MR Brain Image Segmentation: A Framework to Compare Different Clustering Techniques
Abstract
:1. Introduction
2. The Image Segmentation Framework
- (a)
- Partitioning: divide into regions/sequences with coherent internal properties;
- (b)
- Grouping: identify sets of coherent tokens in the image.
- choosing the image
- choosing the color space
- choosing the segmentation method
- setting the running parameters of the specific method
- visualizing the segmented image
- computing the evaluation metrics
- User work section: This component gives the user the possibility to access the primary functions of the tool, which are the clustering method section and the evaluation section.
- Method section: For each implemented method, the user can configure its parameters such as color space conversion, number of cluster, maximum number of iterations, stopping condition and visualization mode.
- Evaluation section: The segmentation results can be evaluated both qualitatively and quantitatively. Qualitative evaluation is made by visualization of the segmented image compared to the ground truth image. Quantitative evaluation is made by computing the metrics described in Section 4.
- by the cluster centroid color: Each point of a cluster is labeled with the color of its centroid (in the case of color conversion, the color space is converted back to RGB);
- by a gray level: Each pixel is labeled with the number of the cluster it belongs to, and the range is stretched in 0–255;
- by a random RGB color: A random RGB value is generated for each cluster;
- by a binary stack: The clustering is represented as a stack of binary images. Each binary image represents a cluster; each pixel shows a hard cluster membership. Thus, it is possible to extract cluster regions from the original image by performing an AND operation between a slide of the stack and the original image.
- by using a fuzzy stack: A stack of gray level images is used to show the membership values of each pixel to each cluster. Each pixel represents the soft cluster membership value of that pixel in the original image according to the currently selected cluster.
3. Clustering Methods
3.1. K-means
- Fix the number of clusters K, and initialize the cluster centers (), either randomly or based on some heuristic;
- Assign each pixel to the cluster that minimizes the distance between the pixel and the cluster center;
- Re-compute the cluster centers by averaging all of the pixels in the cluster, namely:
- Repeat Steps 2 and 3 until convergence is attained (i.e., the assignment of pixels to clusters does not change)
3.2. Fuzzy C-Means
- Fix the number of clusters K and initialize the cluster centers (), either randomly or based on some heuristic;
- Compute membership values using Equation (4)
- Re-compute the cluster centers using Equation (5)
- Repeat Steps 2 and 3 until convergence is attained (i.e., the assignment of pixels to clusters does not change)
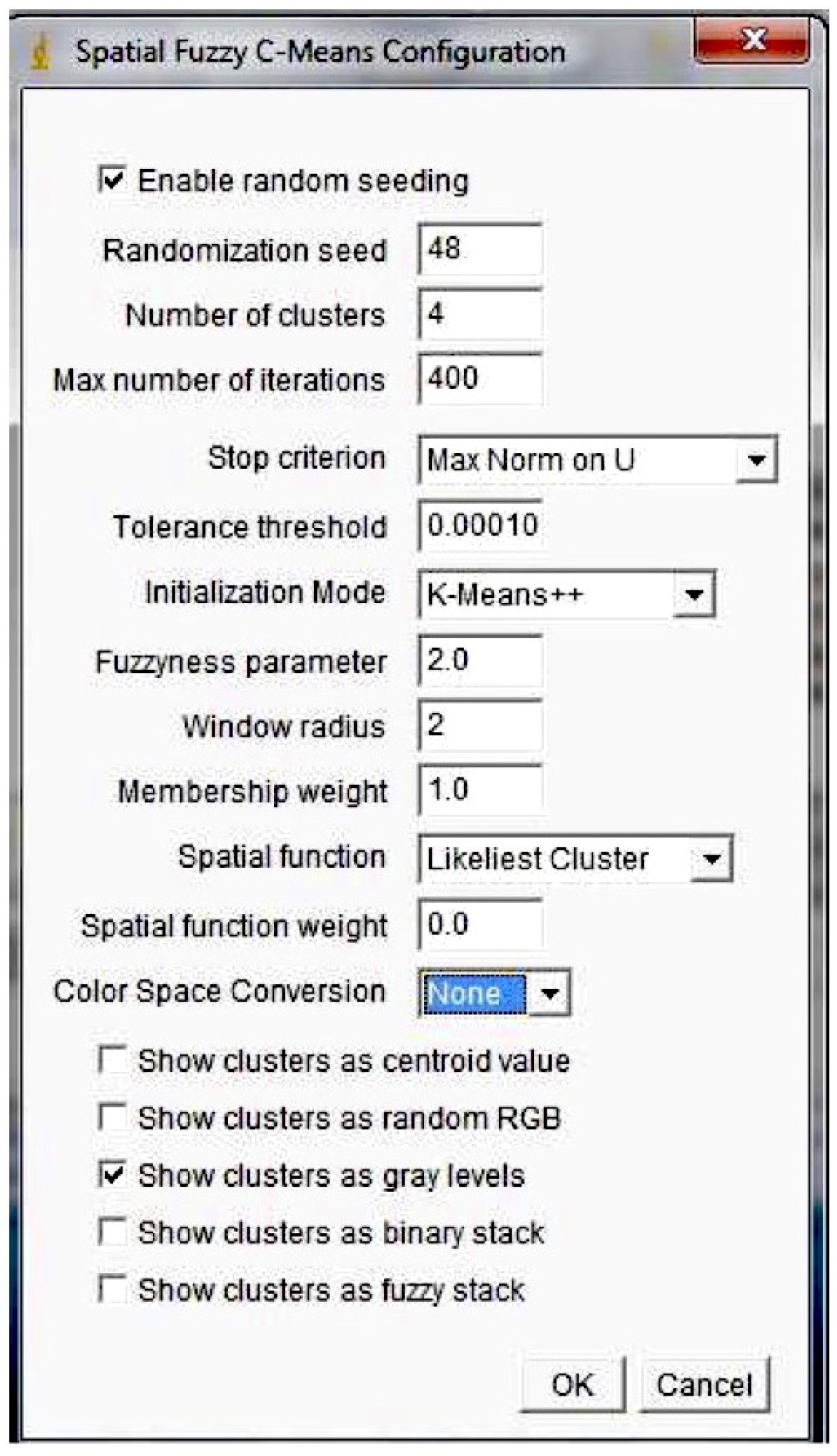
3.3. Spatial FCM
- : fuzziness parameter used to control the fuzziness; if m is near one, the results are similar to those obtained by K-means
- p and q: parameters used to control the relative importance of membership and spatial functions
- Radius r: the spatial function is evaluated on a window centered on the pixel
3.4. Kernelized FCM
4. Evaluation Metrics
- (True Positive) is the number of pixels that belong to tissue T in the ground truth image and are correctly classified as tissue T in the segmented image;
- (False Negative) is the number pixels that are classified as tissue T in the ground truth image, but classified as different tissues in the segmented image;
- (True Negatives) is the number of pixels that are classified as different tissues both in the segmented and ground truth images;
- (False Positives) is the number of pixels incorrectly classified as tissue T in the segmented image compared to the ground truth image.
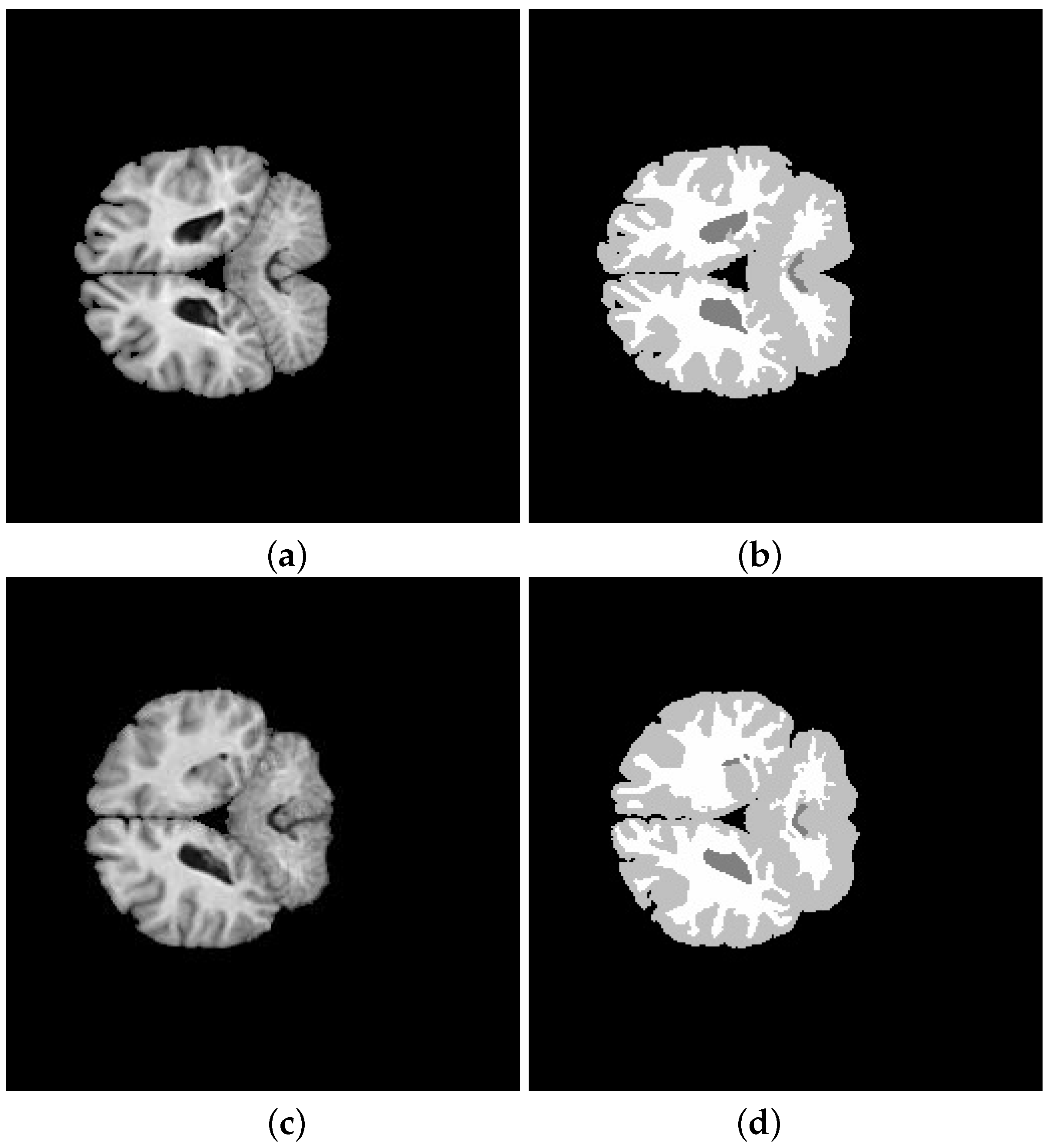
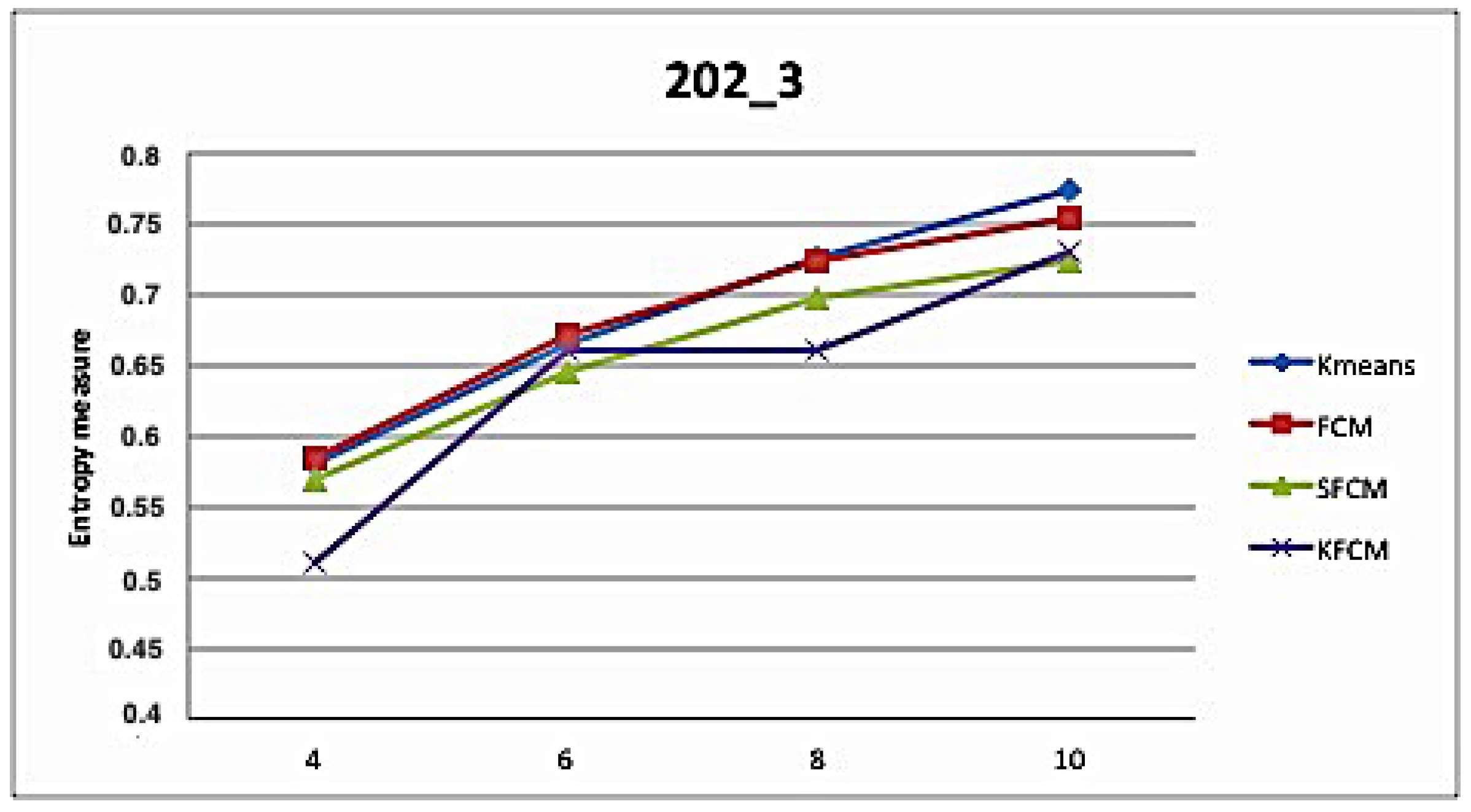
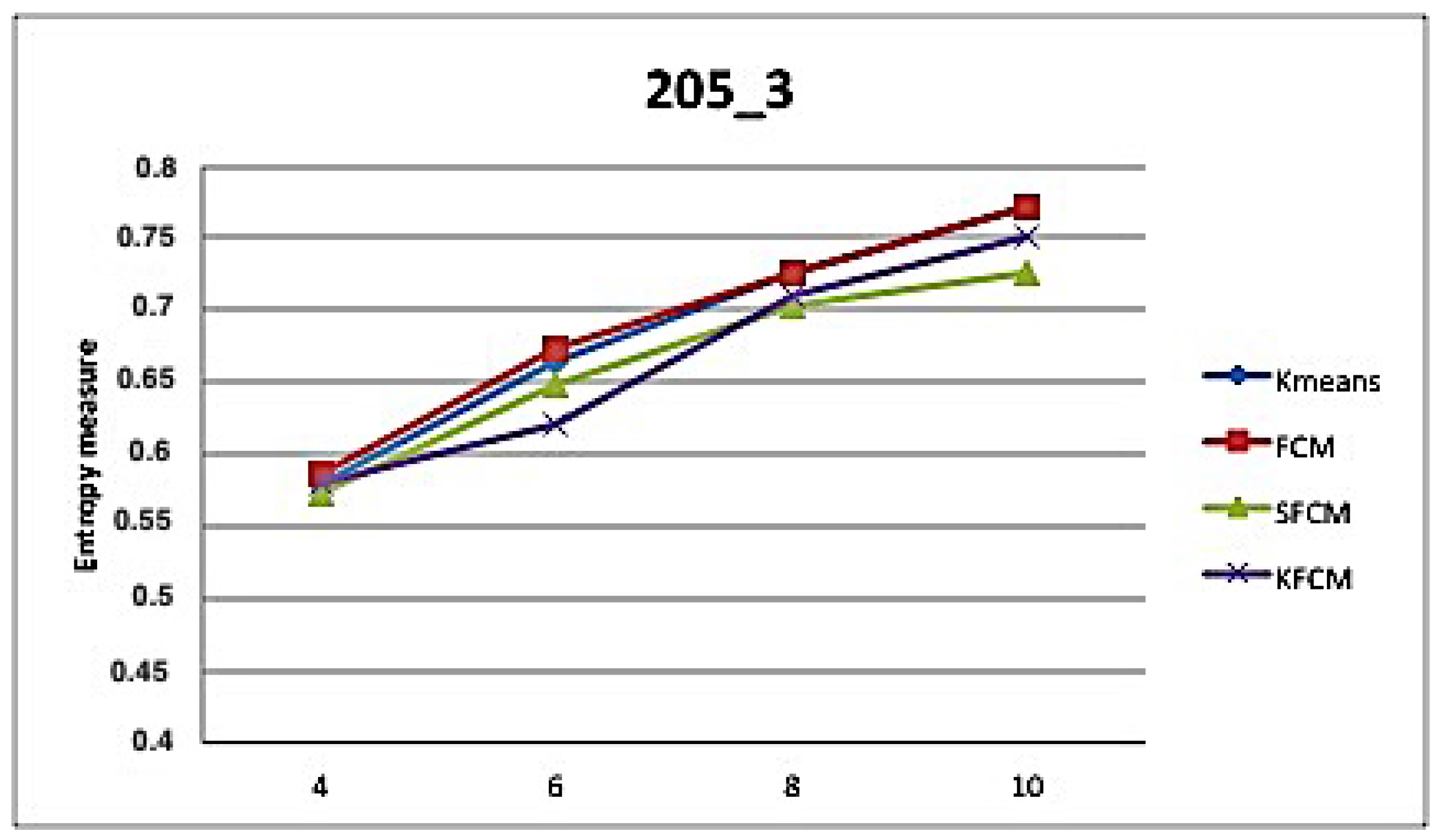
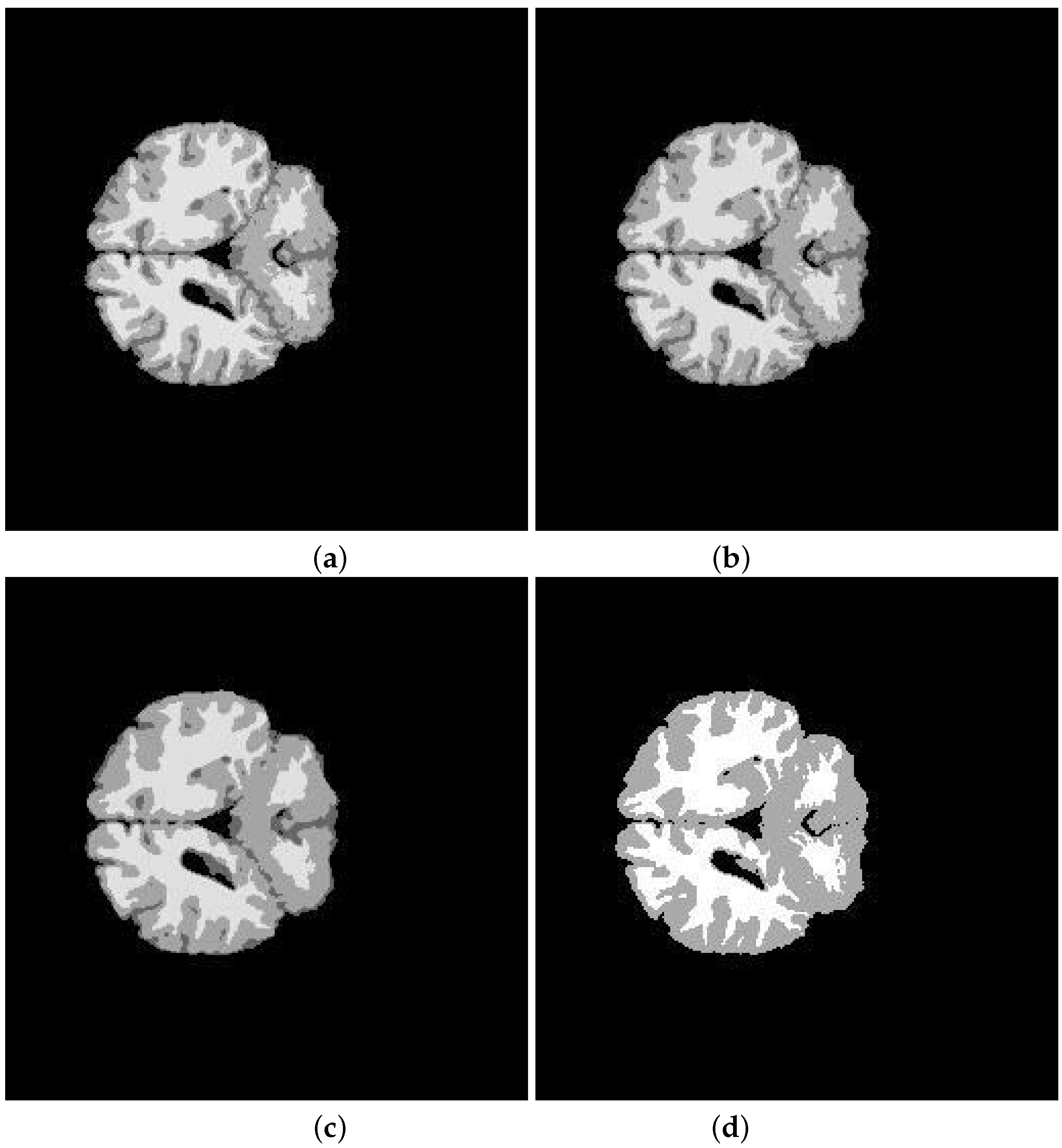
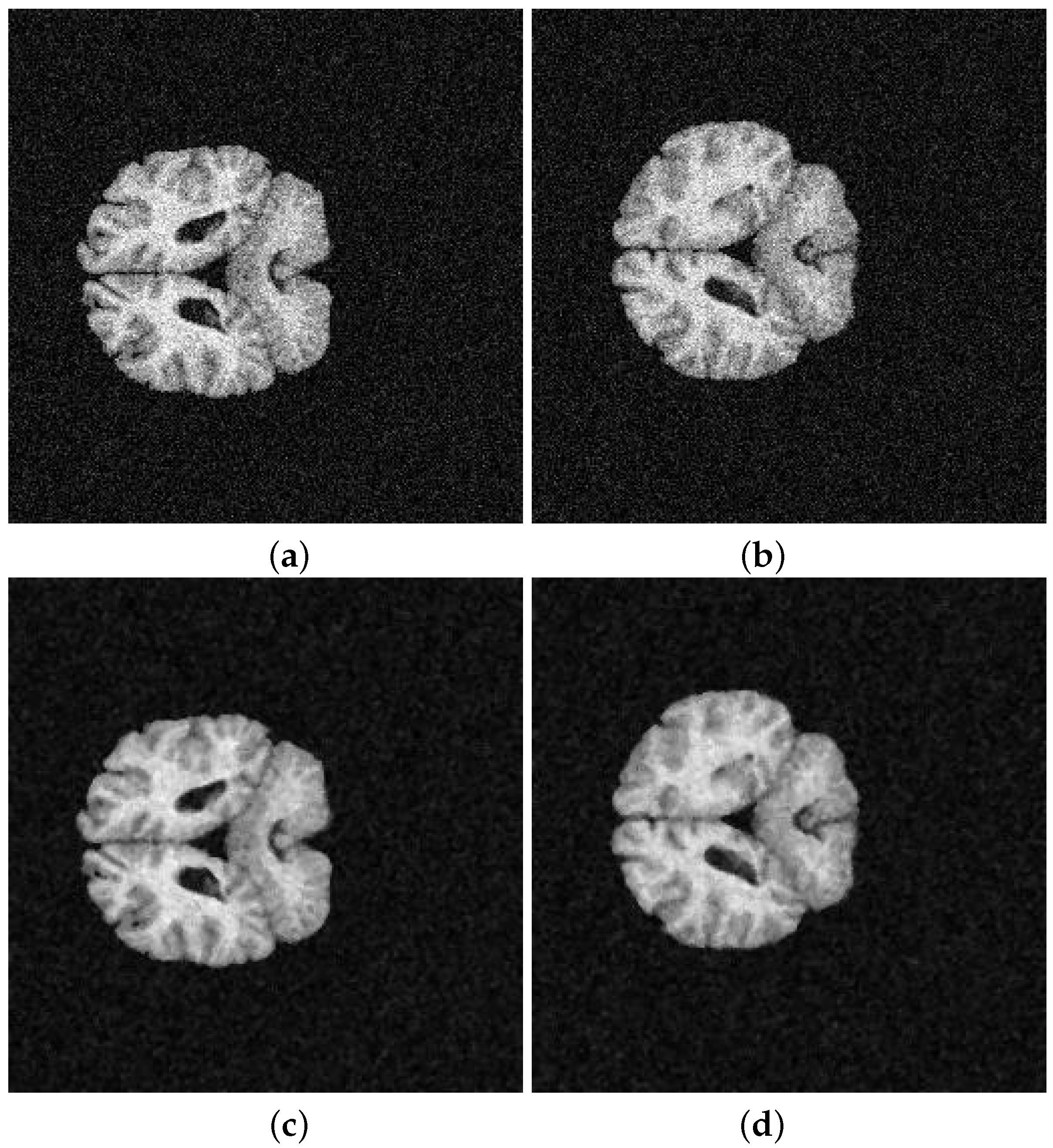
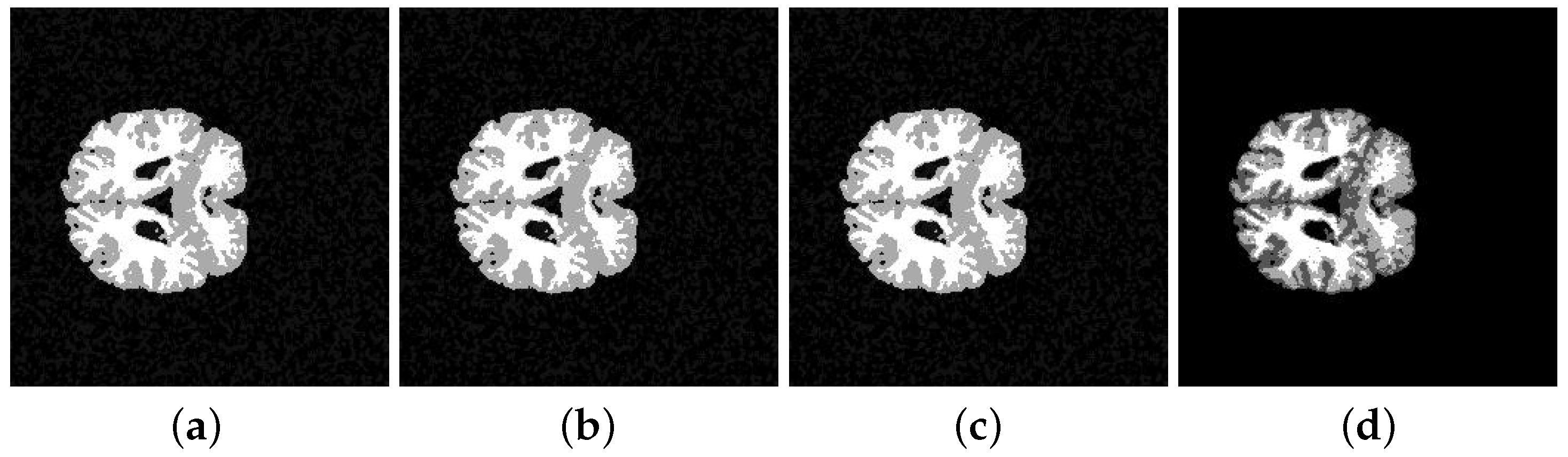
5. Application and Results
6. Conclusions
Author Contributions
Conflicts of Interest
References
- Wells, W.M.; Grimson, W.E.L.; Kikinis, R.; Jolesz, F.A. Adaptive segmentation of MRI data. IEEE Trans. Med. Imaging 1996, 15, 429–442. [Google Scholar] [CrossRef] [PubMed]
- Forouzanfar, M.; Forghani, N.; Teshnehlab, M. Parameter optimization of improved fuzzy c-means clustering algorithm for brain MR image segmentation. Eng. Appl. Artif. Intell. 2010, 23, 160–168. [Google Scholar] [CrossRef]
- Counsell, S.J.; Boardman, J.P. Differential brain growth in the infant born preterm: Current knowledge and future developments from brain imaging. Sem. Fetal Neonatal Med. 2005, 10, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Nishida, M.; Makris, N.; Kennedy, D.N.; Vangel, M.; Fischl, B.; Krishnamoorthy, K.S.; Caviness, V.S.; Grant, P.E. Detailed semiautomated MRI based morphometry of the neonatal brain: Preliminary results. Neuroimage 2006, 32, 1041–1049. [Google Scholar] [CrossRef] [PubMed]
- Cordato, N.J.; Duggins, A.J.; Halliday, G.M.; Morris, J.G.; Pantelis, C. Clinical deficits correlate with regional cerebral atrophy in progressive supranuclear palsy. Brain 2005, 128, 1259–1266. [Google Scholar] [CrossRef] [PubMed]
- Sepulcre, J.; Sastre-Garriga, J.; Cercignani, M.; Ingle, G.T.; Miller, D.H.; Thompson, A.J. Regional gray matter atrophy in early primary progressive multiple sclerosis: A voxel-based morphometry study. Arch. Neurol. 2006, 8, 1175–1180. [Google Scholar] [CrossRef] [PubMed]
- Fotenos, A.F.; Snyder, A.Z.; Girton, L.E.; Morris, J.C.; Buckner, R.L. Normative estimates of cross-sectional and longitudinal brain volume decline in aging and AD. Neurology 2005, 64, 1032–1039. [Google Scholar] [CrossRef] [PubMed]
- Ridha, B.H.; Barnes, J.; Bartlett, J.W.; Godbolt, A.; Pepple, T.; Rossor, M.N.; Fox, N.C. Tracking atrophy progression in familial Alzheimer’s disease: A serial MRI study. Lanc. Neurol. 2006, 5, 828–834. [Google Scholar] [CrossRef]
- Meyer-Lindenberg, A.; Mervis, C.B.; Sarpal, D.; Koch, P.; Steele, S.; Kohn, P.; Marenco, S.; Morris, C.A.; Das, S.; Kippenhan, S.; et al. Functional, structural, and metabolic abnormalities of the hippocampal formation in Williams syndrome. J. Clin. Investig. 2005, 115, 1888–1895. [Google Scholar] [CrossRef] [PubMed]
- Ciumas, C.; Savic, I. Structural changes in patients with primary generalized tonic and clonic seizures. Neurology 2006, 67, 683–686. [Google Scholar] [CrossRef] [PubMed]
- Henriksson, K.M.; Wickstrom, K.; Maltesson, N.; Ericsson, A.; Karlsson, J.; Lindgren, F.; Astrom, K.; McNeil, T.F.; Agartz, I. A pilot study of facial, cranial and brain MRI morphometry in men with schizophrenia. Psychiatr. Res. Neuroimaging 2006, 147, 187–195. [Google Scholar] [CrossRef] [PubMed]
- Honea, R.; Crow, T.J.; Passingham, D.; Mackay, C.E. Regional deficits in brain volume in schizophrenia: A meta-analysis of voxel-based morphometry studies. Am. J. Psychiatr. 2005, 162, 2233–2245. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.O.; Pfefferbaum, A. Segmentation of MR brain images into cerebrospinal fluid spaces, white and gray matter. J. Comput. Assist. Tomogr. 1989, 13, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Toriwaki, J. Automatic segmentation of head MRI images by knowledge guided thresholding. Comput. Med. Imaging Graph. 1991, 15, 233–240. [Google Scholar] [CrossRef]
- Canny, J.A. A computational approach to edge detection. IEEE Trans. Pattern Anal. Mach. Intell. 1986, 6, 679–698. [Google Scholar] [CrossRef]
- Pohle, R.; Toennies, K.D. Segmentation of medical images using adaptive region growing. In Proceedings of the International Society for Optics and Photonics (SPIE), San Diego, CA, USA, 3 July 2001; p. 4322. [Google Scholar] [CrossRef]
- Li, C.; Huang, R.; Ding, Z.; Gatenby, J.C.; Metaxas, D.N.; Gore, J.C. A level set method for image segmentation in the presence of intensity inhomogeneities with application to MRI. IEEE Trans. Image Process. 2011, 20, 2007–2016. [Google Scholar] [PubMed]
- Ciecholewski, M. An edge-based active contour model using an inflation/deflation force with a damping coefficient. Expert Syst. Appl. 2016, 44, 22–36. [Google Scholar] [CrossRef]
- Zhang, K.; Song, H.; Zhang, L. Active contours driven by local image fitting energy. Pattern Recognit. 2010, 43, 1199–1206. [Google Scholar] [CrossRef]
- Dhanachandra, N.; Chanu, Y.J. A Survey on Image Segmentation Methods using Clustering Techniques. Eur. J. Eng. Res. Sci. 2017, 2, 15–20. [Google Scholar] [CrossRef]
- Jain, A.K.; Dubes, R.C. Algorithms for Clustering Data; Prentice Hall: Englewood Cliffs, NJ, USA, 1988. [Google Scholar]
- Li, C.L.; Goldgof, D.B.; Hall, L.O. Knowledge-based classification and tissue labeling of MR images of human brain. IEEE Trans. Med. Imaging 1993, 12, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Panda, R.; Dora, L. A study on fuzzy clustering for magnetic resonance brain image segmentation using soft computing approaches. Appl. Soft Comput. 2014, 24, 522–533. [Google Scholar] [CrossRef]
- Bezdek, J.C. Pattern Recognition with Fuzzy Objective Function Algorithms; Kluwer Academic Publishers: New York, NY, USA, 1981. [Google Scholar]
- Patel, B.C.; Sinha, G.R. An adaptive k-means clustering algorithm for breast image segmentation. Int. J. Comput. Appl. 2010, 10, 35–38. [Google Scholar] [CrossRef]
- Chen, W.; Giger, M.L.; Bick, U. A fuzzy c-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR images. Acad. Radiol. 2006, 13, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Ghane, N.; Vard, A.; Talebi, A.; Nematollahy, P. Segmentation of White Blood Cells From Microscopic Images Using a Novel Combination of K-Means Clustering and Modified Watershed Algorithm. J. Med. Signals Sens. 2017, 7, 92–101. [Google Scholar] [PubMed]
- Karthikeyan, T.; Poornima, N. Microscopic Image Segmentation Using Fuzzy C Means for Leukemia Diagnosis. Leukemia 2017, 4, 3136–3142. [Google Scholar]
- Kechagias-Stamatis, O.; Aouf, N.; Nam, D.; Belloni, C. Automatic X-ray Image Segmentation and Clustering for Threat Detection. In Target and Background Signatures III, 104320O; SPIE: Bellingham, WA, USA, 2017. [Google Scholar] [CrossRef]
- Son, L.H.; Tuan, T.M. Dental segmentation from X-ray images using semi-supervised fuzzy clustering with spatial constraints. Eng. Appl. Artif. Intell. 2017, 59, 186–195. [Google Scholar] [CrossRef]
- Ji, Z.; Liu, J.; Cao, G.; Sun, Q.; Chen, Q. Robust spatially constrained fuzzy c-means algorithm for brain MR image segmentation. Pattern Recognit. 2014, 47, 2454–2466. [Google Scholar] [CrossRef]
- Portela, N.M.; Cavalcanti, G.D.C.; Ren, T.I. Semi-supervised clustering for MR brain image segmentation. Expert Syst. Appl. 2014, 41, 1492–1497. [Google Scholar] [CrossRef]
- Zhang, Y.; Ye, S.; Ding, W. Based on rough set and fuzzy clustering of MRI brain segmentation. Int. J. Biomath. 2017, 10, 1750026. [Google Scholar] [CrossRef]
- Kaya, I.E.; Pehlivanlı, A.Ç.; Sekizkardeş, E.G.; Ibrikci, T. PCA based clustering for brain tumor segmentation of T1w MRI images. Comput. Methods Progr. Biomed. 2017, 140, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Novelli, A.; Aguilar, M.A.; Aguilar, F.J.; Nemmaoui, A.; Tarantino, E. AssesSeg—A Command Line Tool to Quantify Image Segmentation Quality: A Test Carried Out in Southern Spain from Satellite Imagery. Remote Sens. 2017, 9, 40. [Google Scholar] [CrossRef]
- Neubert, M.; Herold, H.; Meinel, G. Evaluation of Remote Sensing Image Segmentation Quality–Further Results and Concepts. Available online: http://www.isprs.org/proceedings/xxxvi/4-c42/papers/10_Adaption%20and%20further%20development%20II/OBIA2006_Neubert_Herold_Meinel.pdf (accessed on 4 November 2017).
- Keshavan, A.; Datta, E.; McDonough, I.M.; Madan, C.R.; Jordan, K.; Henry, R.G. Mindcontrol: A web application for brain segmentation quality control. NeuroImage 2017. [Google Scholar] [CrossRef] [PubMed]
- Udupa, J.K.; LeBlanc, V.R.; Zhuge, Y.; Imielinska, C.; Schmidt, H.; Currie, L.M.; Hirsch, B.E.; Woodburn, J. A framework for evaluating image segmentation algorithms. Comput. Med. Imaging Gr. 2006, 30, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Fritts, J.E.; Goldman, S.A. An entropy-based objective evaluation method for image segmentation. In Electronic Imaging; International Society for Optics and Photonics: Bellingham, WA, USA, 2004; pp. 38–49. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shapiro, L.G. Image segmentation techniques. Comput. Vision Gr. Image Proc. 1985, 29, 100–132. [Google Scholar] [CrossRef]
- Gonzalez, R.C.; Woods, R.E. Digital Image Processing; Addison-Wesley: Reading, MA, USA, 1992. [Google Scholar]
- Pal, N.R.; Pal, S.K. A review on image segmentation techniques. Pattern Recognit. 1993, 26, 1277–1294. [Google Scholar] [CrossRef]
- Langan, D.A.; Modestino, J.W.; Zhang, J. Cluster validation for unsupervised stochastic model-based image segmentation. IEEE Trans. Image Proc. 1998, 7, 180–195. [Google Scholar] [CrossRef] [PubMed]
- MacQueen, J.B. Some Methods for classification and analysis of multivariate observations. In Proceedings of the 5-th Berkeley Symposium on Mathematical Statistics and Probability; Le Cam, L.M., Neyman, J., Eds.; University of California Press: Berkeley, CA, USA, 1967; pp. 281–297. [Google Scholar]
- Bezdek, J.C.; Hall, L.; Clarke, L. Review of MR image segmentation using pattern recognition. Med. Phys. 1993, 20, 1033–1048. [Google Scholar] [CrossRef] [PubMed]
- Udupa, J.K.; Wei, L.; Samarasekera, S.; Miki, Y.; Van Buchem, M.A.; Grossman, R.I. Multiple sclerosis lesion quantification using fuzzy-connectedness principles. IEEE Trans. Med. Imaging 1997, 16, 598–609. [Google Scholar] [CrossRef] [PubMed]
- Masoole, M.G.; Moosavi, A.S. An improved fuzzy algorithm for image segmentation. World Acad. Sci. Eng. Technol. 2008, 38, 400–404. [Google Scholar]
- Shen, S.; Sandham, W.; Granat, M.; Sterr, A. MRI fuzzy segmentation of brain tissue using neighborhood attraction with neural-network optimization. IEEE Trans. Inf. Technol. Biomed. 2005, 9, 459–467. [Google Scholar] [CrossRef]
- Jiang, L.; Wenhui, W. A modified fuzzy c-means algorithm for segmentation of magnetic resonance images. In Proceedings of the VIIth Digital Image Computing: Techniques and Applications, Sydney, Australia, 10–12 December 2003. [Google Scholar]
- Pham, D.L.; Prince, J.L. Adaptive fuzzy segmentation of magnetic resonance images. IEEE Trans. Med. Imaging 1999, 18, 737–752. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Pham, D.L.; Prince, J.L. Finding the brain cortex using fuzzy segmentation, isosurfaces, and deformable surfaces. In Information Processing in Medical Imaging. IPMI 1997; Duncan, J., Gindi, G., Eds.; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 1997; Volume 1230, pp. 399–404. [Google Scholar] [CrossRef]
- Kannan, S.R.; Ramathilagam, S.; Sathya, A.; Pandiyarajan, R. Effective fuzzy c-means based kernel function in segmenting medical images. Comput. Biol. Med. 2010, 40, 572–579. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, S.; Ghazanfari, M.; Fathian, M. Using data mining for learning and clustering FCM. Int. J. Comput. Intell. 2008, 4, 118–125. [Google Scholar]
- Dawant, B.M.; Zijidenbos, A.P.; Margolin, R.A. Correction of intensity variations in MR image for computer-aided tissue classification. IEEE Trans. Med. Imaging 1993, 12, 770–781. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.; Atkins, M.S.; Mackiewich, B.; Andson, M. Segmentation of multiple sclerosis lesions in intensity corrected multispectral MRI. IEEE Trans. Med. Imaging 1996, 15, 154–169. [Google Scholar] [CrossRef] [PubMed]
- Tolias, Y.A.; Panas, S.M. Image segmentation by a fuzzy clustering algorithm using adaptive spatially constrained membership function. IEEE Trans. Syst. Man Cybern. Part A Syst. Hum. 1998, 28, 359–369. [Google Scholar] [CrossRef]
- Liew, A.W.C.; Leung, S.H.; Lau, W.H. Fuzzy image clustering incorporating spatial continuity. IEEE Proc. Vision Image Signal Proc. 2000, 147, 185–192. [Google Scholar] [CrossRef]
- Chuang, K.S.; Tzeng, H.L.; Chen, S.; Wu, J.; Chen, T.J. Fuzzy c-means clustering with spatial information for image segmentation. Comput. Med. Imaging Gr. 2006, 30, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Caponetti, L.; Castellano, G.; Corsini, V.; Basile, T.M.A. Cytoplasm image segmentation by spatial fuzzy clustering. In Fuzzy Logic and Applications; Fanelli, A.M., Pedrycz, W., Petrosino, A., Eds.; Lecture Notes in Computer Science (LNCS); Springer: New York, NY, USA, 2011; Volume 6857, pp. 253–260. ISBN 978-3-642-23712-6. [Google Scholar] [CrossRef]
- Muller, K.R.; Mika, S.; Ratsch, G.; Tsuda, K.; Scholkopf, B. An Introduction to Kernel-based Learning algorithms. IEEE Trans. Neural Netw. 2001, 12, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-Q.; Chen, S.-C. A novel kernelized fuzzy C-means algorithm with application in medical image segmentation. Artificial Intell. Med. 2004, 32, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Valverde, S.; Oliver, A.; Cabezas, M.; Roura, E.; Lladò, X. Comparison of 10 brain tissue segmentation methods using revisited ibsr annotations. J. Magn. Reson. Imaging 2015, 41, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Image Plugin Spatial Fuzzy c-Means. Available online: https://sites.google.com/site/cilabuniba/research/sfcm (accessed on 3 November 2017).
- The Internet Brain Segmentation Repository (IBSR). Available online: http://www.cma.mgh.harvard.edu/ibsr/ (accessed on 3 November 2017).
- Kennedy, D.N.; Filipek, P.A.; Caviness, V.S. Anatomic segmentation and volumetric calculations in nuclear magnetic resonance imaging. IEEE Trans. Med. Imaging 1989, 8, 1–7. [Google Scholar] [CrossRef] [PubMed]

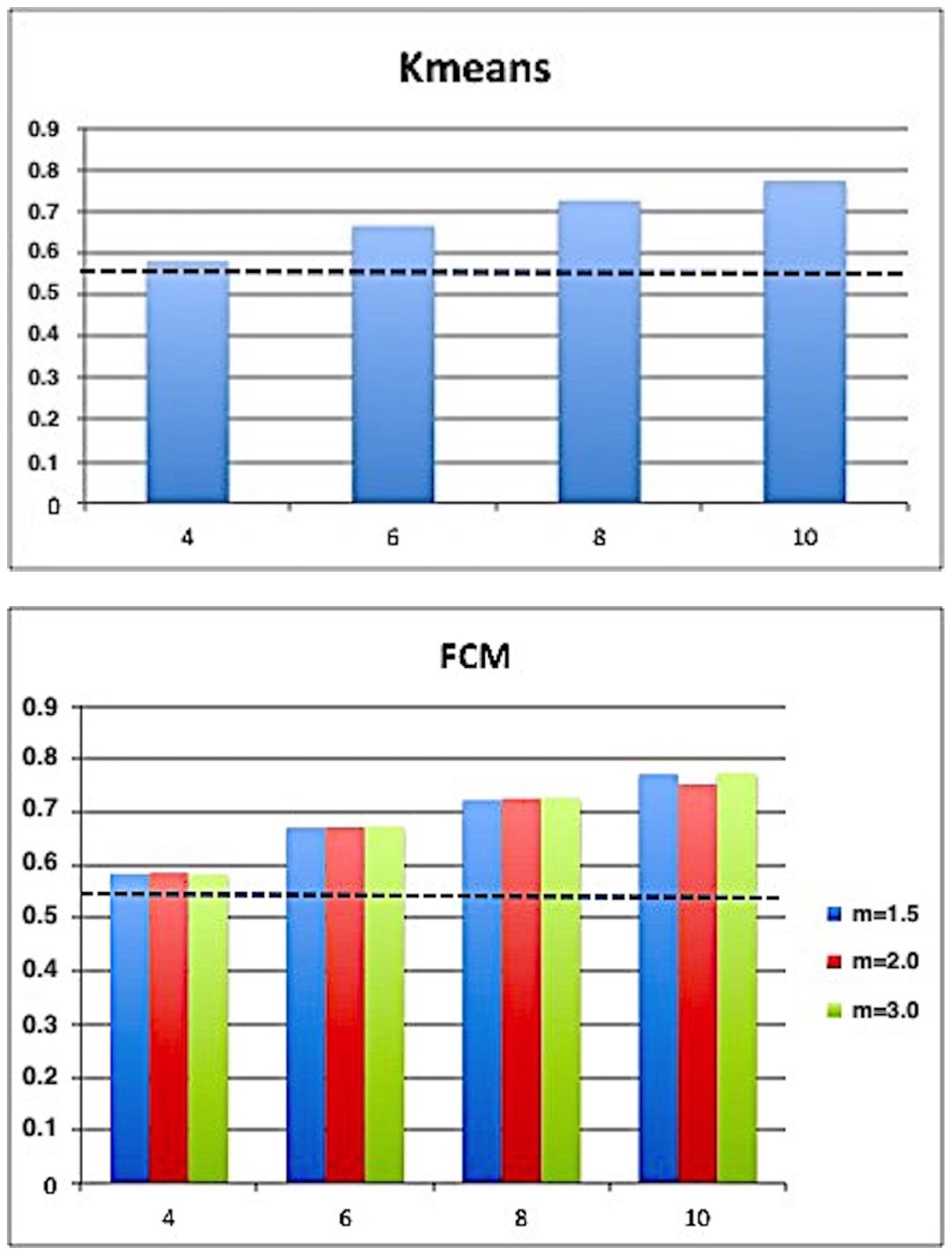
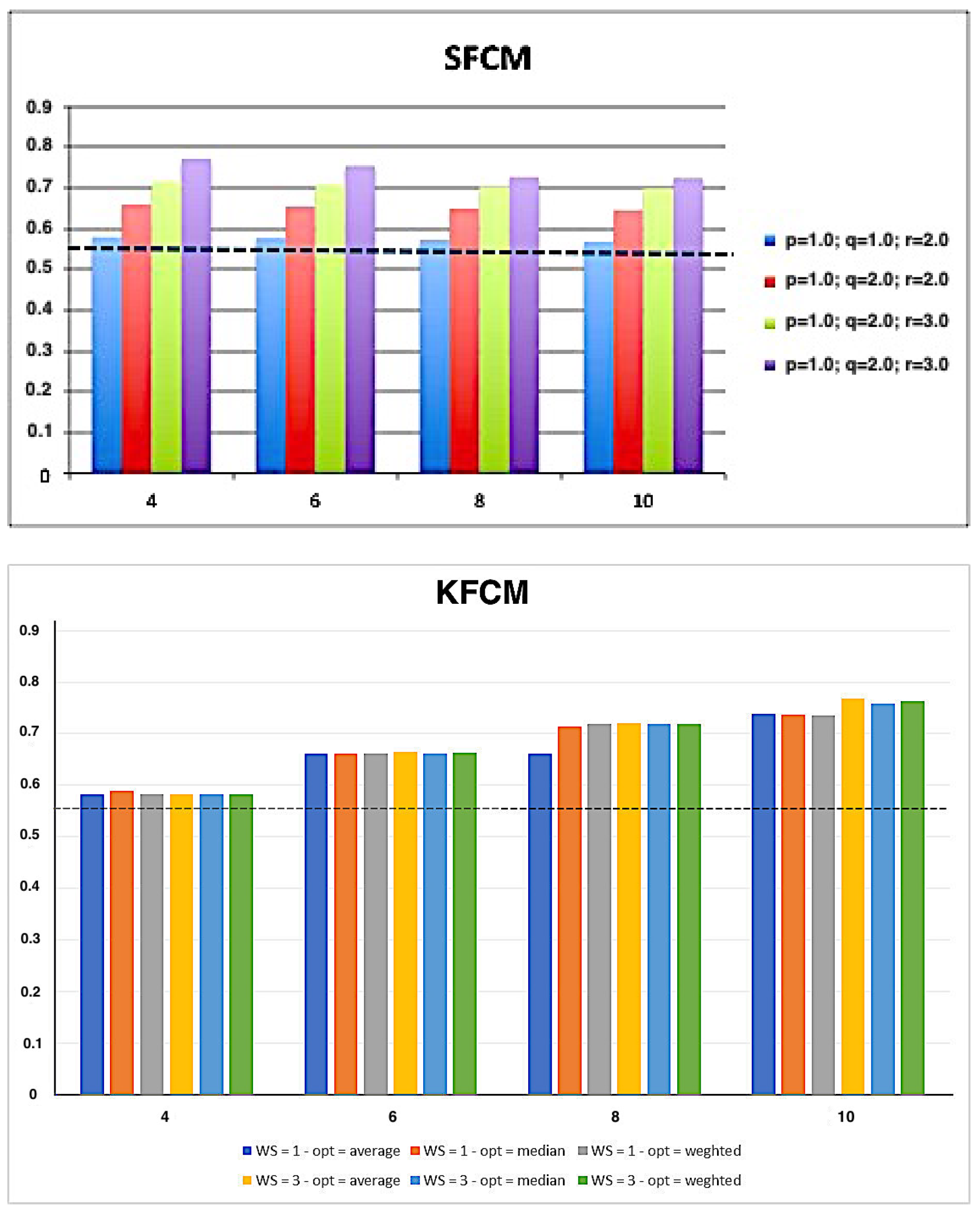


| K-means | FCM | SFCM | KFCM | |
|---|---|---|---|---|
| K | 4, 6, 8, 10 | 4, 6, 8, 10 | 4, 6, 8, 10 | 4, 6, 8, 10 |
| m | - | 1.0, 1.5, 2.0 | 1.0, 1.5, 2.0 | 1.0, 1.5, 2.0 |
| p | - | - | 1.0, 2.0 | - |
| q | - | - | 1.0, 2.0 | - |
| r | - | - | 2.0, 3.0, 4.0 | - |
| - | - | - | 1, 3, 5 | |
| - | - | - | average, median, weighted |
| K-means | FCM | SFCM | KFCM | |
|---|---|---|---|---|
| E-measure | 0.5374 ± 0.0763 | 0.5381 ± 0.0750 | 0.5363 ± 0.0742 | 0.5348 ± 0.0761 |
| Subject | Method | JS | DSC | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|---|---|
| K-means | % | |||||
| 202-3 | ||||||
| 0.8938 | 0.9486 | |||||
| K-means | ||||||
| 205-3 | ||||||
| Segmented Image | K-means | FCM | SFCM | KFCM |
|---|---|---|---|---|
| 202-3 original | 0.5842 | 0.5850 | 0.5832 | 0.5889 |
| 202-3 with noise | 0.5859 | 0.5873 | 0.5856 | 0.5845 |
| 205-3 original | 0.5793 | 0.5781 | 0.5758 | 0.5761 |
| 205-3 with noise | 0.5763 | 0.5787 | 0.5761 | 0.5782 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caponetti, L.; Castellano, G.; Corsini, V. MR Brain Image Segmentation: A Framework to Compare Different Clustering Techniques. Information 2017, 8, 138. https://doi.org/10.3390/info8040138
Caponetti L, Castellano G, Corsini V. MR Brain Image Segmentation: A Framework to Compare Different Clustering Techniques. Information. 2017; 8(4):138. https://doi.org/10.3390/info8040138
Chicago/Turabian StyleCaponetti, Laura, Giovanna Castellano, and Vito Corsini. 2017. "MR Brain Image Segmentation: A Framework to Compare Different Clustering Techniques" Information 8, no. 4: 138. https://doi.org/10.3390/info8040138
APA StyleCaponetti, L., Castellano, G., & Corsini, V. (2017). MR Brain Image Segmentation: A Framework to Compare Different Clustering Techniques. Information, 8(4), 138. https://doi.org/10.3390/info8040138





