Abstract
ISGylation is an important process through which interferon-stimulated genes (ISGs) elicit an antiviral response in the host cells. Several viruses, including the SARS-CoV-2, suppress the host immune response by reversing the ISGylation through a process known as de-ISGylation. The PLpro of SARS-CoV-2 interacts with the host ISG15 and brings about de-ISGylation. Hence, inhibiting the de-ISGylation to restore the activity of ISGs can be an attractive strategy to augment the host immune response against SARS-CoV-2. In the present study, we evaluated several phytochemicals from well-known immunomodulatory herbs, viz. Andrographis paniculata (AG), Tinospora cordifolia (GU), and Ocimum sanctum (TU) for their effect on deISGylation that was mediated by the PLpro of SARS-CoV2. For this purpose, we considered the complex 6XA9, which represents the interaction between SARS-CoV-2 PLpro and ISG15 proteins. The phytochemicals from these herbs were first evaluated for their ability to bind to the interface region between PLpro and ISG15. Molecular docking studies indicated that 14-deoxy-15-isopropylidene-11,12-didehydroandrographolide (AG1), Isocolumbin (GU1), and Orientin (TU1) from AG, GU, and TU, respectively possess better binding energy. The molecular dynamic parameters and MMPBSA calculations indicated that AG1, GU1, and TU1 could favorably bind to the interface and engaged key residues between (PLpro-ISG15)-complex. Protein–protein MMPBSA calculations indicated that GU1 and TU1 could disrupt the interactions between ISG15 and PLpro. Our studies provide a novel molecular basis for the immunomodulatory action of these phytochemicals and open up new strategies to evaluate drug molecules for their effect on de-ISGylation to overcome the virus-mediated immune suppression.
1. Introduction
The host innate immune system acts as the first line of defense during viral infections. Following the initial viral infection, a type-I interferon response is activated in the host, and this subsequently leads to the upregulation of several interferon-stimulated genes (ISGs). ISGs act as effectors of the interferon-mediated antiviral host immune response, which leads to an appropriate antiviral host immune response.
Among the ISGs, ISG15 is a well-studied ISG. It is shown to be robustly induced by type-I interferons [1] following various viral infections. ISG15 is a ubiquitin-like protein [2]. ISG15 is conjugated to target proteins upon its induction through a reaction called ISGylation. The ISGylation of target proteins is a key mechanism through which ISG15 mediates its effect. It plays critical roles in various phases of the host innate immune response against viruses [3]. Several important immune regulatory transcription factors and receptors have been established as substrates for ISGylation by ISG15. ISG15 is shown to limit the viral replication in many viruses [4]. Hence, ISG15 is an important component in the host immune response pathway against viruses.
Viruses have developed mechanisms to defend against the ISG-mediated immune response. Many viruses, such as herpes simplex virus, norovirus, chikungunya virus, and HIV evade the host immune system by counteracting the ISG15-mediated pathways [1]. Studies indicate that many viruses express proteins that possess ISG15 de-conjugating activity [5]. De-ISGylation refers to the process of the deconjugation of ISG from target proteins. It is considered an important means through which viruses evade the interferon-mediated innate immune response. Even in the case of SARS-CoV-2, the PLpro protein is shown to exhibit de-ISGylation activity, potentially leading to a diminished early-host immune response [6]. Suppressing the early-phase host immune system by SARS-CoV-2 is proposed as a key mechanism that can further lead to exaggerated viral replication, viral dominance over the host, and cytokine response at the later stages of infection [7,8,9]. Hence, the therapeutic strategies aimed at enhancing the early-stage host immune response against the SARS-CoV-2 or other viruses should consider targeting the de-ISGylation mechanism. Although vaccination is one of the effective strategies to elicit a specific (adaptive), immune response against the viruses, additional interventions to enhance the early-phase host innate immune response will be an added advantage.
Many medicinal herbs and their extracts have been used to treat and manage viral infections. Various phytochemicals that are present in Andrographis paniculata (AG), Tinospora cordifolia (GU), and Ocimum sanctum (TU) have been shown to possess antiviral and immune-potentiating activity [10,11,12]. For example, Andrographolide can modulate the innate and adaptive immune responses by regulating macrophage phenotypic polarization [11]. Furthermore, the phytochemicals from Tinospora cordifolia show association with immune pathways and act as immunomodulators [12,13]. Previous studies have identified phytochemicals from AG, GU, and TU as potent inhibitors of SARS-CoV-2 [14,15]. Treatment with these herbs in the initial phases of viral infection is shown to have beneficial effects. Most of these studies describe the immune modulatory mechanism of these herbs at the cellular level, involving immune cells and their respective pathways. However, the nature of the molecular targets that are engaged by these phytochemicals is unclear from the literature. A molecular-level understanding of the targets that are engaged by the bioactive compounds will add value for evaluating the immunomodulatory herbs as broad spectrum antiviral immunomodulators. Since de-ISGylation is an important mechanism that is employed by viruses to suppress the host immunity, we hypothesized that phytochemicals from these herbs might act on this arm to bring about their immune-potentiating activity.
In order to evaluate this hypothesis, in the current study, we considered a protein complex 6XA9, which represents the interaction between the SARS-CoV2 PLpro and C- terminal of ISG15 [16]. We refer to this structure as (PLpro-ISG15)-complex throughout the manuscript. We considered key interacting residues between PLpro and ISG15 in this complex for evaluating the potential phytochemicals for their ability to bind to this region and affect the protein–protein interaction. A systematic approach was followed in which the phytochemicals from AG, GU, and TU herbs were first screened by molecular docking. Following this, the top-ranked phytochemicals from these herbs, as per the docking scores, were further evaluated by MD simulation for 300 nanoseconds. In addition, we analysed MM/PBSA (molecular mechanics/Poisson–Boltzmann Surface Area) for protein-ligand and protein–protein interactions. GRL0617, which is known to inhibit de-ISGylation, was also included in our studies for a comparison of the parameters. Our results indicated that 14-deoxy-15-isopropylidene-11,12-didehydroandrographolide (AG1), Isocolumbin (GU1), Orientin (TU1), and GRL0617 could favorably bind to the interface region between PLpro and ISG15. Our results also indicated that among these ligands, TU1 and GU1 could potentially disrupt the PLpro and ISG15 interactions.
2. Results and Discussion
2.1. Screening of Phytochemicals of Andrographis paniculata (AG), Tinospora cordifolia (GU), and Ocimum sanctum (TU) against SARS-CoV-2 PLpro ISG15 Site at UIM (PDB:6XA9)
The key phytochemical constituents of AG, GU, and TU were selected based on the literature search, viz. PubMed (https://www.ncbi.nlm.nih.gov/pmc/) (accessed on 9 July 2021), Google Scholar (https://scholar.google.com/) (accessed on 9 July 2021), and DOAJ (https://doaj.org/) (accessed on 9 July 2021) and are given in Supplementary Table S1. A total of 90 phytochemicals from AG, GU, and TU were docked against the target protein of SARS-CoV-2 (PDB) ID: 6XA9) using AutoDock Vina [17]. The AutoDock Vina results represent the docking scores as the Gibbs free energy of binding (ΔG (kcal/mol), which approximates the sum of all interactions between ligand and receptor minus the desolvation energies. The binding energies of the top five phytochemicals from AG, GU, and TU in order of increasing docking scores are given in Table 1. The lesser the value, the better the binding affinity.

Table 1.
Docking scores of various phytochemicals from A. paniculata (AG), T. cordifolia (GU), O. sanctum (TU) against SARS-CoV-2 (PLpro-ISG15) complex interacting site (PDB ID:6XA9).
Docking against SARS-CoV-2 PLpro ISG15 Interacting Site
The dual role of PLpro in viral peptide cleavage and immune regulation has made it an important target for inhibiting SARS-CoV-2 infectivity. PLpro inhibits the host innate immune response by reversing ISG15 modifications from the proteins. The PLpro of the coronavirus family has a ubiquitin-interacting motif (UIM) that can recognize and hydrolyze ubiquitin (Ub) and the ubiquitin-like protein ISG15 (interferon-induced gene 15). However, SARS-CoV-2 PLpro preferentially catalyzes de-ISGylation over de-ubiquitylation [18,19]. SARS-CoV-2 PLpro UIM accommodates both ubiquitin and ISG15 binding sites. In the current work, 6XA9 crystal structure (SARS-CoV-2 PLpro in complex with ISG15 C-terminal domain) was taken for the study. This complex represents the interaction between PLpro and ISG15. In this complex, Ser170, Tyr171, Phe216, Gln195, Thr225, Lys232, Asn151, and Asn156 from PLpro and Trp123, and Pro130/Glu132 from ISG15 are key interacting amino acid residues that play critical roles in de-ISGylation. In addition, Met208, Glu167, and Arg166 are also important residues in the S2 palm domain of the PLpro that interact with the substrate [16]. In Supplementary Figure S1, we have mapped the respective amino acid of the interface in a 3D structure format. The rationale of the study was to look for the phytochemicals that bind to the key residues representing the interface between PLpro and ISG15. The effective binding of the phytochemicals to the interface region or interfering with the PLpro and ISG15 interaction would potentially inhibit the de-ISGylation activity.
GRL0617 binds to the ISG15 interacting site of PLpro [20]. Hence, we included this molecule as a positive control for docking studies. Our docking results indicate that GRL0617 shows ∆G of −8.5 kcal/mol. The top three phytochemicals from AG, 14-deoxy-15-isopropylidene-11,12-didehydroandrographolide (−9.4 kcal/mol); Andrographolactone (−9.2 kcal/mol); and Neoandrographolide (−9 kcal/mol) showed the highest binding affinity amongst all the screened phytochemicals and were better than the control drug molecule. The top three phytochemicals from AG are diterpenes which possess immunomodulatory and antiviral activity [21,22]. The top-ranked phytochemicals from GU and TU were Isocolumbin (−9.9) and Orientin (−9.4 kcal/mol), respectively.
The docking of phytochemicals shows that they form hydrogen bonds and other non-covalent and electrostatic interactions with major amino acid residues of PLpro at the ISG15 interacting site. The two-dimensional binding interactions of the top phytochemicals from AG, GU, and TU with the PLpro ISG15 binding site are shown in Figure 1.
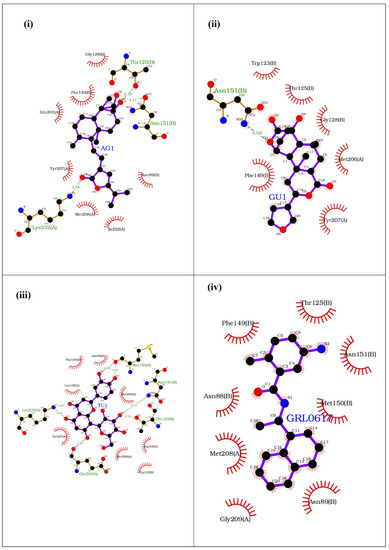
Figure 1.
Binding of (i) AU1 (14-deoxy-15-isopropylidene-11,12-didehydroandrographolide), (ii) GU1 (Isocolumbin), (iii) TU1 (Orientin), and (iv) GRL0617 at the ISG15 interacting site of UIM domain of PLpro of SARS-CoV-2.
To analyze the molecular interactions (H-bond, van der Waals bonds, pi–pi interactions, salt bridges) between various phytochemicals against PLpro ISG15 binding site, LIGPLOT software (Ligplot version 4.15.0-142 generic, E.M.B.L., Hinxton, Cambridgeshire, UK) was used [23]. The 14-deoxy-15-isopropylidene-11,12-didehydroandrographolide phytochemical from AG interacts with the UIM through hydrogen bond interactions with Lys232 (PLpro), Thr125 (ISG15), and Asn151 (ISG15), whereas Met208, an important residue in the S2 palm domain, lies in the binding pocket. (Figure 1i). Isocolumbin from GU interacts with the UIM through hydrogen bond interactions with Asn151 (ISG15) and other important amino acid residues surrounding the binding interface site (Figure 1ii). Orientin from TU forms hydrogen bonds with Lys232 (PLpro), Glu203 (PLpro), Met150 (ISG15), and Asn151 (ISG15), along with other non-covalent interactions that further stabilize the binding (Figure 1iii). The positive control GRL0617 perfectly fits at the ISG15 binding site of UIM, being surrounded by the key residues, such as Met208 (PLpro) and Gly209 (PLpro) that stabilizes the binding of the drug molecule (Figure 1iv). Our results suggest that the top ranked phytochemicals from AG, GU and TU interact with the critical residues of the ISG15 binding site of UIM and, therefore, can destabilize the PLpro and ISG15 interaction, thereby potentially inhibiting the de-ISGylation activity of the PLpro.
2.2. Physiochemical Properties Analysis of the Phytoactives Affirms Their Drug-Likeness
The ADME property of a drug is important in determining its safety and efficacy. The molecular weight, hydrogen bond acceptors and donors, lipophilicity, etc., are important parameters for the generation of an effective and successful drug. The Lipinski rule of five can be applied for filtering out the best potential drug. According to the Lipinski rule, a molecule with a molecular weight of <500 Dalton, a maximum of five hydrogen bond donors, and 10 hydrogen bond acceptors with a log p value of <5 have a better drug-likeness than others that fail these parameters. The drug-likeness and ADMET properties of top-ranked phytochemicals AG1, GU1, TU1, and GRL0617 were calculated using web tools SWISS ADME and PreADMET, and the results are represented in Supplementary Table S2i,ii. The top-ranked phytochemical from AG or GU or GRL0617 showed a drug-likeness property, making them potential lead molecules against the PLpro of SARS-CoV-2. Orientin from TU has the molecular weight and LogP values within the threshold range but violates the rule in terms of hydrogen-bonding properties. However, in vivo studies indicate that Orientin is quickly distributed to the kidney, liver, and lung [24].
2.3. Molecular Dynamics (MD) Simulation Study
The effectiveness of the screened phytochemicals was further analyzed by performing all-atom MD simulations for 300 ns using GROMACS. MD simulation studies provide insights on the dynamic state of the ligands at the interaction site of the target protein in the presence of an ionic aqueous environment. In addition, they provide an elaborate understanding not only of the molecular dynamics of ligand-protein complexes, but also evaluate the crucial interactions during the time scale of few nanoseconds.
2.3.1. Stability and Fluctuations of the Protein: RMSD and RMSF Analysis of the Protein Complex
To understand the predicted binding modes of the candidate phytochemicals that were selected from docking studies, we illustrated the detailed interactions of both the (PLpro-ISG15)-complex and (PLpro-ISG15)-complex with the phytochemicals, viz. AG1, GU1, TU1 or GRL0617 over the 300 ns MD simulation (Figure 2). Figure 2i–iv(A) represents the 3D and 2D images of the phytochemicals with (PLpro-ISG15)-complex at 0 ns and 300 ns. As clear from the Figures, the phytochemicals could bind to the interface at 0 ns and 300 ns. Once the proper binding of the phytochemicals within the interface cavity was confirmed, the other parameters such as RMSD, RMSF, SASA, Rg, hydrogen bond, PCA analysis, and MM-PBSA were evaluated over the entire 300 ns simulation time. PCA analysis data can be found in supplementary information (Figure S2).
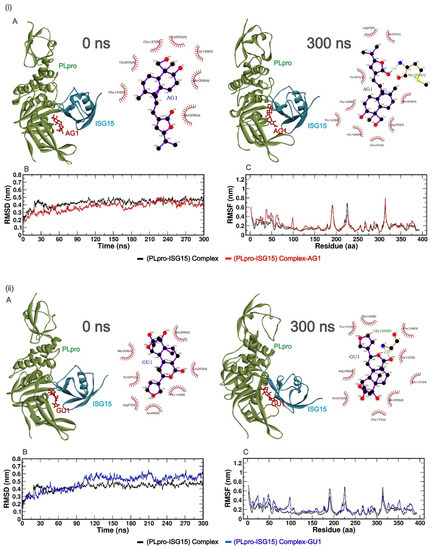
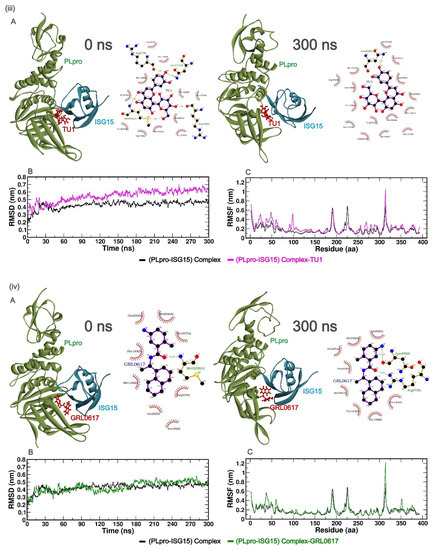
Figure 2.
(i–iv (A))—The best poses of the phytochemicals (AG1 (red), GU1 (blue), TU1 (magenta), and GRL0617 (green)) binding to the interface of (PLpro-ISG15)-complex at 0 ns (left) and 300 ns (right) during molecular dynamics (MD) simulation. Each inset shows the detailed interactions of each drug candidate docked to the ISG15 interacting site of the PLpro, indicating the amino acids involved in the interaction and the type of interaction (hydrogen bonds, hydrophilic interactions, salt bridges, Π-stacking, etc.). (i–iv (B)) show RMSD, and (i–iv (C)) show RMSF values over 300 ns simulation for different complexes with phytochemicals.
In order to evaluate the stability profiles of the (PLpro-ISG15) complex with phytochemicals, RMSD was calculated for Cα backbone atoms of protein over the entire period of 300 ns simulation. RMSD values provide information on the extent of deviation of a given protein–ligand complex, compared to a reference structure over the simulation time. A lower deviation of the given protein–ligand complex from respective reference structures indicates a suitable accommodation of the ligand within the binding pocket.
The time evolution of RMSD for different protein–ligand complexes relative to the initial structure of (PLpro-ISG15)-complex is presented in (Figure 2i–iv(B)). As seen in Figure 2, the RMSD values for the (PLpro-ISG15)-complex increased until 50 ns, after which the values stabilized and leveled off. The same trend was observed for (PLpro-ISG15)-AG1 and (PLpro-ISG15)-GRL0617. The average RMSD values between 50 and 300 ns for (PLpro-ISG15)-complex, (PLpro-ISG15)-AG1, and (PLpro-ISG15)-GRL0617 were 0.44 nm, 0.39 nm, and 0.45 nm, respectively. In the case of (PLpro-ISG15)-GU1 complex, the system equilibrated between 50 and 100 ns (RMSD = 0.40 nm) but later in the trajectory, the RMSD values showed fluctuation. However, this structure maintained an average RMSD of 0.55 nm between 100 and 300 ns. In the case of (PLpro-ISG15)-TU1 complex, the ligand-bound complex deviated from the (Plpro-ISG15)-complex structure, and its average RMSD value was 0.57 nm (between 50 and 300 ns). The higher RMSD values and fluctuations in the RMSD trajectory indicate that the binding of GU1 and TU1 might affect the protein–protein interactions between PLpro and ISG15.
Next, in order to investigate the local fluctuations at the residue level before and after binding with the phytochemicals, the root mean square fluctuation (RMSF) of Cα atom for the entire 300 ns was predicted (Figure 2i–iv(C)). The residues from 1 to 314 correspond to PLpro, and 315 to 400 correspond to ISG15. As indicated in (Figure 2i–iv(C)), there were no significant fluctuations between residues 160–175 and 200–210. This region corresponds to the PLpro and ISG15 interface region. On the other hand, fluctuations were observed in the loop regions for all the complexes. In the case of (PLpro-ISG15)-complex and the (PLpro-ISG15)-complexes with AG1, TU1, and GRL0617, there were fluctuations in the region between 180 and 200 residues. Similarly, (PLpro-ISG15)-complex and (PLpro-ISG15)-complex with GRL0617 showed some fluctuations in the region corresponding to 220–230 residues. However, these fluctuations were not significantly higher than those that were observed for loop regions. The fluctuations were observed between the 310 and 320 residues, between the chains of PLpro and ISG15.
2.3.2. Compactness of Protein Complex: Radius of Gyration (Rg) and Solvent Accessible Surface Area (SASA)
Rg is a parameter that scores for the compactness of protein. We evaluated the compactness of (PLpro-ISG15)-complex upon binding with the phytochemicals over the course of MD simulation Figure 3i(A–D). The average Rg value of (PLpro-ISG15)-complex, (PLpro-ISG15)-complex with AG1, (PLpro-ISG15)-complex with GU1, (PLpro-ISG15)-complex with TU1, and (PLpro-ISG15)-complex with GRL0617 were found to be 2.35 nm, 2.32 nm, 2.33 nm, 2.31 nm, and 2.31 nm, respectively. As shown in Figure 3i(A–D), the Rg values of both (PLpro-ISG15)-complex and (Plpro-ISG15)-complex with phytochemicals did not significantly change over 300 ns simulation, suggesting that all the systems were compact.
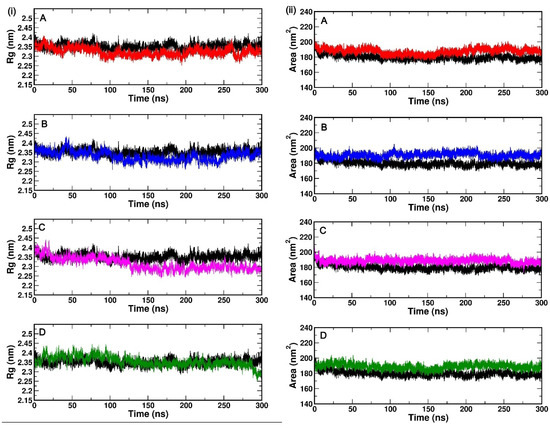
Figure 3.
(i(A–D))—Rg values of backbone atoms over 300 ns simulation of different (Plpro-ISG15)-complex with phytochemicals (AG1 (red), GU1 (blue), TU1 (magenta), and GRL0617 (green)) (left). (ii(A–D))—SASA values of backbone atoms over 300 ns simulation for different (Plpro-ISG15)-complexes with phytochemicals (AG1 (red), GU1 (blue), TU1 (magenta), and GRL0617 (green)) (right). The black line represents (Plpro-ISG15)-complex without phytochemicals.
Flexibility and compactness are correlated with each other. The SASA is a useful parameter for understanding the conformational dynamics of a protein in a solvent environment. In the current study, we evaluated the SASA of the selected docked complexes over 300 ns MD simulation. Figure 3i(A–D) shows the time-dependent SASA plot. The average SASA values of (Plpro-ISG15)-complex, (Plpro-ISG15)-complex with AG1, Plpro-ISG15 complex with GU1, (Plpro-ISG15)-complex with TU1, and (Plpro-ISG15)-complex with GRL0617 were found to be 179.77 nm2, 187.73 nm2, 190.56 nm2, 188.58 nm2 and 188.92 nm2, respectively. All the complexes with phytochemicals showed slightly higher values of SASA compared to (PLpro-ISG15)-complex.
2.3.3. Interactions between the Protein–Ligand Complex: Hydrogen Bond (H-Bond)
Hydrogen bonding plays a critical role in stabilizing the protein–ligand interactions. In our study, the number of H-bonds were calculated over a simulation time of 300 ns for all the complexes. According to Figure 4, (PLpro-ISG15)-complex-AG1, (PLpro-ISG15)-complex-GU1, (PLpro-ISG15)-complex-TU1, and (PLpro-ISG15)-complex-GRL0617 all had an average of four to eight H-bonds each. In addition, the H-bond analysis indicated that the protein–ligand complexes remained stable during simulation.
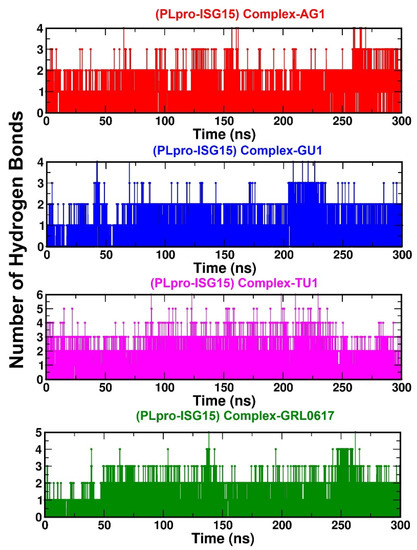
Figure 4.
Number of hydrogen bonds formed between the phytochemicals (AG1 (red) or GU1 (blue) or TU1 (magenta) or GRL0617 (green)) and the (PLpro-ISG15) complex during 300 ns MD simulation.
2.3.4. Binding Affinities of Phytochemicals to the Interface of PLpro-ISG15 Complex: MMPBSA Based Calculations
A detailed understanding of the interactions between (PLpro-ISG15)-complex and the phytochemicals (AG1, GU1, TU1, and GRL0617) is feasible by investigating the thermodynamic parameters that were calculated by the simulations. The MMPBSA method has been widely used to quantify protein–ligand affinities [25]. During 300 ns MD simulation, we extracted an ensemble containing conformations corresponding to the last 50 ns and investigated the details of protein–ligand interactions. The amino acids that were located within the vicinity of 3.5 A distance from the ligand were identified in this average structure. These amino acids represent the critical residues that are involved in binding and interactions, and they were chosen for calculating the free energy of binding. Figure 5 provides more details on these critical residues and their binding energy contributions to each interacting partner of (PLpro-ISG15)-complex.
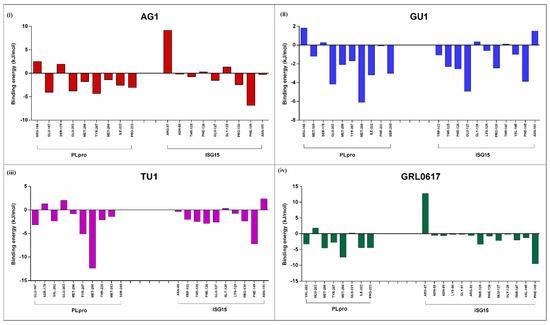
Figure 5.
Binding energies (kJ/mol) at the individual amino acid levels were calculated for the average structure of the last 50 ns for each of the complexes. Key interacting residues from PLpro or ISG15 were considered for calculating binding energy for AG1 (i), GU1 (ii), TU1 (iii) and GRL0617 (iv) in each case.
As detailed in this figure, GRL0617, a known inhibitor of de-ISGylation, engages Val-202 Glu-203 Met-206 Tyr-207 Met-208 Gln-221 Ile-222 Pro-223 from PLpro, and Arg-87, Asn-88, Asn-89, Lys-90, Gly-91, Arg-92, Thr-125, Phe-126, Glu-127, Gly-128, Thr-147, Val-148, Phe-149 from ISG-15. Since GRL0617 is shown to compete with ISG15 for the binding site on PLpro, the amino acids that GRL0617 binds may play an important role in inhibiting de-ISGylation. In order to gain more insight into the similar mechanism of action for AG1, GU1, and TU1 in comparison to GRL0617, we compared the number of amino acids shared between different phytochemicals. As indicated in Table 2, all the phytochemicals (AG1, GU1, and TU1) shared four amino acids, namely Glu-203, Met-206, Tyr-207, and Met-208, in common with GRL0617 for binding to PLpro. Similarly, when compared to GRL0617, all the phytochemicals shared Glu-127, Gly-128, Phe-149, Thr-125, and Phe-126 in common with GRL0617 for binding to ISG15. These analyses indicate that AG1, GU1, and TU1 interact with critical residues from the (PLpro-ISG15)-complex, similar to GRL0617.

Table 2.
Common amino acids engaged by different phytochemicals for binding to PLpro and ISG-15 complex.
Next, to gain insight into the total binding free energy (kJ/mol) for protein–ligand interactions, we analyzed the total binding energy (kJ/mol) of the phytochemicals binding to the key interacting residues from PLpro or ISG15 or (PLpro-ISG15)-complex. As indicated in Figure 6, the binding energy for AG1, GU1, and TU1 were −17.8 kJ/mol, −36.14 kJ/mol, and −41.9 kJ/mol, respectively. GRL0617 showed a value of −33.50 kJ/mol. Overall, GU1 and TU1 had binding affinities that were comparable to or higher than the positive control GRL0617.
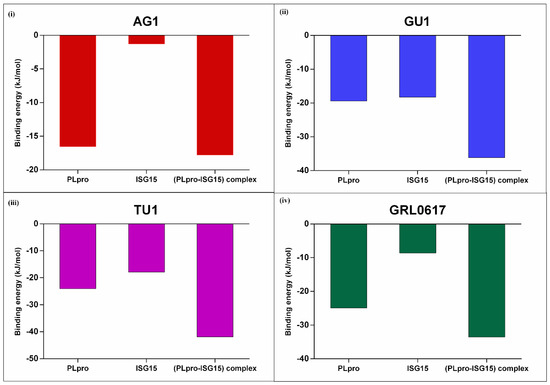
Figure 6.
Binding energies (kJ/mol) of the complexes were calculated by MMPBSA method. Key interacting residues from PLpro or ISG15 or (PLpro-ISG15) complexes were considered for calculating binding energy for AG1 (i), GU1 (ii), TU1 (iii) and GRL0617 (iv).
2.3.5. Effect of Phytochemicals on Protein–Protein Interactions between (PLpro-ISG15)-Complex
As mentioned earlier, PLpro possesses de-ISGylation activity through which it inhibits the immunomodulatory activity of ISG15. For the effective de-ISGylation activity of PLpro, the interaction between PLpro and ISG15 should be maintained stably. Our observations with RMSD values hint that at least GU1 and TU1 may result in disruption of this protein–protein interaction. Work that was carried out by other researchers [26] used the total MMPBSA values to score for protein–protein interactions, and an increase in the MMPBSA for protein–protein complexes was used as an indicator of the destabilization of protein–protein interactions. We employed a similar approach and calculated the MMPBSA for the average structure for the final 50 ns during our simulation periods. Figure 7 provides the details on this. The binding energy for the interaction between PLpro and ISG15 was 437.58 kJ/mol. At the same time, binding the ligands GU1 and TU1 to this complex increased the free energy. This indicates that the binding of GU1 and TU1 to the (PLpro-ISG15)-complex decreased the binding affinity between PLpro and ISG15 proteins. The binding of GRL0617 to (PLpro-ISG15)-complex had a similar trend, but the effect may be less. AG1, on the other hand, did not increase the free energy of binding, instead it slightly decreased it.
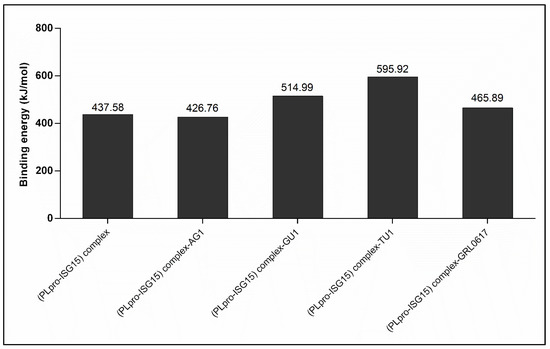
Figure 7.
Protein–protein MMPBSA value (kJ/mol) for PLpro-ISG15 complex, PLpro-ISG15 complex with AG1, PLpro-ISG15 complex with GU1, PLpro-ISG15 complex with TU1 and PLpro-ISG15 complex with GRL0617.
In summary, our studies indicate that 14-deoxy-15-isopropylidene-11,12-didehydroandrographolide from AG, Isocolumbin from GU, and Orientin from TU all accommodated favorably in the interface region of the complex between PLpro and ISG15 proteins. The amino acids that were engaged by these phytochemicals were similar to those that were engaged by GRL0617, indicating that these phytochemicals, similar to GRL0617, interfere with the interaction between PLpro and ISG15 complex. The critical residues of the ISG15 interacting site of the PLpro were well engaged in the interaction with all these ligands, as indicated by docking studies. The efficient binding of the ligands to this interface may potentially interfere with the function of PLpro, i.e., de-ISGylation. In the case of Isocolumbin from GU and Orientin from TU, the protein–protein interaction between PLpro and ISG15 was affected.
3. Materials and Methods
3.1. Target Enzyme Preparation
The PDB structure of a complex between SARS-CoV-2 PLpro and ISG15 (Protein Data Bank (PDB) ID: 6XA9) [16] was obtained from PDB (https://www.rcsb.org/) (accessed on 9 July 2021) and saved as a PDB file (.pdb). AutoDock Tools 1.5.6 was used to prepare the protein targets [17]. Chain A representing PLpro and Chain B representing ISG15 of the 6XA9 complex were retained for the study. Where any water molecules or ligands were removed, polar hydrogen atoms and Kollman charges were added to the protein and saved as a PDBQT file (.pdbqt).
3.2. Ligand Preparation
The 3D structure of the phytochemical compounds (total 90 phytochemicals) was obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov/) (accesses on 9 July 2021) in an SDF file (.sdf) and were converted to a PDB file (.pdb) using Open Babel GUI tool [27]. The ligands were prepared by adding Gasteiger charges, merging non-polar hydrogen, and setting torsion root and then converted to PDBQT (.pdbqt) files using AutoDock Tools 1.5.6 and saved.
3.3. Docking and Visualization
AutoDock Vina 1.1.2 software was used for the molecular docking experiments [17]. Phytochemical compounds were used as ligands. The protein target was prepared as mentioned above. The grid spacing was set to 0.5 Å. The number of grid points along x, y and z dimensions were set as 22 × 40 × 34, respectively, and centered at (x = −30.71, y = −0.002, z = −42.148). The AutoDock Vina output file gives docking scores corresponding to Gibbs free energy of binding (ΔG) (kcal/mol) for each conformation of the ligand. It represents the efficiency of ligand binding to the designated protein–protein interaction interface. Further, the output file of optimal ligand conformations and their 2D interaction with interface residues were visualized using LIGPLOT software [23]. The 3D structure representing the key residues that were involved in PLpro and ISG15 interaction was visualized using a Discovery Studio visualizer version v20.1.0.19295 (BIOVIA, San Diego, CA, USA) [28].
3.4. Drug Likeness Study and ADME Screening
A SwissADME web tool was used to predict the ADME parameters and drug-like nature of GRL0617 and top screened phytochemicals from AG, GU, and TU [29]. A PreADMET web tool was used to predict the ADMET and toxicity parameters of the chemical compound [30]. The ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) properties were determined according to the Lipinski rule, which includes the molecular weight, H-bond acceptors, and H-bond donors, and lipophilicity.
3.5. Molecular Dynamic Simulations and Free Energy Calculation (MM-PBSA)
A molecular dynamics simulation of the selected top protein–ligand complexes was run using Gromacs-2019.4 [31]. For the force field coordinates, the ligand topology was downloaded from the PRODRG server [32]. Using the steepest descent algorithm, 1500 steps were used to prepare the system with the vacuum minimized. Using a water simple point charge (SPC) water model, complex structures were solvated into cubic periodic boxes of 0.5 nm. A salt concentration of 0.15 M was subsequently maintained by adding appropriate numbers of Na+ and Cl− counterions. The system preparation was referred from the previously published paper [33]. Each resultant structure from the NPT equilibration phase was subjected to a final production run in an NPT ensemble for 300 ns of simulation time. The six systems that were considered for simulations include:
- (i)
- (PLpro-ISG15)-complex
- (ii)
- (PLpro-ISG15)-complex with AG1
- (iii)
- (PLpro-ISG15)-complex with GU1
- (iv)
- (PLpro-ISG15)-complex withTU1 and
- (v)
- (PLpro-ISG15)-complex with GRL0617
A trajectory analysis was performed using the GROMACS simulation package for proteins RMSD, RMSF, RG, SASA, H-Bond, and PCA [31]. The molecular mechanics Poisson–Boltzmann surface area (MM-PBSA) approach was employed to understand the free energy of binding (ΔG binding) between the phytochemicals and the target protein complex over the simulation time. A GROMACS utility g_mmpbsa was employed to estimate the binding free energy. To obtain an accurate result, we computed ΔG for the last 50 ns with dt 1000 frames [34].
4. Conclusions
The recent outbreak of SARS-CoV-2 and its subsequent mutations have posed serious problems for disease management. Many therapeutic intervention methods employ strategies to directly inhibit different classes of viral targets, such as proteases and nucleases. However, the host immune system is of equal importance for an effective antiviral response, which in many cases is evaded by viruses using different strategies. ISG15 is established as one of the key mediators of the immunomodulating effects of interferons, and ISGylation is an important host cellular defense against viruses. This is further evident by the observations that several viruses evolved enzyme activities to execute the de-ISGylation activity [35]. Therefore, molecules that interfere with the de-ISGylation activity can offer a very good strategy to boost the host immune system against viruses. They can also be effective in overcoming the virus-mediated host immune suppression. The herbs that we considered in our current study (A. paniculata, T. cordifolia, O. sanctum) have been used for decades as immune modulators, and several phytochemicals from these herbs act at various levels as immunomodulators. In the current study, we provide a molecular basis for a specific mechanism through which the phytochemicals from these herbs can augment the host antiviral immune response. De-ISGylation activity is not restricted only to coronaviruses, and it appears to be a general strategy that is employed by many viruses to overcome the host immune system. While highlighting the mechanism behind the action of these immunomodulatory phytochemicals, our studies also provide a computational strategy to screen for molecules interfering with ISGylation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/computation10070109/s1, Figure S1: Key interacting residues between PLpro and ISG15 ; Table S1: Docking scores of various phytochemicals. Table S2 (i), (ii): Drug-likeness and ADMET properties of top-ranked phytochemicals. Figure S2: PCA analysis.
Author Contributions
Conceptualization, P.S. and R.P.R.; Data curation, A.P.; Formal analysis, S.S.B., R.M. and R.P.R.; Investigation, P.S.; Methodology, A.P.; Resources, A.B. and B.U.V.; Supervision, R.P.R.; Validation, S.S.B. and R.M.; Writing—original draft, P.S.; Writing—review & editing, R.P.R. All authors have read and agreed to the published version of the manuscript.
Funding
The research work was supported by Himalaya Wellness Company, Bangalore, India.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Acknowledgments
The authors sincerely thank Sciomics LLP for offering infrastructure support for conducting the simulation studies.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ADME | Absorption, distribution, metabolism, elimination |
| AG | Andrographis paniculata |
| AG1 | 14-deoxy-15-isopropylidene-11,12-didehydroandrographolide |
| GU | Tinospora cordifolia |
| GU1 | Isocolumbin |
| ISGs Interferon-stimulated genes | |
| MM/PBSA | Molecular mechanics/Poisson–Boltzmann surface area |
| MD | Molecular dynamics |
| PDB | Protein data bank |
| SPC | Simple point charge |
| TU | Ocimum sanctum |
| TU1 | Orientin |
| UIM | Ubiquitin-interacting motif |
References
- Jeon, Y.J.; Yoo, H.M.; Chung, C.H. ISG15 and immune diseases. Biochim. Biophys. Acta 2010, 1802, 485–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Zhang, D.E. Interferon-Stimulated Gene 15 and the Protein ISGylation System. J. Interf. Cytokine Res. 2011, 31, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freitas, B.T.; Scholte, F.E.M.; Bergeron, É.; Pegan, S.D. How ISG15 combats viral infection. Virus Res. 2020, 286, 198036. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, J.; Yan, H.; Huang, J.; Wang, F.; Liu, T.; Zeng, L.; Zhou, F. ISGylation in Innate Antiviral Immunity and Pathogen Defense Responses: A Review. Front. Cell Dev. Biol. 2021, 9, 3196. [Google Scholar] [CrossRef] [PubMed]
- Perng, Y.C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018, 16, 423–439. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Luan, J.; Zhang, L. Molecular docking of potential SARS-CoV-2 papain-like protease inhibitors. Biochem. Biophys. Res. Commun. 2021, 538, 72–79. [Google Scholar] [CrossRef]
- Fung, S.Y.; Yuen, K.S.; Ye, Z.W.; Chan, C.P.; Jin, D.Y. A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: Lessons from other pathogenic viruses. Emerg. Microbes Infect. 2020, 9, 558–570. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, M.; Aktepe, T.E.; Deerain, J.M.; McAuley, J.L.; Audsley, M.D.; David, C.T.; Purcell, D.F.J.; Urin, V.; Hartmann, R.; Moseley, G.W.; et al. SARS-CoV-2 suppresses IFNβ production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not suppress the effects of added interferon. PLoS Pathog. 2021, 17, e1009800. [Google Scholar] [CrossRef]
- Kim, Y.M.; Shin, E.C. Type I and III interferon responses in SARS-CoV-2 infection. Exp. Mol. Med. 2021, 53, 750–760. [Google Scholar] [CrossRef]
- Gupta, S.; Mishra, K.P.; Ganju, L. Broad-spectrum antiviral properties of andrographolide. Arch. Virol. 2017, 162, 611–623. [Google Scholar] [CrossRef]
- Wang, W.; Wang, J.; Dong, S.F.; Liu, C.H.; Italiani, P.; Sun, S.H.; Xu, J.; Boraschi, D.; Ma, S.P.; Qu, D. Immunomodulatory activity of andrographolide on macrophage activation and specific antibody response. Acta Pharmacol. Sin. 2010, 31, 191–201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niraj, S.; Varsha, S. A review on scope of immuno-modulatory drugs in Ayurveda for prevention and treatment of COVID-19. Plant. Sci. Today 2020, 7, 417–423. [Google Scholar] [CrossRef]
- Borse, S.; Joshi, M.; Saggam, A.; Bhat, V.; Walia, S.; Marathe, A.; Sagar, S.; Chavan-Gautam, P.; Girme, A.; Hingorani, L.; et al. Ayurveda botanicals in COVID-19 management: An in silico multi-target approach. PLoS ONE 2021, 16, e0248479. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.S.; Panda, S.K.; Luyten, W. Phytochemicals against SARS-CoV as potential drug leads. Biomed. J. 2021, 44, 74–85. [Google Scholar] [CrossRef] [PubMed]
- Krupanidhi, S.; Peele, K.A.; Venkateswarulu, T.C.; Ayyagari, V.S.; Bobby, M.N.; Babu, D.J.; Narayana, A.V.; Aishwarya, G. Screening of phytochemical compounds of Tinospora cordifolia for their inhibitory activity on SARS-CoV-2: An in silico study. J. Biomol. Struct. Dyn. 2021, 39, 5799–5803. [Google Scholar] [CrossRef]
- Klemm, T.; Ebert, G.; Calleja, D.J.; Allison, C.C.; Richardson, L.W.; Bernardini, J.P.; Lu, B.G.; Kuchel, N.W.; Grohmann, C.; Shibata, Y.; et al. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J. 2020, 39, e106275. [Google Scholar] [CrossRef]
- Anil, K.T.J.W. Autodock vina: Improving the speed and accuracy of docking. J. Comput. Chem. 2019, 31, 455–461. [Google Scholar] [CrossRef]
- Shin, D.; Mukherjee, R.; Grewe, D.; Bojkova, D.; Baek, K.; Bhattacharya, A.; Schulz, L.; Widera, M.; Mehdipour, A.R.; Tascher, G.; et al. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 2020, 587, 657–662. [Google Scholar] [CrossRef]
- Freitas, B.T.; Durie, I.A.; Murray, J.; Longo, J.E.; Miller, H.C.; Crich, D.; Hogan, R.J.; Tripp, R.A.; Pegan, S.D. Characterization and Noncovalent Inhibition of the Deubiquitinase and deISGylase Activity of SARS-CoV-2 Papain-Like Protease. ACS Infect. Dis. 2020, 6, 2099–2109. [Google Scholar] [CrossRef]
- Fu, Z.; Huang, B.; Tang, J.; Liu, S.; Liu, M.; Ye, Y.; Liu, Z.; Xiong, Y.; Zhu, W.; Cao, D.; et al. The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery. Nat. Commun. 2021, 12, 488. [Google Scholar] [CrossRef]
- Islam, M.T.; Bardaweel, S.K.; Mubarak, M.S.; Koch, W.; Gaweł-Beben, K.; Antosiewicz, B.; Sharifi-Rad, J. Immunomodulatory Effects of Diterpenes and Their Derivatives Through NLRP3 Inflammasome Pathway: A Review. Front. Immunol. 2020, 11, 2234. [Google Scholar] [CrossRef] [PubMed]
- Wardana, A.P.; Aminah, N.S.; Rosyda, M.; Abdjan, M.I.; Kristanti, A.N.; Tun, K.N.W.; Choudhary, M.I.; Takaya, Y. Potential of diterpene compounds as antivirals, a review. Heliyon 2021, 7, e07777. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein. Eng. Des. Sel. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, Q.; Yuan, Z.F.; Zhang, L.; Xu, L.; Cui, Y.; Duan, K. Pharmacokinetics and tissue distribution study of orientin in rat by liquid chromatography. J. Pharm. Biomed. Anal. 2008, 47, 429–434. [Google Scholar] [CrossRef]
- Wang, C.; Nguyen, P.H.; Pham, K.; Huynh, D.; Le, T.B.N.; Wang, H.; Ren, P.; Luo, R. Calculating protein-ligand binding affinities with MMPBSA: Method and error analysis. J. Comput. Chem. 2016, 37, 2436–2446. [Google Scholar] [CrossRef] [Green Version]
- Martin, W.R.; Lightstone, F.C.; Cheng, F. In Silico Insights into Protein–Protein Interaction Disruptive Mutations in the PCSK9-LDLR Complex. Int. J. Mol. Sci. 2020, 21, 1550. [Google Scholar] [CrossRef] [Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An Open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Biovia: Discovery Studio Modeling Environment-Google Scholar. Available online: https://scholar.google.com/scholar?cluster=17675170202455151209&hl=en&as_sdt=2005&sciodt=0,5 (accessed on 8 June 2022).
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [Green Version]
- PreADME in EuroQSAR 2004–PreADMET|Prediction of ADME/Tox. 2004. Available online: https://preadmet.webservice.bmdrc.org/2004/09/27/200409-preadme-in-euroqsar-2004/ (accessed on 7 June 2022).
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Schüttelkopf, A.W.; Van Aalten, D.M.F. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 1355–1363. [Google Scholar] [CrossRef] [Green Version]
- Gangadharappa, B.S.; Sharath, R.; Revanasiddappa, P.D.; Chandramohan, V.; Balasubramaniam, M.; Vardhineni, T.P. Structural insights of metallo-beta-lactamase revealed an effective way of inhibition of enzyme by natural inhibitors. J. Biomol. Struct. Dyn. 2020, 38, 3757–3771. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa–a GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Villarroya-Beltri, C.; Guerra, S.; Sánchez-Madrid, F. ISGylation-a key to lock the cell gates for preventing the spread of threats. J. Cell Sci. 2017, 130, 2961–2969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).