian Sadabad, M.; Bourgonje, A.R.; Blokzijl, T.; Dijkstra, G.; Faber, K.N.; Harmsen, H.J.M. The role of gut microbiota in health and disease: In vitro modeling of host-microbe interactions at the aerobe-anaerobe interphase of the human gut. Anaerobe 2017, 44, 3–12. [Google Scholar] [CrossRef]
- Clift, M.J.; Raemy, D.O.; Endes, C.; Ali, Z.; Lehmann, A.D.; Brandenberger, C.; Petri-Fink, A.; Wick, P.; Parak, W.J.; Gehr, P.; et al. Can the Ames test provide an insight into nano-object mutagenicity? Investigating the interaction between nano-objects and bacteria. Nanotoxicology 2013, 7, 1373–1385. [Google Scholar] [CrossRef] [PubMed]
- Barzan, E.; Mehrabian, S.; Irian, S. Antimicrobial and Genotoxicity Effects of Zero-valent Iron Nanoparticles. Jundishapur J. Microbiol. 2014, 7, e10054. [Google Scholar] [CrossRef] [PubMed]
- Barenholz, Y. Doxil(R)—The first FDA-approved nano-drug: Lessons learned. J. Control. Release 2012, 160, 117–134. [Google Scholar] [CrossRef] [PubMed]
- Keatley, K.L. A comparison of the U.S. EPA FIFRA GLP standards with the U.S. FDA GLP standards for nonclinical laboratory studies. Qual. Assur. 1999, 7, 147–154. [Google Scholar] [CrossRef]
- Collins, T.F.; Sprando, R.L.; Shackelford, M.E.; Hansen, D.K.; Welsh, J.J. Food and Drug Administration proposed testing guidelines for developmental toxicity studies. Revision Committee. FDA Guidelines for Developmental Toxicity and Reproduction, Food and Drug Administration. Regul. Toxicol. Pharm. 1999, 30, 39–44. [Google Scholar] [CrossRef]
- Henney, J.E. Toxicity testing: The FDA perspective. Ann. N. Y. Acad. Sci. 2000, 919, 75–78. [Google Scholar] [CrossRef]
- Arvidson, K.B. FDA toxicity databases and real-time data entry. Toxicol. Appl. Pharm. 2008, 233, 17–19. [Google Scholar] [CrossRef]
- Acosta, C.; Barat, J.M.; Martinez-Manez, R.; Sancenon, F.; Llopis, S.; Gonzalez, N.; Genoves, S.; Ramon, D.; Martorell, P. Toxicological assessment of mesoporous silica particles in the nematode Caenorhabditis elegans. Environ. Res. 2018, 166, 61–70. [Google Scholar] [CrossRef]
- Charao, M.F.; Souto, C.; Brucker, N.; Barth, A.; Jornada, D.S.; Fagundez, D.; Avila, D.S.; Eifler-Lima, V.L.; Guterres, S.S.; Pohlmann, A.R.; et al. Caenorhabditis elegans as an alternative in vivo model to determine oral uptake, nanotoxicity, and efficacy of melatonin-loaded lipid-core nanocapsules on paraquat damage. Int. J. Nanomed. 2015, 10, 5093–5106. [Google Scholar] [CrossRef]
- Qu, Y.; Li, W.; Zhou, Y.; Liu, X.; Zhang, L.; Wang, L.; Li, Y.F.; Iida, A.; Tang, Z.; Zhao, Y.; et al. Full assessment of fate and physiological behavior of quantum dots utilizing Caenorhabditis elegans as a model organism. Nano Lett. 2011, 11, 3174–3183. [Google Scholar] [CrossRef]
- Bergman, P.; Seyedoleslami Esfahani, S.; Engstrom, Y. Drosophila as a Model for Human Diseases-Focus on Innate Immunity in Barrier Epithelia. Curr. Top. Dev. Biol. 2017, 121, 29–81. [Google Scholar] [CrossRef] [PubMed]
- Behrman, E.L.; Howick, V.M.; Kapun, M.; Staubach, F.; Bergland, A.O.; Petrov, D.A.; Lazzaro, B.P.; Schmidt, P.S. Rapid seasonal evolution in innate immunity of wild Drosophila melanogaster. Proc. Biol. Sci. 2018, 285. [Google Scholar] [CrossRef] [PubMed]
- Troha, K.; Buchon, N. Methods for the study of innate immunity in Drosophila melanogaster. Wiley Interdiscip. Rev. Dev. Biol. 2019, 8, e344. [Google Scholar] [CrossRef] [PubMed]
- Can, H.; Chanumolu, S.K.; Gonzalez-Munoz, E.; Prukudom, S.; Otu, H.H.; Cibelli, J.B. Comparative analysis of single-cell transcriptomics in human and Zebrafish oocytes. BMC Genom. 2020, 21, 471. [Google Scholar] [CrossRef]
- Sander, V.; Patke, S.; Lee, J.Y.; Chang, Y.T.; Davidson, A.J. The Vital Dye CDr10b Labels the Zebrafish Mid-Intestine and Lumen. Molecules 2017, 22, 454. [Google Scholar] [CrossRef]
- Duan, J.; Liang, S.; Yu, Y.; Li, Y.; Wang, L.; Wu, Z.; Chen, Y.; Miller, M.R.; Sun, Z. Inflammation-coagulation response and thrombotic effects induced by silica nanoparticles in zebrafish embryos. Nanotoxicology 2018, 12, 470–484. [Google Scholar] [CrossRef] [PubMed]
- Merrifield, D.L.; Shaw, B.J.; Harper, G.M.; Saoud, I.P.; Davies, S.J.; Handy, R.D.; Henry, T.B. Ingestion of metal-nanoparticle contaminated food disrupts endogenous microbiota in zebrafish (Danio rerio). Environ. Pollut. 2013, 174, 157–163. [Google Scholar] [CrossRef]
- Mathai, B.J.; Meijer, A.H.; Simonsen, A. Studying Autophagy in Zebrafish. Cells 2017, 6, 21. [Google Scholar] [CrossRef]
- Fodor, E.; Sigmond, T.; Ari, E.; Lengyel, K.; Takacs-Vellai, K.; Varga, M.; Vellai, T. Methods to Study Autophagy in Zebrafish. Methods Enzym. 2017, 588, 467–496. [Google Scholar] [CrossRef]
- Collett, A.; Tanianis-Hughes, J.; Carlson, G.L.; Harwood, M.D.; Warhurst, G. Comparison of P-glycoprotein-mediated drug-digoxin interactions in Caco-2 with human and rodent intestine: Relevance to in vivo prediction. Eur. J. Pharm. Sci. 2005, 26, 386–393. [Google Scholar] [CrossRef]
- Storch, J.; Veerkamp, J.H.; Hsu, K.T. Similar mechanisms of fatty acid transfer from human anal rodent fatty acid-binding proteins to membranes: Liver, intestine, heart muscle, and adipose tissue FABPs. Mol. Cell Biochem. 2002, 239, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Perlman, R.L. Mouse models of human disease: An evolutionary perspective. Evol. Med. Public Health 2016, 2016, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Bosgra, S.; van Eijkeren, J.C.; van der Schans, M.J.; Langenberg, J.P.; Slob, W. Toxicodynamic analysis of the combined cholinesterase inhibition by paraoxon and methamidophos in human whole blood. Toxicol. Appl. Pharm. 2009, 236, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Eiki, J.; Nagata, Y.; Futamura, M.; Sasaki-Yamamoto, K.; Iino, T.; Nishimura, T.; Chiba, M.; Ohyama, S.; Yoshida-Yoshimioto, R.; Fujii, K.; et al. Pharmacokinetic and pharmacodynamic properties of the glucokinase activator MK-0941 in rodent models of type 2 diabetes and healthy dogs. Mol. Pharm. 2011, 80, 1156–1165. [Google Scholar] [CrossRef]
- Yokota, T.; Struzik, Z.R.; Jurica, P.; Horiuchi, M.; Hiroyama, S.; Li, J.; Takahara, Y.; Ogawa, K.; Nishitomi, K.; Hasegawa, M.; et al. Semi-Automated Biomarker Discovery from Pharmacodynamic Effects on EEG in ADHD Rodent Models. Sci. Rep. 2018, 8, 5202. [Google Scholar] [CrossRef]
- Bahamonde, J.; Brenseke, B.; Chan, M.Y.; Kent, R.D.; Vikesland, P.J.; Prater, M.R. Gold Nanoparticle Toxicity in Mice and Rats: Species Differences. Toxicol. Pathol. 2018, 46, 431–443. [Google Scholar] [CrossRef]
- Phillips, K.A.; Bales, K.L.; Capitanio, J.P.; Conley, A.; Czoty, P.W.; t Hart, B.A.; Hopkins, W.D.; Hu, S.L.; Miller, L.A.; Nader, M.A.; et al. Why primate models matter. Am. J. Primatol. 2014, 76, 801–827. [Google Scholar] [CrossRef]
- Yong, K.T.; Law, W.C.; Hu, R.; Ye, L.; Liu, L.; Swihart, M.T.; Prasad, P.N. Nanotoxicity assessment of quantum dots: From cellular to primate studies. Chem. Soc. Rev. 2013, 42, 1236–1250. [Google Scholar] [CrossRef]
- Ye, L.; Yong, K.T.; Liu, L.; Roy, I.; Hu, R.; Zhu, J.; Cai, H.; Law, W.C.; Liu, J.; Wang, K.; et al. A pilot study in non-human primates shows no adverse response to intravenous injection of quantum dots. Nat. Nanotechnol. 2012, 7, 453–458. [Google Scholar] [CrossRef]
- Farcal, L.; Torres Andon, F.; Di Cristo, L.; Rotoli, B.M.; Bussolati, O.; Bergamaschi, E.; Mech, A.; Hartmann, N.B.; Rasmussen, K.; Riego-Sintes, J.; et al. Comprehensive In Vitro Toxicity Testing of a Panel of Representative Oxide Nanomaterials: First Steps towards an Intelligent Testing Strategy. PLoS ONE 2015, 10, e0127174. [Google Scholar] [CrossRef]
- Nel, A.; Xia, T.; Meng, H.; Wang, X.; Lin, S.; Ji, Z.; Zhang, H. Nanomaterial toxicity testing in the 21st century: Use of a predictive toxicological approach and high-throughput screening. Acc. Chem. Res. 2013, 46, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Stone, V.; Johnston, H.; Schins, R.P.F. Development of in vitro systems for nanotoxicology: Methodological considerations. Crit. Rev. Toxicol. 2009, 39, 613–626. [Google Scholar] [CrossRef]
- Larsson, P.; Engqvist, H.; Biermann, J.; Werner Rönnerman, E.; Forssell-Aronsson, E.; Kovács, A.; Karlsson, P.; Helou, K.; Parris, T.Z. Optimization of cell viability assays to improve replicability and reproducibility of cancer drug sensitivity screens. Sci. Rep. 2020, 10, 5798. [Google Scholar] [CrossRef]
- Peters, K.; Unger, R.E.; Kirkpatrick, C.J.; Gatti, A.M.; Monari, E. Effects of nano-scaled particles on endothelial cell function in vitro: Studies on viability, proliferation and inflammation. J. Mater. Sci. Mater. Med. 2004, 15, 321–325. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.G.; O’Claonadh, N.; Casey, A.; Chambers, G. Comparative in vitro cytotoxicity study of silver nanoparticle on two mammalian cell lines. Toxicol. In Vitro 2012, 26, 238–251. [Google Scholar] [CrossRef]
- Fisichella, M.; Berenguer, F.; Steinmetz, G.; Auffan, M.; Rose, J.; Prat, O. Intestinal toxicity evaluation of TiO2 degraded surface-treated nanoparticles: A combined physico-chemical and toxicogenomics approach in caco-2 cells. Part. Fibre Toxicol. 2012, 9, 18. [Google Scholar] [CrossRef]
- Yao, K.; Huang, D.; Xu, B.; Wang, N.; Wang, Y.; Bi, S. A sensitive electrochemical approach for monitoring the effects of nano-Al2O3 on LDH activity by differential pulse voltammetry. Analyst 2010, 135, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, G.K.; Cardoso, E.; Vuolo, F.S.; Galant, L.S.; Michels, M.; Goncalves, C.L.; Rezin, G.T.; Dal-Pizzol, F.; Benavides, R.; Alonso-Nunez, G.; et al. Effect of acute and long-term administration of gold nanoparticles on biochemical parameters in rat brain. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 79, 748–755. [Google Scholar] [CrossRef]
- van de Sandt, J.J.; Rutten, A.A.; Koeter, H.B. Cutaneous toxicity testing in organ culture: Neutral red uptake and reduction of tetrazolium salt (MTT). Toxicol. In Vitro 1993, 7, 81–86. [Google Scholar] [CrossRef]
- Goodwin, C.J.; Holt, S.J.; Downes, S.; Marshall, N.J. Microculture tetrazolium assays: A comparison between two new tetrazolium salts, XTT and MTS. J. Immunol. Methods 1995, 179, 95–103. [Google Scholar] [CrossRef]
- Eilenberger, C.; Kratz, S.R.A.; Rothbauer, M.; Ehmoser, E.K.; Ertl, P.; Kupcu, S. Optimized alamarBlue assay protocol for drug dose-response determination of 3D tumor spheroids. MethodsX 2018, 5, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of Cell Viability by the alamarBlue Assay. Cold Spring Harb. Protoc. 2018, 2018. [Google Scholar] [CrossRef]
- Kramer, C.E.; Singh, A.; Helfrich, S.; Grunberger, A.; Wiechert, W.; Noh, K.; Kohlheyer, D. Non-Invasive Microbial Metabolic Activity Sensing at Single Cell Level by Perfusion of Calcein Acetoxymethyl Ester. PLoS ONE 2015, 10, e0141768. [Google Scholar] [CrossRef]
- Kwizera, R.; Akampurira, A.; Kandole, T.K.; Nabaggala, M.S.; Williams, D.A.; Kambugu, A.; Meya, D.B.; Rhein, J.; Boulware, D.R.; Team, A. Evaluation of trypan blue stain in the TC20 automated cell counter as a point-of-care for the enumeration of viable cryptococcal cells in cerebrospinal fluid. Med. Mycol. 2018, 56, 559–564. [Google Scholar] [CrossRef]
- McArdle, A.; Pollock, N.; Staunton, C.A.; Jackson, M.J. Aberrant redox signalling and stress response in age-related muscle decline: Role in inter- and intra-cellular signalling. Free Radic. Biol. Med. 2019, 132, 50–57. [Google Scholar] [CrossRef]
- Rutley, N.; Miller, G. Large-Scale Analysis of Pollen Viability and Oxidative Level Using H2DCFDA-Staining Coupled with Flow Cytometry. Methods Mol. Biol. 2020, 2160, 167–179. [Google Scholar] [CrossRef]
- Oparka, M.; Walczak, J.; Malinska, D.; van Oppen, L.; Szczepanowska, J.; Koopman, W.J.H.; Wieckowski, M.R. Quantifying ROS levels using CM-H2DCFDA and HyPer. Methods 2016, 109, 3–11. [Google Scholar] [CrossRef]
- Ju, J.; Liu, X.; Yu, J.J.; Sun, K.; Fathi, F.; Zeng, X. Electrochemistry at Bimetallic Pd/Au Thin Film Surfaces for Selective Detection of Reactive Oxygen Species and Reactive Nitrogen Species. Anal. Chem. 2020, 92, 6538–6547. [Google Scholar] [CrossRef]
- Garcia, Y.J.; Rodriguez-Malaver, A.J.; Penaloza, N. Lipid peroxidation measurement by thiobarbituric acid assay in rat cerebellar slices. J. Neurosci. Methods 2005, 144, 127–135. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Q.; Xie, Z.; Fu, X.A. Characterization of DNPH-coated microreactor chip for analysis of trace carbonyls with application for breath analysis. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2019, 1106–1107, 58–63. [Google Scholar] [CrossRef]
- Soglia, F.; Petracci, M.; Ertbjerg, P. Novel DNPH-based method for determination of protein carbonylation in muscle and meat. Food Chem. 2016, 197, 670–675. [Google Scholar] [CrossRef]
- Van den Bergh, V.; Coeckelberghs, H.; Vankerckhoven, H.; Compernolle, F.; Vinckier, C. Study of the carbonyl products of terpene/OH radical reactions: Detection of the 2,4-DNPH derivatives by HPLC-MS. Anal. Bioanal. Chem. 2004, 379, 484–494. [Google Scholar] [CrossRef]
- Ighodaro, O.M.; Akinloye, O.A. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex. J. Med. 2018, 54, 287–293. [Google Scholar] [CrossRef]
- Weydert, C.J.; Cullen, J.J. Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nat. Protoc. 2010, 5, 51–66. [Google Scholar] [CrossRef] [PubMed]
- Gosalvez, J.; Tvrda, E.; Agarwal, A. Free radical and superoxide reactivity detection in semen quality assessment: Past, present, and future. J. Assist. Reprod. Genet. 2017, 34, 697–707. [Google Scholar] [CrossRef]
- Kim, J.-D.; Lee, C.-H.; Park, J.H.; Cho, J.-H.; Kim, I.-H.; Ahn, J.-H.; Lee, J.-C.; Chen, B.; Shin, B.-N.; Tae, H.-J.; et al. Effect of Oenanthe Javanica Extract on Antioxidant Enzyme in the Rat Liver. Chin. Med. J. 2015, 128, 1649. [Google Scholar] [CrossRef]
- Tsang, C.K.; Liu, Y.; Thomas, J.; Zhang, Y.; Zheng, X.F.S. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 2014, 5, 3446. [Google Scholar] [CrossRef] [PubMed]
- Iskusnykh, I.Y.; Popova, T.N.; Agarkov, A.A.; Pinheiro de Carvalho, M.Â.A.; Rjevskiy, S.G. Expression of Glutathione Peroxidase and Glutathione Reductase and Level of Free Radical Processes under Toxic Hepatitis in Rats. J. Toxicol. 2013, 2013, 870628. [Google Scholar] [CrossRef]
- Jones, D.P. [11] Redox potential of GSH/GSSG couple: Assay and biological significance. In Methods in Enzymology; Sies, H., Packer, L., Eds.; Academic Press: Cambridge, MA, USA, 2002; Volume 348, pp. 93–112. [Google Scholar]
- McMahon, B.K.; Gunnlaugsson, T. Selective Detection of the Reduced Form of Glutathione (GSH) over the Oxidized (GSSG) Form Using a Combination of Glutathione Reductase and a Tb(III)-Cyclen Maleimide Based Lanthanide Luminescent ‘Switch On’ Assay. J. Am. Chem. Soc. 2012, 134, 10725–10728. [Google Scholar] [CrossRef]
- Katerji, M.; Filippova, M.; Duerksen-Hughes, P. Approaches and Methods to Measure Oxidative Stress in Clinical Samples: Research Applications in the Cancer Field. Oxid. Med. Cell. Longev. 2019, 2019, 1279250. [Google Scholar] [CrossRef]
- Jiang, X.; Yu, Y.; Chen, J.; Zhao, M.; Chen, H.; Song, X.; Matzuk, A.J.; Carroll, S.L.; Tan, X.; Sizovs, A.; et al. Quantitative imaging of glutathione in live cells using a reversible reaction-based ratiometric fluorescent probe. ACS Chem. Biol. 2015, 10, 864–874. [Google Scholar] [CrossRef] [PubMed]
- Canzler, S.; Schor, J.; Busch, W.; Schubert, K.; Rolle-Kampczyk, U.E.; Seitz, H.; Kamp, H.; von Bergen, M.; Buesen, R.; Hackermüller, J. Prospects and challenges of multi-omics data integration in toxicology. Arch. Toxicol. 2020, 94, 371–388. [Google Scholar] [CrossRef] [PubMed]
- González-Ruiz, V.; Schvartz, D.; Sandström, J.; Pezzatti, J.; Jeanneret, F.; Tonoli, D.; Boccard, J.; Monnet-Tschudi, F.; Sanchez, J.-C.; Rudaz, S. An Integrative Multi-Omics Workflow to Address Multifactorial Toxicology Experiments. Metabolites 2019, 9, 79. [Google Scholar] [CrossRef]
- O’Donnell, S.T.; Ross, R.P.; Stanton, C. The Progress of Multi-Omics Technologies: Determining Function in Lactic Acid Bacteria Using a Systems Level Approach. Front. Microbiol. 2020, 10. [Google Scholar] [CrossRef]
- Chen, C.; Gao, J.; Wang, T.-S.; Guo, C.; Yan, Y.-J.; Mao, C.-Y.; Gu, L.-W.; Yang, Y.; Li, Z.-F.; Liu, A. NMR-based Metabolomic Techniques Identify the Toxicity of Emodin in HepG2 Cells. Sci. Rep. 2018, 8, 9379. [Google Scholar] [CrossRef] [PubMed]
- Verrastro, I.; Pasha, S.; Jensen, K.T.; Pitt, A.R.; Spickett, C.M. Mass spectrometry-based methods for identifying oxidized proteins in disease: Advances and challenges. Biomolecules 2015, 5, 378–411. [Google Scholar] [CrossRef] [PubMed]
- Bollineni, R.; Hoffmann, R.; Fedorova, M. Identification of protein carbonylation sites by two-dimensional liquid chromatography in combination with MALDI- and ESI-MS. J. Proteom. 2011, 74, 2338–2350. [Google Scholar] [CrossRef]
- Ji, X.; Xu, H.; Zhang, H.; Hillery, C.A.; Gao, H.-Q.; Pritchard, K.A., Jr. Anion exchange HPLC isolation of high-density lipoprotein (HDL) and on-line estimation of proinflammatory HDL. PLoS ONE 2014, 9, e91089. [Google Scholar] [CrossRef]
- Koivusalmi, E.; Haatainen, E.; Root, A. Quantitative RP-HPLC Determination of Some Aldehydes and Hydroxyaldehydes as Their 2,4-Dinitrophenylhydrazone Derivatives. Anal. Chem. 1999, 71, 86–91. [Google Scholar] [CrossRef]
- Barden, A.; Mori, T.A. GC-MS Analysis of Lipid Oxidation Products in Blood, Urine, and Tissue Samples. Methods Mol. Biol. 2018, 1730, 283–292. [Google Scholar] [CrossRef]
- Kerns, E.H.; Di, L.; Petusky, S.; Farris, M.; Ley, R.; Jupp, P. Combined Application of Parallel Artificial Membrane Permeability Assay and Caco-2 Permeability Assays in Drug Discovery. J. Pharm. Sci. 2004, 93, 1440–1453. [Google Scholar] [CrossRef] [PubMed]
- DiMarco, R.L.; Hunt, D.R.; Dewi, R.E.; Heilshorn, S.C. Improvement of paracellular transport in the Caco-2 drug screening model using protein-engineered substrates. Biomaterials 2017, 129, 152–162. [Google Scholar] [CrossRef]
- Manabe, A.; Furukawa, C.; Endo, S.; Marunaka, K.; Nishiyama, T.; Fujii, N.; Tabuchi, Y.; Matsunaga, T.; Ikari, A. Chlorpheniramine Increases Paracellular Permeability to Marker Fluorescein Lucifer Yellow Mediated by Internalization of Occludin in Murine Colonic Epithelial Cells. Biol. Pharm. Bull. 2017, 40, 1299–1305. [Google Scholar] [CrossRef]
- Franken, N.A.P.; Rodermond, H.M.; Stap, J.; Haveman, J.; van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 2006, 1, 2315–2319. [Google Scholar] [CrossRef]
- Toomeh, D.; Gadoue, S.M.; Yasmin-Karim, S.; Singh, M.; Shanker, R.; Pal Singh, S.; Kumar, R.; Sajo, E.; Ngwa, W. Minimizing the potential of cancer recurrence and metastasis by the use of graphene oxide nano-flakes released from smart fiducials during image-guided radiation therapy. Phys. Med. 2018, 55, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-J.; Park, J.-H.; Han, J.W.; Kim, E.; Jae-Wook, O.; Lee, S.Y.; Kim, J.-H.; Gurunathan, S. Differential Cytotoxic Potential of Silver Nanoparticles in Human Ovarian Cancer Cells and Ovarian Cancer Stem Cells. Int. J. Mol. Sci. 2016, 17, 2077. [Google Scholar] [CrossRef] [PubMed]
- Plotnikov, E.; Silnikov, V.; Gapeyev, A.; Plotnikov, V. Investigation of DNA-damage and Chromosomal Aberrations in Blood Cells under the Influence of New Silver-based Antiviral Complex. Adv. Pharm. Bull. 2016, 6, 71–74. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yılmaz, S.; Ünal, F.; Yüzbaşıoğlu, D. The in vitro genotoxicity of benzoic acid in human peripheral blood lymphocytes. Cytotechnology 2009, 60, 55. [Google Scholar] [CrossRef] [PubMed]
- Blajeski, A.L.; Phan, V.A.; Kottke, T.J.; Kaufmann, S.H. G(1) and G(2) cell-cycle arrest following microtubule depolymerization in human breast cancer cells. J. Clin. Investig. 2002, 110, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Hillegass, J.M.; Shukla, A.; Lathrop, S.A.; MacPherson, M.B.; Fukagawa, N.K.; Mossman, B.T. Assessing nanotoxicity in cells in vitro. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2010, 2, 219–231. [Google Scholar] [CrossRef]
- Pacurari, M.; Castranova, V. Single-Cell Gel Electrophoresis (Comet) Assay in Nano-genotoxicology. In Nanotoxicity: Methods and Protocols; Reineke, J., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 57–67. [Google Scholar] [CrossRef]
- Vandghanooni, S.; Eskandani, M. Comet assay: A method to evaluate genotoxicity of nano-drug delivery system. Bioimpacts 2011, 1, 87–97. [Google Scholar] [CrossRef]
- Olive, P.L.; Banáth, J.P. The comet assay: A method to measure DNA damage in individual cells. Nat. Protoc. 2006, 1, 23–29. [Google Scholar] [CrossRef]
- Birk, M.; Bürkle, A.; Pekari, K.; Maier, T.; Schmidt, M. Cell cycle-dependent cytotoxicity and mitotic spindle checkpoint dependency of investigational and approved antimitotic agents. Int. J. Cancer 2012, 130, 798–807. [Google Scholar] [CrossRef] [PubMed][Green Version]
- De Fries, R.; Mitsuhashi, M. Quantification of mitogen induced human lymphocyte proliferation: Comparison of alamarbluetm assay to 3h-thymidine incorporation assay. J. Clin. Lab. Anal. 1995, 9, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Schall, K.A.; Holoyda, K.A.; Grant, C.N.; Levin, D.E.; Torres, E.R.; Maxwell, A.; Pollack, H.A.; Moats, R.A.; Frey, M.R.; Darehzereshki, A.; et al. Adult zebrafish intestine resection: A novel model of short bowel syndrome, adaptation, and intestinal stem cell regeneration. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G135–G145. [Google Scholar] [CrossRef] [PubMed]
- Muir, D.; Varon, S.; Manthorpe, M. An enzyme-linked immunosorbent assay for bromodeoxyuridine incorporation using fixed microcultures. Anal. Biochem. 1990, 185, 377–382. [Google Scholar] [CrossRef]
- Lan, M.-Y.; Hsu, Y.-B.; Hsu, C.-H.; Ho, C.-Y.; Lin, J.-C.; Lee, S.-W. Induction of apoptosis by high-dose gold nanoparticles in nasopharyngeal carcinoma cells. Auris Nasus Larynx 2013, 40, 563–568. [Google Scholar] [CrossRef]
- Ferlini, C.; Kunkl, A.; Scambia, G.; Fattorossi, A. The use of Apostain in identifying early apoptosis. J. Immunol. Methods 1997, 205, 95–101. [Google Scholar] [CrossRef]
- Ganguly, P.; Breen, A.; Pillai, S.C. Toxicity of Nanomaterials: Exposure, Pathways, Assessment, and Recent Advances. ACS Biomater. Sci. Eng. 2018, 4, 2237–2275. [Google Scholar] [CrossRef]
- Ju, S.; Mu, J.; Dokland, T.; Zhuang, X.; Wang, Q.; Jiang, H.; Xiang, X.; Deng, Z.-B.; Wang, B.; Zhang, L.; et al. Grape Exosome-like Nanoparticles Induce Intestinal Stem Cells and Protect Mice From DSS-Induced Colitis. Mol. Ther. 2013, 21, 1345–1357. [Google Scholar] [CrossRef]
- Belev, B.; Brčić, I.; Prejac, J.; Golubić, Z.A.; Vrbanec, D.; Božikov, J.; Alerić, I.; Boban, M.; Razumović, J.J. Role of Ki-67 as a prognostic factor in gastrointestinal stromal tumors. World J. Gastroenterol. 2013, 19, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-W.; Cambre, M.; Lee, H.-J. The Toxicity of Nanoparticles Depends on Multiple Molecular and Physicochemical Mechanisms. Int. J. Mol. Sci. 2017, 18, 2702. [Google Scholar] [CrossRef]
- Deng, F.; He, S.; Cui, S.; Shi, Y.; Tan, Y.; Li, Z.; Huang, C.; Liu, D.; Zhi, F.; Peng, L. A Molecular Targeted Immunotherapeutic Strategy for Ulcerative Colitis via Dual-targeting Nanoparticles Delivering miR-146b to Intestinal Macrophages. J. Crohn’s Colitis 2019, 13, 482–494. [Google Scholar] [CrossRef]
- Streit, S.; Michalski, C.W.; Erkan, M.; Kleeff, J.; Friess, H. Northern blot analysis for detection and quantification of RNA in pancreatic cancer cells and tissues. Nat. Protoc. 2009, 4, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Ton-That, H.; Kaestner, K.H.; Shields, J.M.; Mahatanankoon, C.S.; Yang, V.W. Expression of the gut-enriched Krüppel-like factor gene during development and intestinal tumorigenesis. FEBS Lett. 1997, 419, 239–243. [Google Scholar] [CrossRef]
- Mo, Y.; Wan, R.; Zhang, Q. Application of reverse transcription-PCR and real-time PCR in nanotoxicity research. Methods Mol. Biol. 2012, 926, 99–112. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.; Kaushik, A.; Zhu, X.; Zhang, C.; Li, C.-Z. Chip based single cell analysis for nanotoxicity assessment. Analyst 2014, 139, 2088–2098. [Google Scholar] [CrossRef]
- Giorgio, M.; Dellino, G.I.; Gambino, V.; Roda, N.; Pelicci, P.G. On the epigenetic role of guanosine oxidation. Redox Biol. 2020, 29, 101398. [Google Scholar] [CrossRef]
- Kino, K.; Hirao-Suzuki, M.; Morikawa, M.; Sakaga, A.; Miyazawa, H. Generation, repair and replication of guanine oxidation products. Genes Environ. 2017, 39, 21. [Google Scholar] [CrossRef]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA damage: Mechanisms, mutation, and disease. FASEB J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef]
- Andersen, M.E.; Krewski, D. Toxicity Testing in the 21st Century: Bringing the Vision to Life. Toxicol. Sci. 2009, 107, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Wani, M.Y.; Hashim, M.A.; Nabi, F.; Malik, M.A. Nanotoxicity: Dimensional and Morphological Concerns. Adv. Phys. Chem. 2011, 2011, 450912. [Google Scholar] [CrossRef]
- Shvedova, A.; Pietroiusti, A.; Kagan, V. Nanotoxicology ten years later: Lights and shadows. Toxicol. Appl. Pharmacol. 2016, 299, 1–2. [Google Scholar] [CrossRef]
- Singh, D.K.; Iyer, P.K.; Giri, P.K. Role of molecular interactions and structural defects in the efficient fluorescence quenching by carbon nanotubes. Carbon 2012, 50, 4495–4505. [Google Scholar] [CrossRef]
- Aranda, A.; Sequedo, L.; Tolosa, L.; Quintas, G.; Burello, E.; Castell, J.V.; Gombau, L. Dichloro-dihydro-fluorescein diacetate (DCFH-DA) assay: A quantitative method for oxidative stress assessment of nanoparticle-treated cells. Toxicol. In Vitro 2013, 27, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Kroll, A.; Pillukat, M.H.; Hahn, D.; Schnekenburger, J. Current in vitro methods in nanoparticle risk assessment: Limitations and challenges. Eur. J. Pharm. Biopharm. 2009, 72, 370–377. [Google Scholar] [CrossRef]
- Dobrovolskaia, M.A.; Aggarwal, P.; Hall, J.B.; McNeil, S.E. Preclinical Studies To Understand Nanoparticle Interaction with the Immune System and Its Potential Effects on Nanoparticle Biodistribution. Mol. Pharm. 2008, 5, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Ong, K.J.; MacCormack, T.J.; Clark, R.J.; Ede, J.D.; Ortega, V.A.; Felix, L.C.; Dang, M.K.; Ma, G.; Fenniri, H.; Veinot, J.G.; et al. Widespread nanoparticle-assay interference: Implications for nanotoxicity testing. PLoS ONE 2014, 9, e90650. [Google Scholar] [CrossRef]
- Han, X.; Gelein, R.; Corson, N.; Wade-Mercer, P.; Jiang, J.; Biswas, P.; Finkelstein, J.N.; Elder, A.; Oberdörster, G. Validation of an LDH assay for assessing nanoparticle toxicity. Toxicology 2011, 287, 99–104. [Google Scholar] [CrossRef]
- Xiao, Y.; Tang, Z.; Wang, J.; Liu, C.; Kong, N.; Farokhzad, O.; Tao, W. Oral insulin delivery platforms: Strategies to address the biological barriers. Angew. Chem. Int. Ed. Engl. 2020. [Google Scholar] [CrossRef]
- Mahjub, R.; Najafabadi, F.K.; Dehkhodaei, N.; Kheiripour, N.; Ahmadabadi, A.N.; Asl, S.S.; Taheri-Azandariani, M.; Ranjbar, A. Eudragit L-100 capsules/aromatize and quaternerize chitosan for insulin nanoparticle oral delivery on toxic oxidative stress in rat liver and kidney. Pharm. Nanotechnol. 2020. [Google Scholar] [CrossRef]
- Robla, S.; Alonso, M.J.; Csaba, N.S. Polyaminoacid-based nanocarriers: A review of the latest candidates for oral drug delivery. Expert Opin. Drug Deliv. 2020, 17, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, A.; Soleimani, S.; Pezeshgi Modarres, H.; Hojjati Emami, S.; Tondar, M.; Bahlakeh, G.; Hasani-Sadrabadi, M.M. Exosome-inspired targeting of cancer cells with enhanced affinity. J. Mater. Chem. B 2016, 4, 768–778. [Google Scholar] [CrossRef]
- Wang, Q.; Zhuang, X.; Mu, J.; Deng, Z.B.; Jiang, H.; Zhang, L.; Xiang, X.; Wang, B.; Yan, J.; Miller, D.; et al. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat. Commun. 2013, 4, 1867. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Deng, Z.B.; Mu, J.; Zhang, L.; Yan, J.; Miller, D.; Feng, W.; McClain, C.J.; Zhang, H.G. Ginger-derived nanoparticles protect against alcohol-induced liver damage. J. Extracell. Vesicles 2015, 4, 28713. [Google Scholar] [CrossRef]
- Teng, Y.; Ren, Y.; Sayed, M.; Hu, X.; Lei, C.; Kumar, A.; Hutchins, E.; Mu, J.; Deng, Z.; Luo, C.; et al. Plant-Derived Exosomal MicroRNAs Shape the Gut Microbiota. Cell Host Microbe 2018, 24, 637–652.e638. [Google Scholar] [CrossRef]
- Dai, W.; Jin, W.; Zhang, J.; Wang, X.; Wang, J.; Zhang, X.; Wan, Y.; Zhang, Q. Spatiotemporally controlled co-delivery of anti-vasculature agent and cytotoxic drug by octreotide-modified stealth liposomes. Pharm. Res. 2012, 29, 2902–2911. [Google Scholar] [CrossRef]
- Amoozgar, Z.; Yeo, Y. Recent advances in stealth coating of nanoparticle drug delivery systems. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2012, 4, 219–233. [Google Scholar] [CrossRef]
- Henn, A.; Mattinen, M.L. Chemo-enzymatically prepared lignin nanoparticles for value-added applications. World J. Microbiol. Biotechnol. 2019, 35, 125. [Google Scholar] [CrossRef]
- Kwon, D.; Park, J.; Park, J.; Choi, S.Y.; Yoon, T.H. Effects of surface-modifying ligands on the colloidal stability of ZnO nanoparticle dispersions in in vitro cytotoxicity test media. Int. J. Nanomed. 2014, 9 (Suppl. 2), 57–65. [Google Scholar] [CrossRef][Green Version]
- Florence, A.T. Nanoparticle uptake by the oral route: Fulfilling its potential? Drug Discov. Today Technol. 2005, 2, 75–81. [Google Scholar] [CrossRef]
- Wolfram, J.; Zhu, M.; Yang, Y.; Shen, J.; Gentile, E.; Paolino, D.; Fresta, M.; Nie, G.; Chen, C.; Shen, H.; et al. Safety of Nanoparticles in Medicine. Curr. Drug Targets 2015, 16, 1671–1681. [Google Scholar] [CrossRef]
- Ciappellano, S.G.; Tedesco, E.; Venturini, M.; Benetti, F. In vitro toxicity assessment of oral nanocarriers. Adv. Drug Deliv. Rev. 2016, 106, 381–401. [Google Scholar] [CrossRef] [PubMed]
- Wilhelmi, V.; Fischer, U.; van Berlo, D.; Schulze-Osthoff, K.; Schins, R.P.F.; Albrecht, C. Evaluation of apoptosis induced by nanoparticles and fine particles in RAW 264.7 macrophages: Facts and artefacts. Toxicol. In Vitro 2012, 26, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Scherbart, A.M.; Langer, J.; Bushmelev, A.; van Berlo, D.; Haberzettl, P.; van Schooten, F.-J.; Schmidt, A.M.; Rose, C.R.; Schins, R.P.F.; Albrecht, C. Contrasting macrophage activation by fine and ultrafine titanium dioxide particles is associated with different uptake mechanisms. Part. Fibre Toxicol. 2011, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Warheit, D.B.; Hoke, R.A.; Finlay, C.; Donner, E.M.; Reed, K.L.; Sayes, C.M. Development of a base set of toxicity tests using ultrafine TiO2 particles as a component of nanoparticle risk management. Toxicol. Lett. 2007, 171, 99–110. [Google Scholar] [CrossRef] [PubMed]
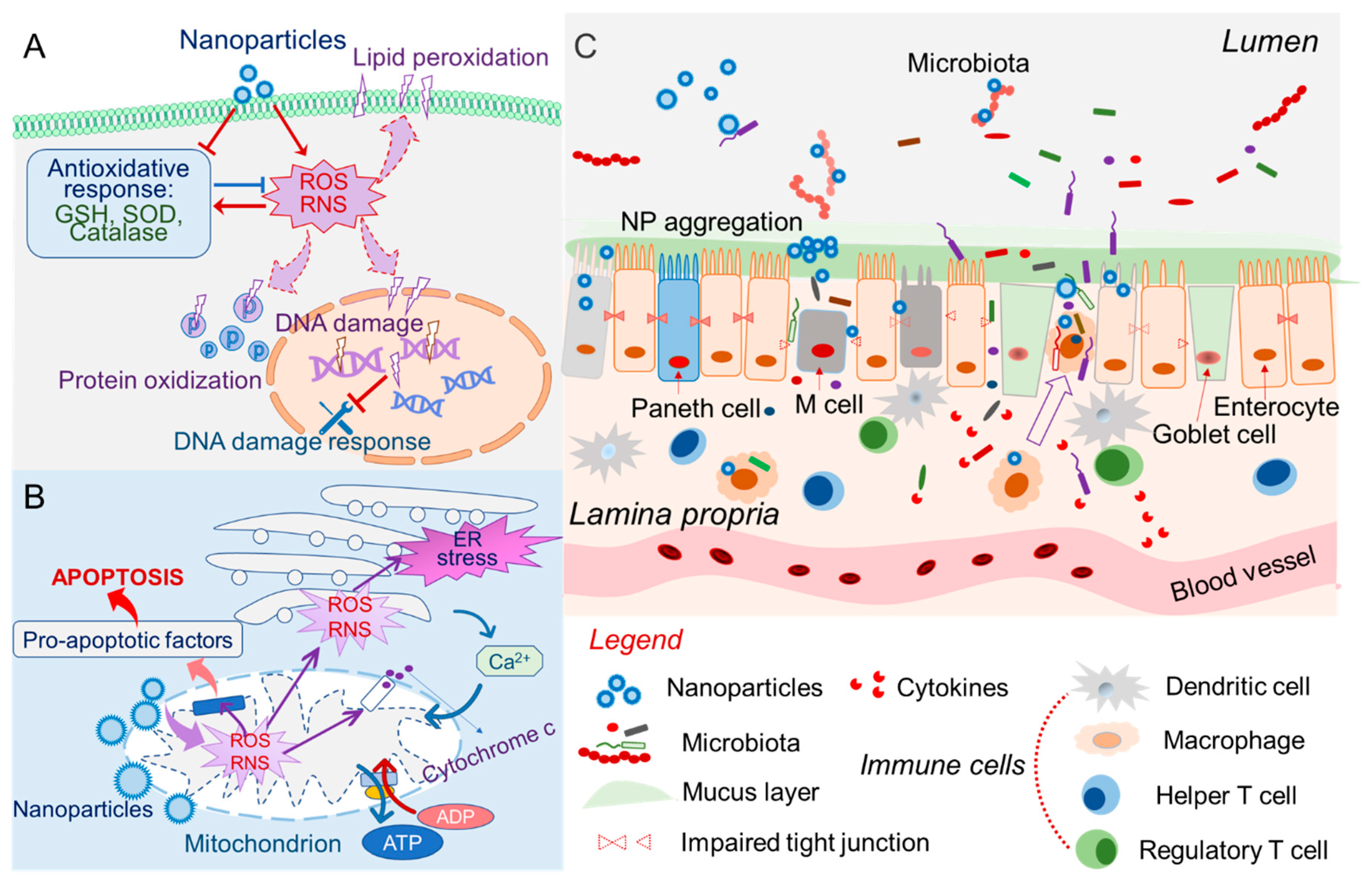
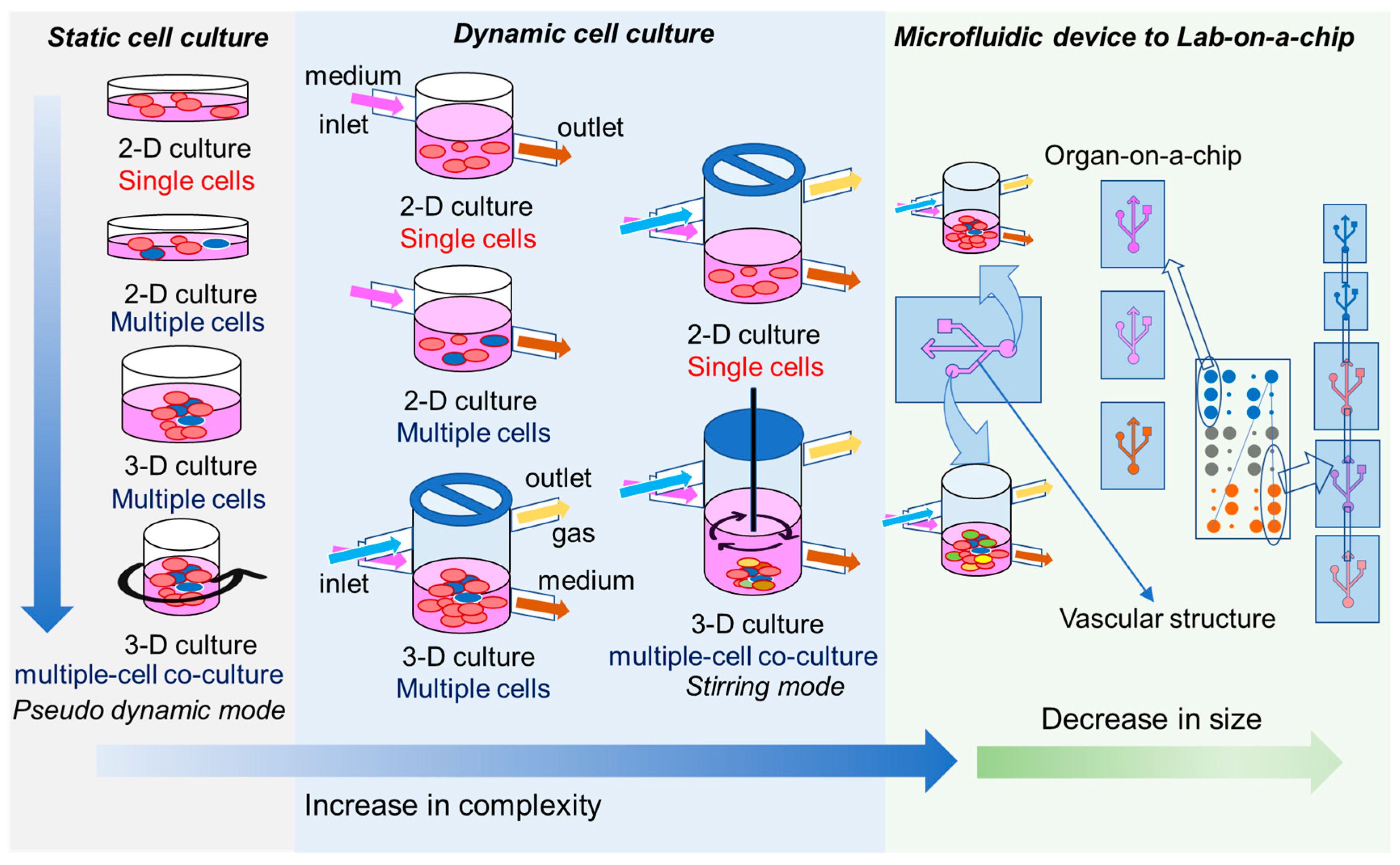
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lama, S.; Merlin-Zhang, O.; Yang, C. In Vitro and In Vivo Models for Evaluating the Oral Toxicity of Nanomedicines. Nanomaterials 2020, 10, 2177. https://doi.org/10.3390/nano10112177
Lama S, Merlin-Zhang O, Yang C. In Vitro and In Vivo Models for Evaluating the Oral Toxicity of Nanomedicines. Nanomaterials. 2020; 10(11):2177. https://doi.org/10.3390/nano10112177
Chicago/Turabian StyleLama, Sudeep, Olivier Merlin-Zhang, and Chunhua Yang. 2020. "In Vitro and In Vivo Models for Evaluating the Oral Toxicity of Nanomedicines" Nanomaterials 10, no. 11: 2177. https://doi.org/10.3390/nano10112177
APA StyleLama, S., Merlin-Zhang, O., & Yang, C. (2020). In Vitro and In Vivo Models for Evaluating the Oral Toxicity of Nanomedicines. Nanomaterials, 10(11), 2177. https://doi.org/10.3390/nano10112177




