Explainable Ensemble-Based Machine Learning Models for Detecting the Presence of Cirrhosis in Hepatitis C Patients
Abstract
1. Introduction
Contribution
- Compare the performance of ensemble learners in diagnosing cirrhosis in hepatitis C patients;
- Apply SFS to minimize the number of features required to form the diagnosis;
- Utilize XAI techniques to explain the outcomes of the best-performing model;
- Utilize XAI techniques to identify the most significant attributes for diagnosing cirrhosis in hepatitis C patients.
2. Literature Review
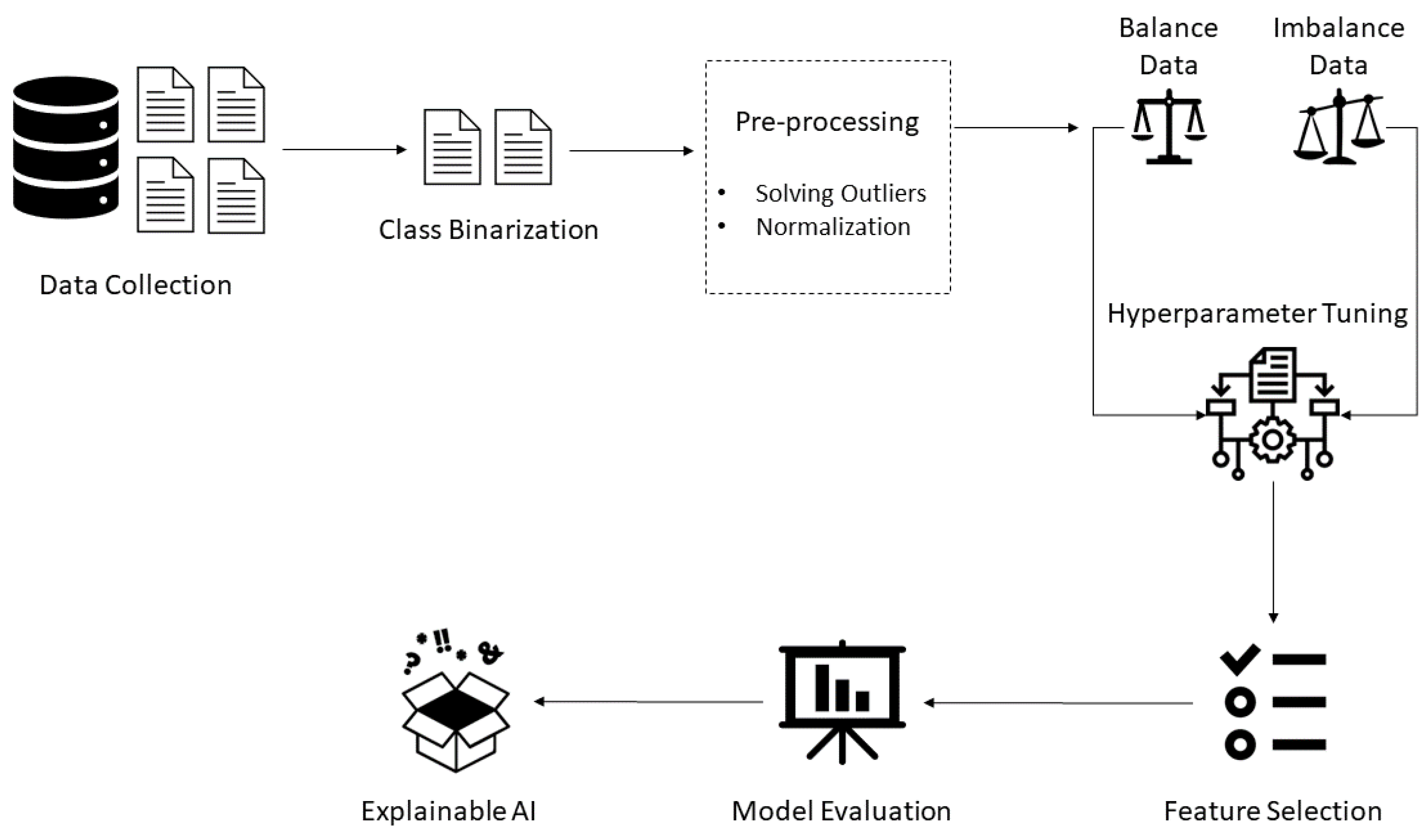
3. Materials and Methods
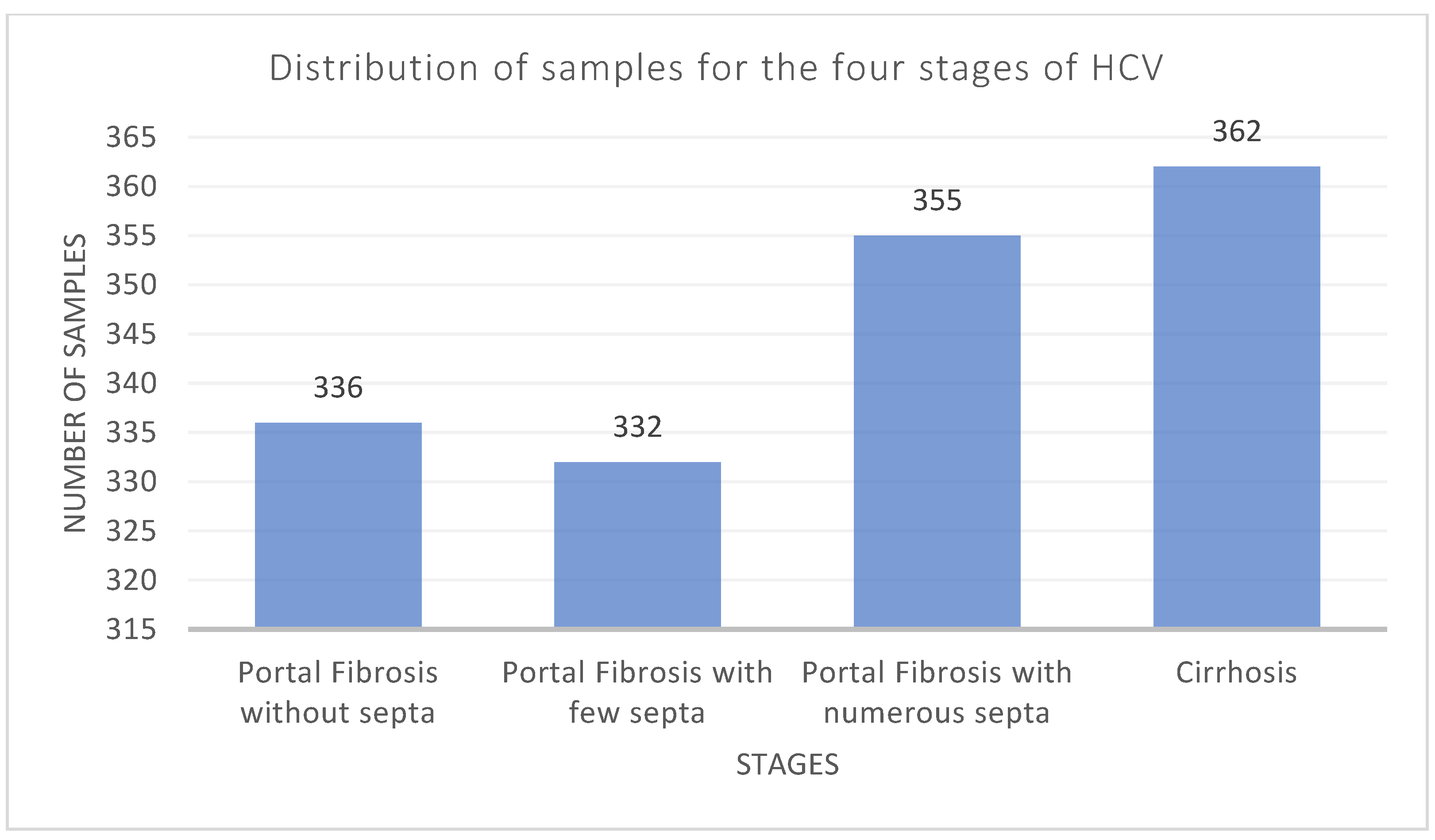
3.1. Dataset Description and Analysis
3.2. Description of the Utilized Machine Learning Techniques
3.2.1. Random Forest
3.2.2. Gradient Boosting Machine
3.2.3. Extreme Gradient Boosting
3.2.4. Extra Trees
3.3. Performance Measures
3.4. Optimization Strategy
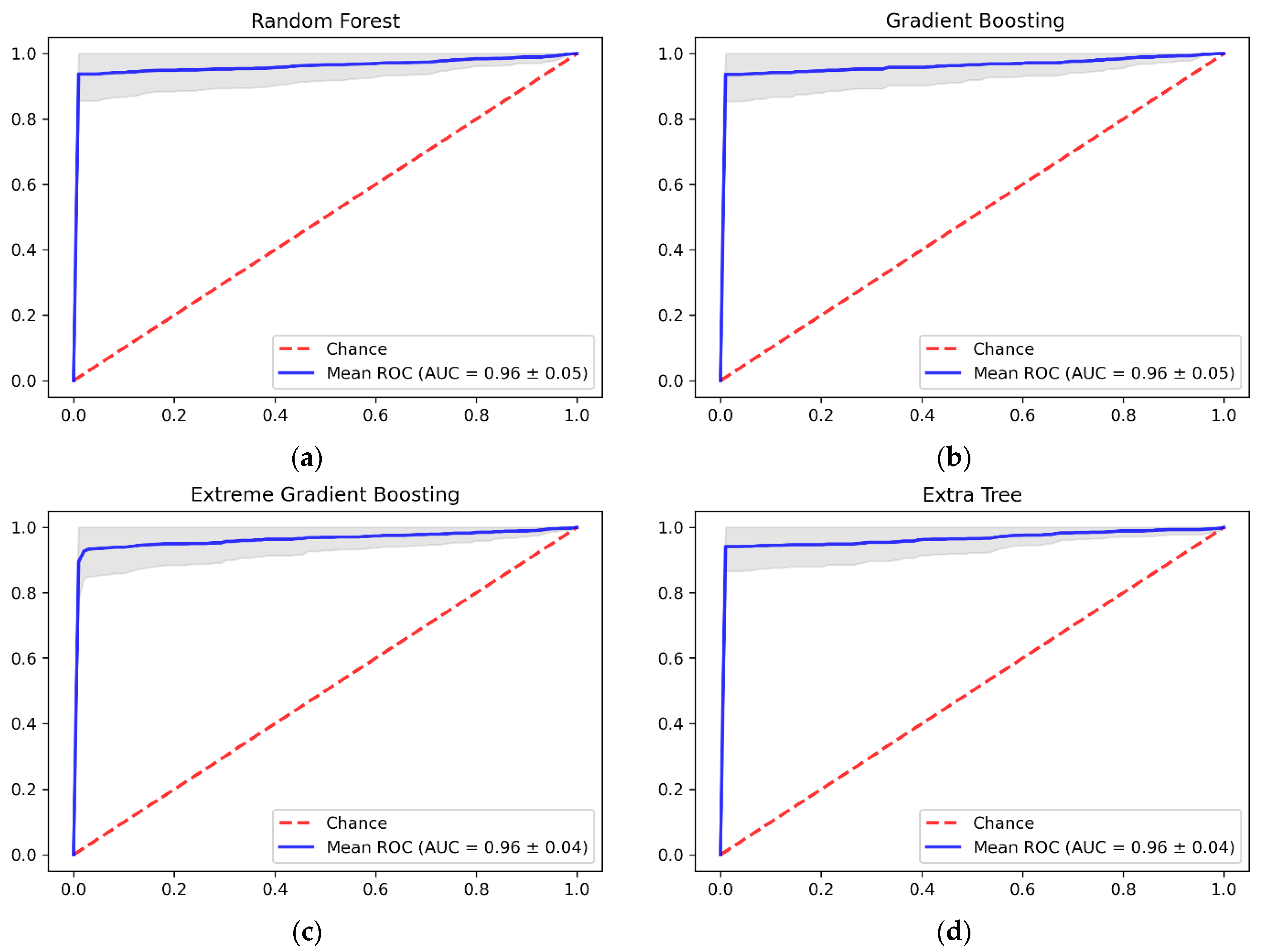
4. Results and Discussion
4.1. Feature Selection
4.2. Further Discussion of the Results
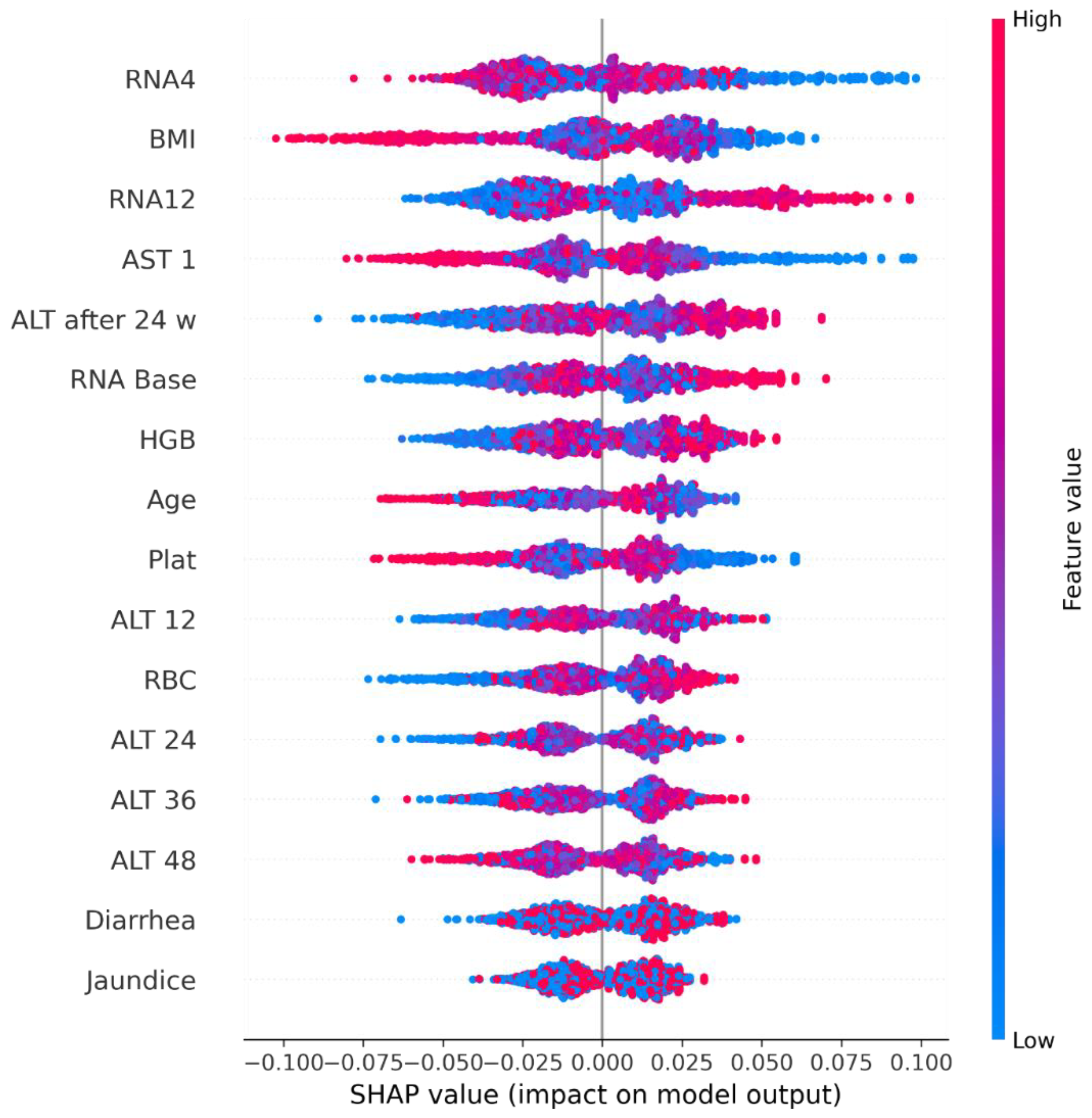
5. Explainable Artificial Intelligence
5.1. Shapley Additive Explanations
5.2. Local Interpretable Model-Agnostic Explanations
6. Conclusions and Recommendations
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hepatitism, C. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c (accessed on 10 November 2022).
- Kouyoumjian, S.P.; Chemaitelly, H.; Abu-Raddad, L.J. Characterizing hepatitis C virus epidemiology in Egypt: Systematic reviews, meta-analyses, and meta-regressions. Sci. Rep. 2018, 8, 1661. [Google Scholar] [CrossRef]
- Elgharably, A.; Gomaa, A.I.; Crossey, M.M.E.; Norsworthy, P.J.; Waked, I.; Taylor-Robinson, S.D. Hepatitis C in Egypt—Past, present, and future. Int. J. Gen. Med. 2016, 10, 1–6. [Google Scholar] [CrossRef]
- Pinzani, M.; Rosselli, M.; Zuckermann, M. Liver cirrhosis. Best Pract. Res. Clin. Gastroenterol. 2011, 25, 281–290. [Google Scholar] [CrossRef]
- Sepanlou, S.G.; Safiri, S.; Bisignano, C.; Ikuta, K.S.; Merat, S.; Saberifiroozi, M.; Poustchi, H.; Tsoi, D.; Colombara, D.V.; Abdoli, A.; et al. The global, regional, and national burden of cirrhosis by cause in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 245–266. [Google Scholar] [CrossRef]
- Muir, A.J. Understanding the Complexities of Cirrhosis. Clin. Ther. 2015, 37, 1822–1836. [Google Scholar] [CrossRef]
- Jain, P.; Tripathi, B.K.; Gupta, B.; Bhandari, B.; Jalan, D. Evaluation of Aspartate Aminotransferase-to-Platelet Ratio Index as a Non-Invasive Marker for Liver Cirrhosis. J. Clin. Diagn. Res. 2015, 9, OC22–OC24. [Google Scholar] [CrossRef]
- Heidelbaugh, J.J.; Bruderly, M. Cirrhosis and Chronic Liver Failure: Part I. Diagnosis and Evaluation. Am. Fam. Physician 2006, 74, 756–762. Available online: https://www.aafp.org/pubs/afp/issues/2006/0901/p756.html (accessed on 9 May 2023).
- Patel, K.; Sebastiani, G. Limitations of non-invasive tests for assessment of liver fibrosis. JHEP Rep. 2020, 2, 100067. [Google Scholar] [CrossRef]
- Vali, Y.; Lee, J.; Boursier, J.; Spijker, R.; Verheij, J.; Brosnan, M.J.; Anstee, Q.M.; Bossuyt, P.M.; Zafarmand, M.H. Fibrotest for evaluating fibrosis in non-alcoholic fatty liver disease patients: A systematic review and meta-analysis. J. Clin. Med. 2021, 10, 2415. [Google Scholar] [CrossRef]
- Afdhal, N.H. Fibroscan (Transient Elastography) for the Measurement of Liver Fibrosis. Gastroenterol. Hepatol. 2012, 8, 605. [Google Scholar]
- Ahuja, A.S. The impact of artificial intelligence in medicine on the future role of the physician. PeerJ 2019, 7, e7702. [Google Scholar] [CrossRef]
- Saleem, S.; Slehria, A.U.R.; Rauf, M.H.; Sohail, M.; Taufiq, N.; Khan, M.U. The Assessment of Diagnostic Accuracy of Real Time Shear Wave Elastography in Detecting Liver Cirrhosis Keeping Histopathology as Reference Standard. Pak. Armed Forces Med. J. 2022, 72, 590–593. [Google Scholar] [CrossRef]
- Krajna, A.; Kovac, M.; Brcic, M.; Sarcevic, A. Explainable Artificial Intelligence: An Updated Perspective. In Proceedings of the 2022 45th Jubilee International Convention on Information, Communication and Electronic Technology (MIPRO), Opatija, Croatia, 23–27 May 2022; pp. 859–864. [Google Scholar] [CrossRef]
- Dosilovic, F.K.; Brcic, M.; Hlupic, N. Explainable artificial intelligence: A survey. In Proceedings of the 2018 41st International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), Opatija, Croatia, 21–25 May 2018; pp. 210–215. [Google Scholar] [CrossRef]
- Mostafa, F.; Hasan, E.; Williamson, M.; Khan, H.; Statistical, H.; Jaeschke, H.W. Statistical Machine Learning Approaches to Liver Disease Prediction. Livers 2021, 1, 294–312. [Google Scholar] [CrossRef]
- UCI Machine Learning Repository: HCV Data Data Set. Available online: https://archive.ics.uci.edu/ml/datasets/HCV+data (accessed on 9 May 2023).
- Oladimeji, O.O.; Oladimeji, A.; Olayanju, O. Machine Learning Models for Diagnostic Classification of Hepatitis C Tests. Front. Health Inform. 2021, 10, 70. [Google Scholar] [CrossRef]
- Safdari, R.; Deghatipour, A.; Gholamzadeh, M.; Maghooli, K. Applying data mining techniques to classify patients with suspected hepatitis C virus infection. Intell. Med. 2022, 2, 193–198. [Google Scholar] [CrossRef]
- Septina, P.L.; Sihotang, J.I. A Comparative Study on Hepatitis C Predictions Using Machine Learning Algorithms. 8ISC Proc. Technol. 2022, 33–42. [Google Scholar]
- Li, T.H.S.; Chiu, H.J.; Kuo, P.H. Hepatitis C Virus Detection Model by Using Random Forest, Logistic-Regression and ABC Algorithm. IEEE Access 2022, 10, 91045–91058. [Google Scholar] [CrossRef]
- Ghazal, T.M.; Anam, M.; Hasan, M.K.; Hussain, M.; Farooq, M.S.; Ali, H.M.A.; Ahmad, M.; Soomro, T.R. Hep-Pred: Hepatitis C Staging Prediction Using Fine Gaussian SVM. Comput. Mater. Contin. 2021, 69, 191–203. [Google Scholar] [CrossRef]
- UCI Machine Learning Repository: Hepatitis C Virus (HCV) for Egyptian patients Data Set. Available online: https://archive.ics.uci.edu/ml/datasets/Hepatitis+C+Virus+%28HCV%29+for+Egyptian+patients (accessed on 9 May 2023).
- Butt, M.B.; Alfayad, M.; Saqib, S.; Khan, M.A.; Ahmad, M.; Khan, M.A.; Elmitwally, N.S. Diagnosing the Stage of Hepatitis C Using Machine Learning. J. Healthc. Eng. 2021, 2021, 8062410. [Google Scholar] [CrossRef]
- Mamdouh, H.; Shams, M.Y.; Abd El-Hafeez, T. Hepatitis C Virus Prediction Based on Machine Learning Framework: A Real-world Case Study in Egypt Hepatitis C Virus Prediction based on Machine Learning Framework: A Real-World Case Study in Egypt. Knowl. Inf. Syst. 2022, 65, 2595–2617. [Google Scholar] [CrossRef]
- Barakat, N.H.; Barakat, S.H.; Ahmed, N. Prediction and Staging of Hepatic Fibrosis in Children with Hepatitis C Virus: A Machine Learning Approach. Healthc. Inform. Res. 2019, 25, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, V.; Tokin, I.; Lioznov, D. Machine Learning Model for Diagnosing the Stage of Liver Fibrosis in Patients With Chronic Viral Hepatitis C. Preprints.org 2021, 2021020488. [Google Scholar] [CrossRef]
- Nasr, M.; El-Bahnasy, K.; Hamdy, M.; Kamal, S.M. A novel model based on non invasive methods for prediction of liver fibrosis. In Proceedings of the 2017 13th International Computer Engineering Conference (ICENCO), Cairo, Egypt, 27–28 December 2017; pp. 276–281. [Google Scholar] [CrossRef]
- Breiman, B.; Greenwell, B. Random Forests; Chapman and Hall/CRC: Boca Raton, FL, USA, 2019; pp. 1–122. [Google Scholar] [CrossRef]
- Artificial Neural Networks for Machine Learning—Every Aspect You Need to Know About—DataFlair. Available online: https://data-flair.training/blogs/artificial-neural-networks-for-machine-learning/amp/ (accessed on 9 May 2023).
- Morvant, E.; Habrard, A.; Ayache, S. Majority vote of diverse classifiers for late fusion. Lect. Notes Comput. Sci. Subser. Lect. Notes Artif. Intell. Lect. Notes Bioinform. 2014, 8621, 153–162. [Google Scholar] [CrossRef]
- Story of Gradient Boosting: How It Evolved over Years. Available online: https://analyticsindiamag.com/story-of-gradient-boosting-how-it-evolved-over-years/ (accessed on 9 May 2023).
- Cheng, J.; Li, G.; Chen, X. Research on travel time prediction model of freeway based on gradient boosting decision tree. IEEE Access 2019, 7, 7466–7480. [Google Scholar] [CrossRef]
- Chen, T.; He, T. xgboost: EXtreme Gradient Boosting. 2022. Available online: https://cran.r-project.org/web/packages/xgboost/vignettes/xgboost.pdf (accessed on 18 June 2022).
- Zhang, D.; Chen, H.D.; Zulfiqar, H.; Yuan, S.S.; Huang, Q.L.; Zhang, Z.Y.; Deng, K.J. IBLP: An XGBoost-Based Predictor for Identifying Bioluminescent Proteins. Comput. Math. Methods Med. 2021, 2021, 6664362. [Google Scholar] [CrossRef]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach Learn 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Bui, X.N.; Nguyen, H.; Soukhanouvong, P. Extra Trees Ensemble: A Machine Learning Model for Predicting Blast-Induced Ground Vibration Based on the Bagging and Sibling of Random Forest Algorithm. Lect. Notes Civ. Eng. 2022, 228, 643–652. [Google Scholar] [CrossRef]
- SequentialFeatureSelector: The Popular forward and Backward Feature Selection Approaches Incl. Floating Variants—Mlxtend. Available online: http://rasbt.github.io/mlxtend/user_guide/feature_selection/SequentialFeatureSelector/ (accessed on 8 May 2022).
- Molnar, C.; Casalicchio, G.; Bischl, B. Interpretable Machine Learning—A Brief History, State-of-the-Art and Challenges. Commun. Comput. Inf. Sci. 2020, 1323, 417–431. [Google Scholar] [CrossRef]
- Zafar, M.R.; Khan, N.M. DLIME: A Deterministic Local Interpretable Model-Agnostic Explanations Approach for Computer-Aided Diagnosis Systems. arXiv 2019, arXiv:1906.10263. [Google Scholar]


| Feature | Mean | std | min | 25% | 50% | 75% | max |
|---|---|---|---|---|---|---|---|
| Age | 46.32 | 8.78 | 32.00 | 39.00 | 46.00 | 54.00 | 61.00 |
| Body mass index (BMI) | 28.61 | 4.08 | 22.00 | 25.00 | 29.00 | 32.00 | 35.00 |
| White blood cells (WBC) | 7533.39 | 2668.22 | 2991.00 | 5219.00 | 7498.00 | 9902.00 | 12,101.00 |
| Red blood cells (RBC) | 4,422,129.61 | 346,357.71 | 3,816,422.00 | 4,121,374.00 | 4,438,465.00 | 4,721,279.00 | 5,018,451.00 |
| Hemoglobin (HGB) | 12.59 | 1.71 | 10.00 | 11.00 | 13.00 | 14.00 | 15.00 |
| Platelets (Plat) | 158,348.06 | 38,794.79 | 93,013.00 | 124,479.00 | 157,916.00 | 190,314.00 | 226,464.00 |
| Aspartate transaminase ratio (AST 1) | 82.77 | 25.99 | 39.00 | 60.00 | 83.00 | 105.00 | 128.00 |
| Alanine transaminase ratio 1 week (ALT 1) | 83.92 | 25.92 | 39.00 | 62.00 | 83.00 | 106.00 | 128.00 |
| Alanine transaminase ratio 4 week (ALT4) | 83.41 | 26.53 | 39.00 | 61.00 | 82.00 | 107.00 | 128.00 |
| Alanine transaminase ratio 12 week (ALT 12) | 83.51 | 26.06 | 39.00 | 60.00 | 84.00 | 106.00 | 128.00 |
| Alanine transaminase ratio 24 week (ALT 24) | 83.71 | 26.21 | 39.00 | 61.00 | 83.00 | 107.00 | 128.00 |
| Alanine transaminase ratio 36 week (ALT 36) | 83.12 | 26.40 | 5.00 | 61.00 | 84.00 | 106.00 | 128.00 |
| Alanine transaminase ratio 48 week (ALT 48) | 83.63 | 26.22 | 5.00 | 61.00 | 83.00 | 106.00 | 128.00 |
| Alanine transaminase after 24 weeks (ALT after 24 w) | 33.44 | 7.07 | 5.00 | 28.00 | 34.00 | 40.00 | 45.00 |
| RNA Base | 590,951.22 | 353,935.36 | 11.00 | 269,253.00 | 593,103.00 | 886,791.00 | 1,201,086.00 |
| RNA after 4 weeks (RNA4) | 600,895.65 | 362,315.13 | 5.00 | 270,893.00 | 597,869.00 | 909,093.00 | 1,201,715.00 |
| RNA after 12 weeks (RNA12) | 288,753.61 | 285,350.67 | 5.00 | 5.00 | 234,359.00 | 524,819.00 | 3,731,527.00 |
| RNA end-of-treatment (RNAEOT) | 287,660.34 | 264,559.53 | 5.00 | 5.00 | 251,376.00 | 517,806.00 | 808,450.00 |
| RNA elongation factor (RNAEF) | 291,378.29 | 267,700.69 | 5.00 | 5.00 | 244,049.00 | 527,864.00 | 810,333.00 |
| Baseline histological Grading | 9.76 | 4.02 | 3.00 | 6.00 | 10.00 | 13.00 | 16.00 |
| Feature | Value | Count |
|---|---|---|
| Gender | 1 | 707 |
| 2 | 678 | |
| Fever | 1 | 671 |
| 2 | 714 | |
| Nausea/Vomiting | 1 | 689 |
| 2 | 696 | |
| Headache | 1 | 698 |
| 2 | 687 | |
| Diarrhea | 1 | 689 |
| 2 | 696 | |
| Fatigue and generalized bone ache | 1 | 694 |
| 2 | 691 | |
| Epigastric pain | 1 | 687 |
| 2 | 698 | |
| Jaundice | 1 | 691 |
| 2 | 694 |
| Stage | Number of Samples | Number of Samples after Removing Outliers | Number of Samples after Using Random Oversampling |
|---|---|---|---|
| 0 | 1023 | 1021 | 1021 |
| 1 | 362 | 360 | 1021 |
| Classifier | Hyperparameter | Without Oversampling | With Oversampling |
|---|---|---|---|
| RF | n_estimators | 100 | 450 |
| max_depth | 23 | 24 | |
| max_features | sqrt | log2 | |
| max_samples_leaf | 1 | 1 | |
| GBM | n_estimators | 50 | 110 |
| learning_rate | 0.01 | 0.3 | |
| max_depth | 1 | 10 | |
| loss | log_loss | exponential | |
| XGBoost | n_estimators | 50 | 170 |
| booster | gblinear | gbtree | |
| learning_rate | 0.01 | 0.1 | |
| gamma | 0 | 0.4 | |
| ET | n_estimators | 50 | 200 |
| max_depth | None | 11 | |
| max_features | log2 | log2 | |
| min_samples_leaf | 1 | 1 |
| Classifier | Dataset | Mean of Accuracy | Std of Accuracy | Precision | Recall | AUC-ROC |
|---|---|---|---|---|---|---|
| RF | Original | 74.22% | 0.0047 | 3.00% | 1.00% | 0.49 |
| Oversampled | 96.48% | 0.0417 | 99.27% | 93.64% | 0.96 | |
| GBM | Original | 73.93% | 0.0006 | 0.00% | 0.00% | 0.45 |
| Oversampled | 95.70% | 0.0439 | 97.49% | 93.74% | 0.97 | |
| XGBoost | Original | 73.93% | 0.0006 | 0.00% | 0.00% | 0.52 |
| Oversampled | 90.99% | 0.0396 | 88.00% | 94.81% | 0.96 | |
| ET | Original | 74.22% | 0.0060 | 45.00% | 1.66% | 0.51 |
| Oversampled | 96.82% | 0.0413 | 100% | 93.64% | 0.97 |
| Classifier | Number of Features Selected | Features Selected | Mean of Accuracy |
|---|---|---|---|
| RF | 27 | {Jaundice, Age, Gender, Fatigue & generalized bone ache, ALT 48, RNA 48, RNA base, BMI, HGB, ALT 12, RNA EOT, ALT 4, Nausea/Vomiting, ALT 1, Epigastric pain, Fever, Plat, AST 1, RNA EF, Headache, Baseline histological grading, ALT 24, RBC, ALT after 24 w, WBC, ALT 36, RNA 4, Diarrhea} | 96.58% |
| GBM | 28 | All features | 95.70% |
| XGBoost | 28 | All features | 90.99% |
| ET | 16 | {Age, BMI, Diarrhea, Jaundice, RBC, HGB, Plat, AST 1, ALT 12, ALT 24, ALT 36, ALT 48, ALT after 24 w, RNA base, RNA 4, RNA 12} | 96.92% |
| Classifier | Mean of Accuracy | Std of Accuracy | Recall | Precision |
|---|---|---|---|---|
| RF | 96.58% | 0.0399 | 93.74% | 99.39% |
| GBM | 95.70% | 0.0439 | 93.74% | 97.49% |
| XGBoost | 90.99% | 0.0396 | 94.81% | 88.00% |
| ET | 96.92% | 0.0380 | 94.00% | 99.81% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alotaibi, A.; Alnajrani, L.; Alsheikh, N.; Alanazy, A.; Alshammasi, S.; Almusairii, M.; Alrassan, S.; Alansari, A. Explainable Ensemble-Based Machine Learning Models for Detecting the Presence of Cirrhosis in Hepatitis C Patients. Computation 2023, 11, 104. https://doi.org/10.3390/computation11060104
Alotaibi A, Alnajrani L, Alsheikh N, Alanazy A, Alshammasi S, Almusairii M, Alrassan S, Alansari A. Explainable Ensemble-Based Machine Learning Models for Detecting the Presence of Cirrhosis in Hepatitis C Patients. Computation. 2023; 11(6):104. https://doi.org/10.3390/computation11060104
Chicago/Turabian StyleAlotaibi, Abrar, Lujain Alnajrani, Nawal Alsheikh, Alhatoon Alanazy, Salam Alshammasi, Meshael Almusairii, Shoog Alrassan, and Aisha Alansari. 2023. "Explainable Ensemble-Based Machine Learning Models for Detecting the Presence of Cirrhosis in Hepatitis C Patients" Computation 11, no. 6: 104. https://doi.org/10.3390/computation11060104
APA StyleAlotaibi, A., Alnajrani, L., Alsheikh, N., Alanazy, A., Alshammasi, S., Almusairii, M., Alrassan, S., & Alansari, A. (2023). Explainable Ensemble-Based Machine Learning Models for Detecting the Presence of Cirrhosis in Hepatitis C Patients. Computation, 11(6), 104. https://doi.org/10.3390/computation11060104







