Abstract
Social media platforms are emerging digital communication channels that provide an easy way for common people to share their health and medication experiences online. With more people discussing their health information online publicly, social media platforms present a rich source of information for exploring adverse drug reactions (ADRs). ADRs are major public health problems that result in deaths and hospitalizations of millions of people. Unfortunately, not all ADRs are identified before a drug is made available in the market. In this study, an ADR event monitoring system is developed which can recognize ADR mentions from a tweet and classify its assertion. We explored several entity recognition features, feature conjunctions, and feature selection and analyzed their characteristics and impacts on the recognition of ADRs, which have never been studied previously. The results demonstrate that the entity recognition performance for ADR can achieve an F-score of 0.562 on the PSB Social Media Mining shared task dataset, which outperforms the partial-matching-based method by 0.122. After feature selection, the F-score can be further improved by 0.026. This novel technique of text mining utilizing shared online social media data will open an array of opportunities for researchers to explore various health related issues.
1. Introduction
An adverse drug reaction (ADR) is an unexpected occurrence of a harmful response as a result of consumption or administration of a pharmaceutical drug at a known normal prophylactic, diagnostic, or therapeutic dose. Even though drugs are monitored in clinical trials for safety prior to approval and marketing, not all ADRs are reported due to the short duration and number of patients registered in clinical trials. Therefore, post marketing surveillance of ADRs is of utmost importance [1,2]. Reporting of ADRs is commonly done by medical practitioners. However, the relevance of reports given by individual drug users or patients has also been emerging [3]. For example, MedWatch (http://www.fda.gov/Safety/MedWatch/) allows both patients and drug providers to submit ADRs manually. Although there are diverse surveillance programs developed to mine ADRs, only a very small fraction of ADRs was reported. Immediate observation of adverse events help not only the drug regulators, but also the manufacturers for pharmacovigilance. Therefore, currently existing methods rely on patients’ spontaneous self-reports that attest problems. On the other hand, with more and more people using social media to discuss health information, there are millions of messages on Twitter that discuss drugs and their side-effects. These messages contain data on drug usage in much larger test sets than any clinical trial will ever have [4]. Although leading drug administrative agencies do not make use of online social media user reviews because of the highly time consuming and expensive process for manual ADR identification from unstructured and noisy data, the social media platforms presents a new information source for searching potential adverse events [5]. Researchers have begun diving into this resource to monitor or detect health conditions on a population level.
Text mining can be employed to automatically classify texts or posts that are assertive of ADRs. However, mining information from social media is not straightforward and often complex. Social media data in general is short and noisy. It is common to notice misspellings, abbreviations, symbols, and acronyms in Twitter posts. Tweets usually contain a special character. For example, in the tweet “Shouldn’t have taken 80 mg of vyvanse today … #cantsleep”, the word “cantsleep” is preceded with the “#” symbol. The sign is called a hashtag, which is used to mark keywords or topics in a tweet. The symbol was used by twitter users to categorize messages. In this example, the hash-tagged word (can’t sleep) is an ADR. In addition, the terms used for describing ADR events in social medial are usually informal and do not match clinical terms found in medical lexicons. Moreover, beneficial effects or other general mention types are usually ambiguous with ADR mentions.
In this study, an ADR event monitoring system that can classify Twitter posts regarding ADRs from Twitter is developed. The system includes an ADR mention recognizer that can recognize ADR mentions from a given Twitter post. In addition, because tweets mentioning ADRs may not always be ADR assertive posts, an ADR post classifier that can classify the given post for indication of ADR events is included in the system. The two systems were developed by using supervised learning approaches based on conditional random fields (CRFs) [6] and support vector machines (SVMs) [7], respectively. A variety of features have been proposed for supervised named entity recognition (NER) systems [8,9,10] in the newswire and biomedical domains. Supervised learning is extremely sensitive to the selection of an appropriate feature set. However, only limited studies focus on the impact of these features and their combinations on the effectiveness of mining ADRs from Twitter. In light of this, our study emphasizes the feature engineering for mining ADR events by analyzing the impact of various features taken from previous supervised NER systems. This study selected features widely used in various NER tasks to individually investigate their effectiveness for ADR mining, and conducted a feature selection algorithm to remove improper feature combinations to identify the optimal feature sets. Some previous works [11,12] demonstrated that the results of NER can be exploited to improve the performance of the classification task. Therefore, the output of the NER system is integrated with the features extracted for the ADR post classifier. The performance of both systems is finally reported on the manually annotated dataset released by the Pacific Symposium on Biocomputing (PSB) Social Media Mining (SMM) shared task [13].
2. Related Work
Identifying ADRs is an important task for drug manufacturers, government agencies, and public health. Although there are diverse surveillance programs developed to mine ADRs, only a very small fraction of ADRs was submitted. On the other hand, there are millions of messages on Twitter that discuss drugs and their side-effects. These messages contain data on drug usage in much larger test sets than any clinical trial will ever have [4]. Unfortunately, mining information related to ADRs from big social media reveals a great challenge. A series of papers has demonstrated how state-of-the-art natural language processing (NLP) systems perform significantly worse on social media text [14]. For example, Ritter et al. [15] presented that the Stanford NER system achieved an F-score of only 0.42 on the Twitter data, which is significantly lower than 0.86 on the CoNLL test set [16]. The challenges of mining information from Twitter can be summarized as follows [13,15,17,18]. (1) Length limits: Twitter’s 140 character limit leads to insufficient contextual information for text analysis without the aid of background knowledge. The limit may somewhat lead to the use of shortened forms that leads to the second challenge; (2) The non-standard use of language, which includes shortened forms such as “ur” which can represent both “your” and “you’re”, misspellings and abbreviations like lol (laugh out loud) and ikr (i know, right?), expressive lengthening (e.g., sleeeeep), and phrase construction irregularities; (3) The final challenge is the lack of the ability to computationally distinguish true personal experiences of ADRs from hearsay or media-stimulated reports [19].
NER is one of the most essential tasks in mining information from unstructured data. Supervised NER that uses CRFs has been demonstrated to be especially effective in a variety of domains [20,21,22]. Several types of features have been established and widely used in various applications. Some features capture only one linguistic characteristic of a token. For example, the context information surrounding a word and its morphologic or part-of-speech (PoS) information. Zhang and Johnson [23] indicated that these basic features alone can achieve competitive levels of accuracy in the general domain. Conjunction features, on the other hand, consist of multiple linguistic properties, such as the combination of words within a context window. They are usually more sophisticated linguistic features and can also be helpful after feature selection [24]. Syntactic information, such as the shallow parsing (chunk), is usually considered a very useful feature in recognizing named entities since in most cases either the left or right boundary of an entity is aligned with either edge of a noun phrase. NER is also a knowledge-extensive task. Therefore, domain-specific features such as the lexicon (or gazetteers) feature [25] turned out to be a critical resource to improve recognition performance. For instance, Kazama and Torisawa [22] used the IOB tags to represent their lexicon features and showed an improvement of F-score by 0.03 in the task of recognizing four common entity categories. In addition, semi-supervised approaches based on unlabeled data have attracted lots of attentions recently, especially after the great success of employing word representation features in NLP tasks [26]. The idea of the feature could contribute to the pioneering n-gram model proposed by Brown et al. [27], which provides an abstraction of words that could address the data sparsity problem in NLP tasks [28]. Turian et al. [26] showed that the use of unsupervised word representations as extra word features could improve the quality of NER and chunking. The results of NER can also be exploited to improve the performance of the article classification task [11,12].
Based on the aforementioned works, several studies had adapted NLP techniques for utilizing social media data to detect ADRs. Pioneering studies [18,29] and systems developed in the recent PSB SMM workshop [13] implemented some of the conventional features described in their ADR mining systems. Nikfarjam et al. [18] introduced ADRMine, a CRF-based NER system that can recognize ADR-related concepts mentioned in data from DailyStrength and Twitter. In addition to the surrounding word, PoS, and lexicon features, they implemented a negation feature which indicates whether or not the current word is negated. Furthermore, they utilized word2vec [30] to generate 150-dimensional word vectors from data about drugs. Afterwards, the K-means clustering algorithm was performed to group the vectors into 150 clusters. The generated clusters were then used in the implementation of their word representation features. Lin et al. [29] studied the effect of different context representation methods, including normalization and word vector representations based on word2vec and global vector [31]. They observed that using either of them could reduce feature spaces and improve the recall and overall F-measure. Yates et al. [32] employed CRF model with two tag sets to recognize ADRs. They implemented surrounding word, PoS, lexicon, and syntactic features. The Stanford parser was employed to provide the syntactic dependency information. The orthographic features commonly used in biomedical NER were ignored because they believe that ADR expressions do not frequently follow any orthographic patterns.
Automatic classification of ADR containing user posts is a crucial task, since most posts on social media are not associated with ADRs [2]. Sarker and Gonzalez [33] considered the task as a binary classification problem and implemented a variety of features, including the n-gram features in which n was set from one to three, lexicon features, polarity features, sentimental score features, and topic modeling features. The system was then trained by using a data combining three different corpora. They observed that based on their features the SVM algorithm had the best performance. However, when multi-corpus training was applied, the performance cannot be further improved if dissimilar datasets are combined. Sarker et al. [34] manually annotated Twitter data and performed analyses to determine whether posts on Twitter contain signals of prescription medication abuse. By using the annotated corpus, they implemented the same n-gram features, lexicon features, and word representation features. The results once again demonstrated that the SVM algorithm achieved the highest F-score for the binary classification task of medication abuse. Paul and Dredze [35] improved their ailment topic aspect model by incorporating prior knowledge about diseases, and found that the new model outperformed the previous one without prior knowledge in the applications of syndromic surveillance.
3. Materials and Methods
Figure 1 shows the final flowchart of the developed systems for the task of ADR post classification (ADR-C) and the task of recognizing ADR mention (ADR-R) in the form of a pipeline. Because tweets in general are noisy, a few preprocessing steps were developed to address this issue. After preprocessing, we extracted various features to train the machine learning models for the ADR mention recognizer and the ADR post classifier. With the generated models, the same preprocessing steps and machine learning algorithms were used to classify the given Twitter post and recognize described ADR mentions.
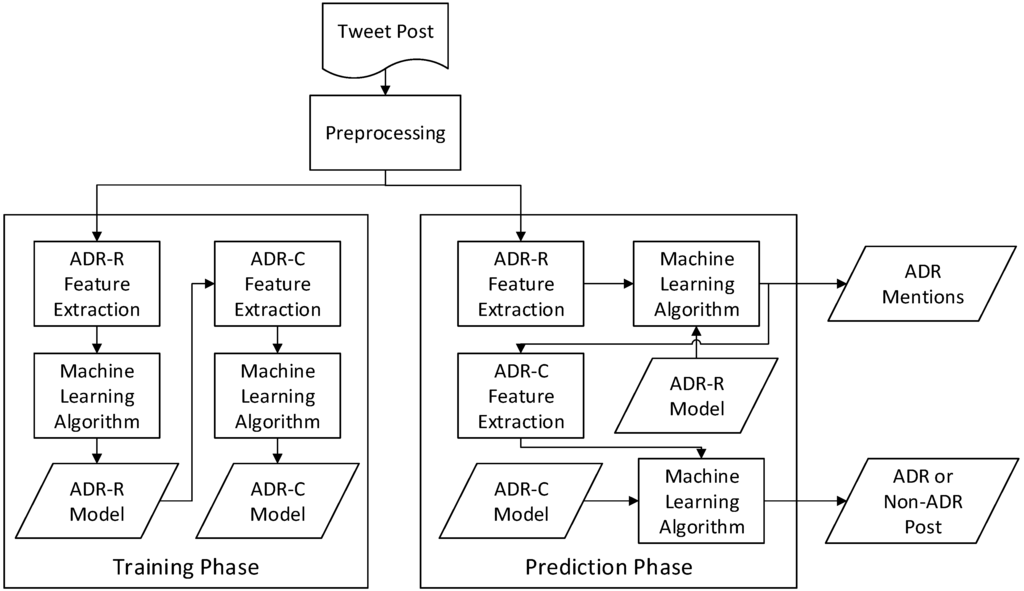
Figure 1.
High level flowchart of the developed ADR mining system.
3.1. Preprocessing
Twokenizer [36] is used to tokenize the Twitter post into tokens and generate the PoS information for each of them. Each token is then processed by Hunspell (version 1.2.5554.16953, CRAWler-Lib, Neu-Ulm, Germany, http://hunspell.github.io/) to correct spelling errors. The spell checker is configured to use the English dictionaries for Apache OpenOffice and two other dictionaries. One dictionary contains ADR terms released by Nikfarjam et al. [18], and the other contains drug terms collected from the training set.
For ADR-R, the numerical normalization approach is employed to modify the numeral parts in each token to one representative numeral. The advantages of numerical normalization, including the reduction of the number of features, as well as the possibility of transforming unseen features to seen features, have been portrayed in several NER tasks [24,29] and could further improve the accuracy of feature weight estimation. In addition, the hashtag symbol “#” is deleted from its attached keywords or topics. The token prefixed with the “@” symbol is replaced with @REF. As a result, after the normalization preprocess, the example tweet “Shouldn’t have taken 80 mg of vyvanse today … #cantsleep” is convert into the following tokens “Shouldn't have taken 1mg of vyvanse today … cantsleep”.
For ADR-C, all tokens are lowercased and characters including web links, usernames, punctuations, and Twitter specific characters are deleted by using regular expressions. The Snowball stemmer (version C, open source tool, http://snowball.tartarus.org/) is then used to perform stemming. Finally, a custom stop word list created based on the training set is used to remove noisy tokens in tweets. The list mainly comprised of social media slang terms such as “retweet”, “tweeter” and “tweetation”, and words related to emails, inbox, and messages. For example, the tweet “©C4Dispatches Eeeeek! Just chucked my Victoza in the bin. I will take my chances with the diabetes #diabetes” is transformed to “eek chuck victoza i chanc diabet diabet” after the preprocessing step.
3.2. Development of the ADR Mention Recognizer
3.2.1. Machine Learning Algorithm and Formulation
The CRFs model has been successfully applied in many different NER tasks and showed a great performance. This study formulates the ADR-R task as a sequential labeling task by using the IOBES scheme with the CRF++ toolkit (version 0.58, open source tool, https://taku910.github.io/crfpp/) to develop the ADR mention recognizer. Figure 2 shows two example tweets after formulating ADR-R as the labelling task.
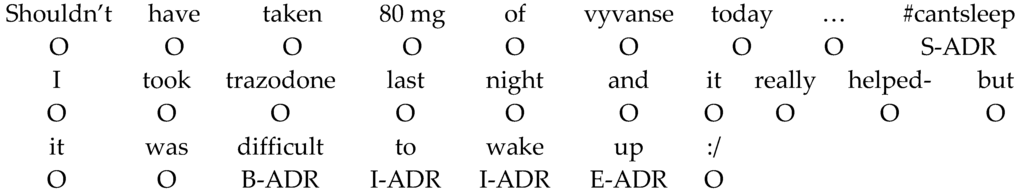
Figure 2.
The sequential labeling formulation with the IOBES scheme for the ADR-R task.
The IOBES scheme suggests the CRFs model to learn and recognize the Beginning, the Inside, the End, and the Outside of a particular category of ADR entities. The S tag is used to specifically represent a single-token entity. There are three ADR entity categories, resulting in a total of 13 tags ({ADR, Indication, Drug} × {B, I, E, S} + {O} = 13 tags.) for the ADR-R task.
3.2.2. Feature Extraction
The features extracted for ADR-R are elaborated as follows.
- Contextual features: For every token, its surrounding token is referred to as its context. For a target token, its context is described as the token itself (denoted as w0) with its preceding tokens (denoted as w−n, w−n + 1, …, w−1) and its following tokens (denoted as w1, w2, …, wn). In our implementation, the contextual features were extracted for the original tokens and the spelling checked tokens. All of the tokens were transformed into more compact representation with the process of normalization and stemming. As described later in the Results section, after the feature selection procedure, the context window was set to three, including w−1, w0, and w1.
- Morphology features: The feature set represents more information extracted from the current token. In our implementation, the prefixes and the suffixes of both the normalized and the spelling checked normalized tokens were extracted as features. The lengths of the prefix/suffix features were set to 3 to 4 within one-length context window.
- PoS features: The PoS information generated by Twokenizer for every token was encoded as features.
- Lexicon features: Three lexicon features were implemented to indicate a matching between the spelling corrected tokens with the entry in a lexicon. The first lexicon feature was implemented as a binary feature to indicate whether or not the current token partially matches with an entry in a given lexicon; the second feature further combines the matched token with the first feature to create a conjunction feature. Note that the conjunct spelling checked token may not be the same as the original token used for matching. The spelling checker may generate several suggestions for a misspelled token. In our implementation, the spelling checked contextual feature always uses the first suggestion generated by the checker, which may not match with the ADR lexicon. However, in the implementation of the lexicon feature, the matching procedure will match all suggestions against the ADR lexicon until a match is found, which may result in unmatched cases. The last lexicon feature encoded a match by using the IOB scheme that represents the matched position of the current token in the employed ADR lexicon. In some circumstances, especially when the post contains unique symbols such as hashtagged terms and nonstandard compound words, the spelling checker used in this study could decompose the tokens from the compound words. For example, “cant sleep” will be decomposed from the compound word “cantsleep”. Each of the token will be matched with all of the entries in a lexicon. The ADR lexicon created by Leaman et al. [37] was employed as the lexicon for matching ADR terms. The sources of the lexicon include the UMLS Metathesaurus [38], the SIDER side effect resource [39], and other databases. The tokens annotated with the “Drug” tag were collected to form the lexicon for drugs. Take the Twitter post “Seroquel left me with sleep paralysis” as an example. The compound noun “sleep paralysis” matched with the ADR lexicon and their corresponding feature values are listed as follows.
- ○
- Binary: 1, 1.
- ○
- Conjunction: sleep/1, paralysis/1.
- ○
- IOB: B-ADR, I-ADR.
- Word representation feature: The large unlabeled data from the Twitter website was utilized to generate word clusters for all of the unique tokens with the vector representation method [30]. The feature value for a token is then assigned based on its associated cluster number. If the current token does not have a corresponding cluster, its normalized and stemmed result will be used. The feature adds a high level abstraction by assigning the same cluster number to similar tokens. In order to create the unlabeled data, we searched the Twitter website for a predefined query to collect 7 days of tweets including 97,249 posts. The query was compiled by collecting each of the entries listed in the lexicon used for generating the lexicon feature, the described ADRs, their related drugs collected from the training set of the SMM shared task, as well as the hashtags annotated as ADRs in the training set. The final query contains 14,608 unique query terms. After the query was defined, the Twitter REST API was used to search for Twitter posts related to the collected ADR-drug pairs and hashtagged terms. Afterwards, Twokenizer was used on the collected dataset to generate tokens. The word2vec toolkit (open source tool, https://code.google.com/archive/p/word2vec/) was then used to learn a vector representation for all tokens based on their contexts in different tweets. The neural network behind the toolkit was set to use the continuous bag of words scheme, which can predict the word given its context. In our implementation, the size of context window was set to 5 with 200 dimension, and a total of 200 clusters were generated.
3.3. Development of the ADR Post Classifier
3.3.1. Machine Learning Algorithm
SVM with the linear kernel is used to develop the ADR post classifier. Due to the large class imbalance in the training set, instead of assigning class weight of 1 for both classes, we adjusted class weights inversely based on the class distribution. The cost parameter of the model is set to 0.5, which was optimized on the training set for better performance during the development.
3.3.2. Feature Extraction
Various feature sets are extracted, which include the linguistic, polarity, lexicon, and topic modelling based features.
- Linguistic features: We extracted common linguistic information like bag of words, bigrams, trigrams, PoS tags, token-PoS pairs, and noun phrases as features.
- Polarity features: The polarity cues developed by Niu et al. [40] were implemented to extract four binary features that can be categorized as “more-good”, “less-good”, “more-bad”, and “less-bad”. The categories are inferred based on the presence of polarity keywords in a tweet, which were then encoded as binary features for a tweet. For example, considering the tweet “could you please address evidence abuutcymbalta being less effective than TCAs”, the value of the feature “less good” would be 1 and the rest would take the value 0 because the token “less” and “effective” matched with the “less-good” polarity cue.
- Lexicon based features: The features were generated by using the recognition results of a string matching algorithm combined with the developed ADR mention recognizer. Tweets were processed to find exact matches of lexical entries from the existing ADR and drug name lexicons [18]. The presence of lexical entries were engineered as two binary features with the value of either 0 or 1. For example, in the Twitter post “Antipsychotic drugs such as Zyprexa, Risperdal & Seroquel place the elderly at increased risk of strokes & death”, both the ADR and the drug name lexical features take the value of 1.
- Topic modeling features: In our system, the topic distribution weights per tweet were extracted as features. The Stanford Topic Modelling Toolbox (version 0.4, The Stanford NLP Group, Stanford, CA, USA, http://nlp.stanford.edu/software/tmt/tmt-0.4/) was used to extract these features. The number of features depends on the number of topics to be obtained from the dataset. For example, if the topic model is configured to extract five topics, then the weights corresponding to the five topics are represented as the topic modeling features.
3.4. Dataset
The training set and development set released by the PSB SMM shared task [13] were used to assess the performance of the developed system. For the ADR-C task, a total of 7574 annotated tweets were made available, which contains binary annotations, ADR and non-ADR, to indicate the relevance of ADR assertive user posts. For the task of ADR-R, 1784 Twitter posts were fully annotated for the following three types of ADR mentions.
- Drug: A medicine or other substance which has a physical effect when ingested or otherwise introduced into the body. For example, “citalopram”, “lexapro”, and “nasal spray”.
- Indication: A specific circumstance that indicates the advisability of a special medical treatment or method to describe the reason to use the drug. For example, “anti-depressant”, “arthritis”, and “autoimmune disease”.
- ADR: A harmful or unpleasant reaction to the use of a drug. For instance, Warfarin (Coumadin, Jantoven) is used to prevent blood clots and is usually well tolerated, but a serious internal hemorrhage may occur. Therefore, the occurrence of serious internal bleeding is an ADR for Warfarin.
Nevertheless, during the preparation of this manuscript, some Twitter users removed their posts or even deactivated their accounts. As a result, some of the tweets from the original corpus are inaccessible. Only 1245 and 5283 tweets can be downloaded from the Twitter website for ADR-R and ADR-C, respectively. Therefore, the experiment results presented in the following section were based on a subset of the original dataset.
3.5. Evaluation Scheme
We devised an ADR mention recognizer which recognizes the text span of reported ADRs from a given Twitter post, and an ADR post classifier which categorizes the given posts as an indication of ADRs or not. Both systems were evaluated by using the following two paired criteria, precision (P) and recall (R), and the combined criterion, F-measure (F).
In the equations, the notations of TP, FP and FN stand for true positives, false positives and false negatives, respectively. In the evaluation of the ADR-R task, the approximate-match criterion [41] is used to determine the TP/FP/FN cases. Therefore, a TP is counted if the recognized text span is a substring of the manually annotated span or vice versa, and its associated entity type is matched with the one given by domain experts. The modified version of the official evaluation tool evalIOB2.pl of the BioNLP/NLPBA 2004 Bio-Entity Recognition Task [42] was used to calculate the PRF scores. ADR-C can be considered as a binary classification task. Hence, an instance is considered as a TP when the predicted class is matched with the class manually determined by domain experts.
4. Results
4.1. Feature Engineering for the ADR Mention Recognizer
Here we report the performance of the developed ADR mention recognizer by different feature combinations. We started by handling the local contextual features, then studied the evaluation of external knowledge features. Tenfold cross validation (CV) was performed on the ADR-R training set to assess the performance during the development phase. Finally, all of the studied features were processed by a feature selection algorithm to sieve the most appropriate feature subsets. The performance of the model based on the selected features was evaluated on the training set and the development set of the SMM shared task.
4.1.1. Local Contextual Features
Table 1 reports the ADR-R performance when only the local information about a current token is used. As shown in configurations 1–7, it is not surprising that the ADR-R performance is poor with only contextual features. The best F-score obtained is the fourth configuration which only considers the normalized and stemmed tokens within three context-window size. Configurations 5 to 7 demonstrate that with larger context, the P of ADR-R can be improved but at the cost of decline in the R.

Table 1.
Local contextual feature comparison on the training set. The best PRF-scores for each configuration set are highlighted in bold.
Configurations 8, 9, and 10 ignored surrounding context information but took the prefixes, suffixes, and shape features of the current token into consideration. The prefix and suffix features provided the recognizer good evidence of a particular token being a part of an ADR mention. However, the shape features did not increase the F-score of ADR-R.
4.1.2. External Knowledge Features
The external knowledge features studied include the spelling checking and PoS information for a token, the chunking information generated by a shallow parser, the lexicon information for ADR mentions, and the word representation information.
Table 2 compares the performance of the spelling checked contextual features with that of the unchecked contextual features. The results obtained in the configurations with spelling checked features such as 2, 4, 8, and 12 demonstrate the need for spelling check on Twitter posts. Precision improved when we replaced the original token with the spelling checked token, and recall can be further improved if the token is stemmed. Similar to the finding of Table 1, the performance drops with larger context, and the best size for the context window observed is three (configuration 8). Finally, by employing spelling check with normalized and stemmed prefixes and suffixes, the best F-score of 0.586 (configuration 12) was achieved.

Table 2.
Impact of the spelling checking for the local contextual feature. The best PRF-scores for each configuration set are highlighted in bold.
Table 3 compares the ADR-R performance when we combined the local contextual features with the PoS information generated by two different PoS taggers—Twokenizer [36] and GENIA tagger [43]. The results shows that with the PoS information the precision of ADR-R can be boosted from 0.377 to 0.781 and 0.784, but the impact of these features on the F-score depends on the underlying PoS tagger.

Table 3.
Comparison of the ADR-R performance based on different PoS information. The best PRF-scores for each configuration set are highlighted in bold.
Table 4 displays the effect after including the parsing results created by the GENIA tagger in which a tweet was divided into a series of chunks that include nouns, verbs, and prepositional phrases. As shown in Table 4, although the P is improved after including the chunk information, the overall F-score was not improved with a larger context window.

Table 4.
Effect of the chunk information on ADR-R. The best PRF-scores for each configuration set are highlighted in bold.
The impacts of the three implemented lexicon features were studied and illustrated in Table 5. In configuration 2, the IOB tag set was used. Configuration 3 represented the matching as a binary feature for the current token. The binary feature was further in conjunction with the matched spelling checked tokens in configuration 4. As indicated in Table 5, adding the three lexicon features improved the overall F-scores when a limited context window was employed. With the conjunct lexicon feature, the model performed better than that with just the binary feature. Considering the larger context window, the lexicon feature implemented by using the BIO tag set is the best choice.

Table 5.
Comparison of the different representations for the lexicon features in the ADR-R task. The best PRF-scores for each configuration set are highlighted in bold.
4.1.3. Word Representation Features
Table 6 exhibits the effect of the word representation features for ADR-R. From the results we can see that with the larger context window, inclusion of the word features can improve the recall and results in the increase of F-score.

Table 6.
Comparison of the different word representation features in the ADR-R task. The best PRF-scores for each configuration set are highlighted in bold.
4.1.4. Backward/Forward Sequential Feature Selection Results
We integrated the features of all of the best configurations shown in the previous tables, and conducted a backward/forward sequential feature selection (BSFS/FSFS) algorithm [44] using tenfold CV of the training set to select the most effective feature sets. The procedure began with a feature space of 3,716,741 features, in which features were iteratively removed to examine whether the average F-score has improved. The algorithm then selected the subset of features that yields the best performance. The BSFS procedure terminated when no improvement of F-score can be obtained from the current subsets or there are no features available in the feature pool. The FSFS procedure then proceeds by adding the second-tier feature sets that could also improve the F-score but were not involved in the BSFS process. In each iteration, the FSFS procedure adds a feature set and selects the one with the best F-score for inclusion in the feature subset. The cycle repeats until no improvement is obtained from extending the current subset. Figure 3 displays the number of selected features and their corresponding F-scores.
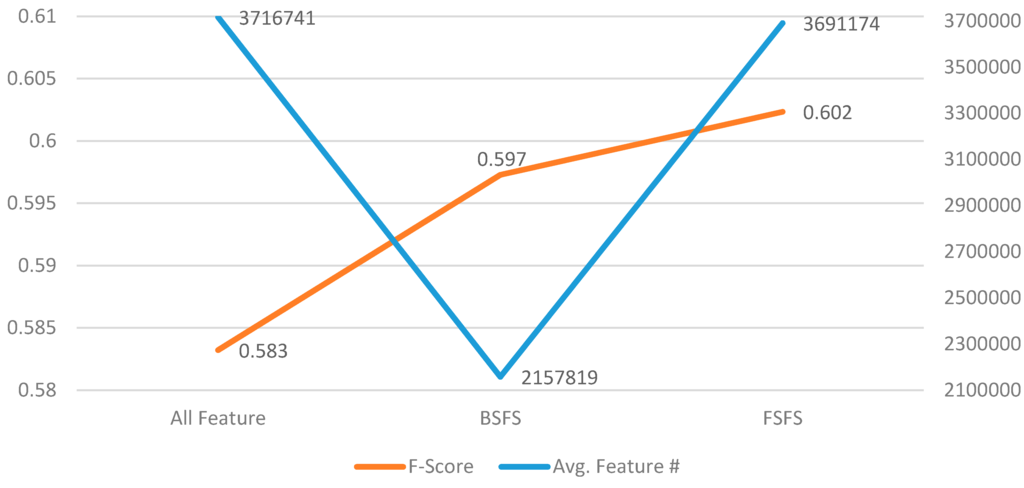
Figure 3.
Comparison of the change in the number of features (the right y-axis) and F-scores (the left y-axis) after applying the feature selection procedure.
After the feature selection process, the F-score improved by 3.26%. The final PRF-scores of the developed ADR mention recognizer on the training set are 0.752, 0.502, and 0.602, respectively. Throughout this study, the organizers of the PSB SMM shared task have not released the gold annotations for their test set. Thus, the development set was used to compare the developed recognizer with a baseline system. The baseline system utilized a partial matching method based on the same lexicon used for extracting lexicon features, and all lexicon entries in the system were normalized for matching with the normalized Twitter posts.
As shown in Table 7, the recognizer with selected features can achieve an F-score of 0.588, which outperforms the same CRF-based recognizer with all features and the baseline system by 0.026 and 0.122, respectively.

Table 7.
Performance comparison on the development set of the PSB SMM shared task.
4.2. Performance of the ADR Post Classifier
Table 8 reports the performance of the developed ADR post classifier on the development set. The first configuration uses a set of baseline features including the polarity, ADR-R, and linguistic features. The second configuration further includes the topic modeling feature that was set to extract three topics per tweet. The results suggest that the performance of the developed ADR post classifier can be improved with the topic modeling features.

Table 8.
Performance of the developed ADR post classifier on the development set.
4.3. Availability
All of the employed tools, datasets, and the compiled resources used in this study, including the stop word list and the word clusters generated from 7-days tweets, are available at https://sites.google.com/site/hjdairesearch/Projects/adverse-drug-reaction-mining.
5. Discussion
5.1. ADR Mention Recognition
We have demonstrated the results and performances of the feature selection based on the BSFS/FSFS algorithm. This approach is usually referred to as the wrapper method because the learning algorithm is wrapped into the selection process [45]. Wrappers are often criticized due to the requirement of intensive computation. On the other hand, the filter method is another feature selection method that makes an independent assessment based only on the characteristics of the data without considering the underlying learning algorithm. Here we implemented a filter-based feature selection algorithm, the simple information gain (IG) algorithm proposed by Klinger and Friedrich [46], to compare its results with that of the BSFS/FSFS algorithm. Figure 4 shows the F-score curves of the developed model on the development set when using different percentages of all features.
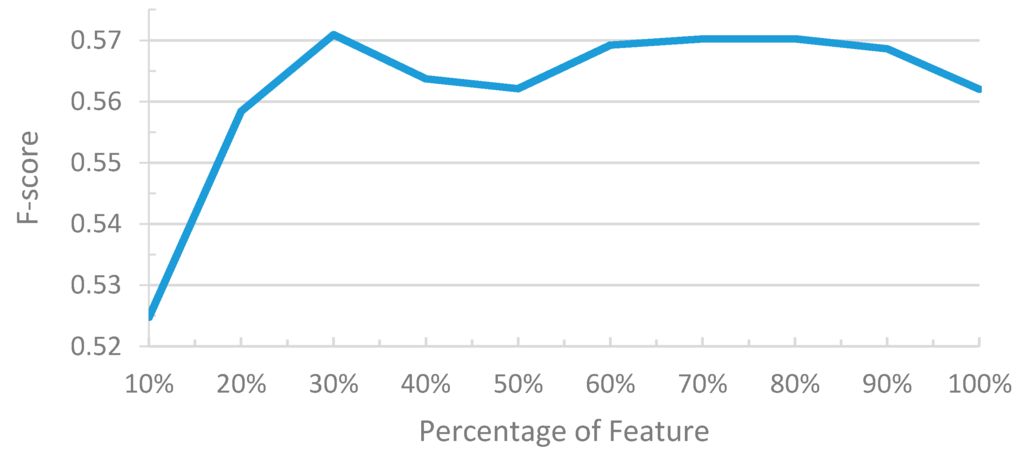
Figure 4.
F-score curves of the filter-based feature selection with different percentages of all features.
It can be observed that when 30% of the features were used, the model achieved the best F-scores of 0.579, which improved the original model with all features by 0.017. When only 10% or 20% of the features were used, the F-scores dropped by 0.06 and 0.03, respectively. The F-scores also decreased when we increased the percentage of the employed features from 30% to 50%, but the scores were still better than that of the model with all features. The F-score curve lifted again after including around 60% to 80% of the features. This phenomena is similar to the results shown in Figure 3, in which including the additional feature sets selected by FSFS can improve the performance of the feature subset selected by BSFS. The results demonstrate that both the FSFS/BSFS algorithm and the IG selection algorithm could be employed for the task of ADR-R feature selection and in general they have compatible performance.
Table 9 lists the features used for ADR-R after applying the BSFS/FSFS feature selection. The developed ADR mention recognizer with the selected features achieved an F-score of 0.631 for ADR mentions, which is significantly lower than the performance of the Stanford NER system in general domain. The Stanford NER system can achieve an F-score of 0.86 on the CoNLL test set [16]. As demonstrated by Ritter et al. [15], in which they reported the same system only achieved an F-score of 0.42 on the Twitter data, the results reveal the great challenge in mining information from big social media.

Table 9.
Features selected for ADR-R.
One of the main reasons leading to the decrease of performance is that social media language is not descriptively accurate [17], which usually contains several non-standard spellings like “fx” for “affect”, and word lengthening such as “killlerrr” for representing their subjectivity or sentiment [47]. We observed that certain ADR mentions are usually lengthening. For example, insomnia (UMLS CUI: C0917801) could be described in a tweet as “can’t sleeeep” or “want to sleeeeep”. The prefix and suffix features can capture the phenomenon and its implications. However, as shown in Table 2, the orthographic feature is less reliable for ADR-R. This is due to wide variety of letter case styles in Twitter posts. In the training set of ADR-R, 5.8% of tweets contain all lower case words, while 0.8% of the posts are all capitalized. Thus, the shape feature is not informative. Finally, the spelling variation leads to out-of-vocabulary (OOV) words, which requires the inclusion of the spell check token feature in the supervised machine learning model. Nonetheless, the suggestions generated by a spelling checker may not be perfect, so the original word is still an important feature for ADR-R. This is also supported by the distribution of the feature sets selected by the IG algorithm shown in Figure 5. We can observe that the top three important feature sets are the original word features, the spelling checked word features and the prefix/suffix features. The shape features only occupy 1% of the features.
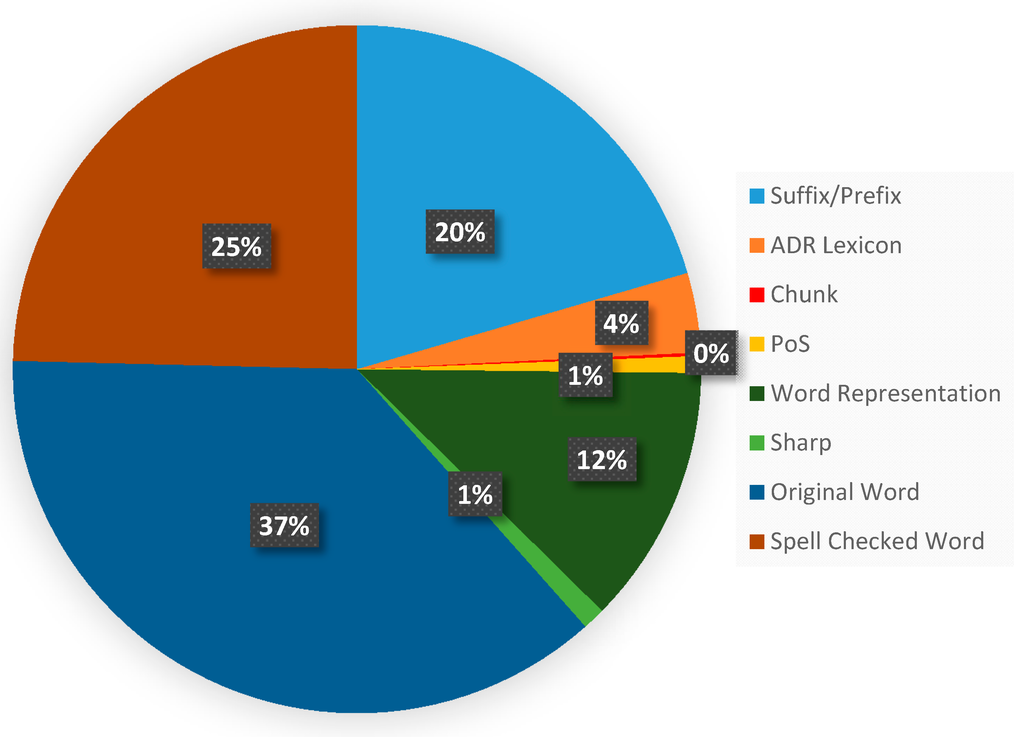
Figure 5.
Feature distribution among the top 30% of features selected by the IG algorithm. Note that some feature sets were merged to simply the pie chart. For instance, the PoS information generated by either Twokenizer or GENIATagger were merged into the PoS feature set, and the original word feature set include the non-normalized, normallized, and stemmed word features.
Generally speaking, named entities such as person names or organization names are usually located in noun phrases. In most cases, named entities rarely exceed phrase boundaries, in which either the left or right boundary of an entity is aligned with either edge of a noun phrase [24]. However, the nomenclature for the entities in the ADR-R task are different from entities in general domains. Some ADR mentions are descriptive, like the ADR mention “feel like I cant even stand”. Furthermore, off the shelf shallow parsers, such as the GENIATagger used in this study, have been observed to perform noticeably worse on tweets. Hence, addition of the chunk feature cannot improve the performance of ADR-R, which can also be interpreted from Figure 5, in which the chunk features occupy less than 1% of the features. Moreover, our results showed that larger context did not benefit the ADR-R either. In fact, during the BSFS procedure, features with larger contextual window except the chunk feature were the first few features to be removed from the feature space. Such behavior indicates that in the ADR-R task, the statistics of the dependency between the local context and the label of the token did not provide sufficient information to infer the current token’s label, which is possibly due to the 140 character limit of Twitter post.
Previous work has shown that unlabeled text can be used to induce unsupervised word clusters which can improve the performance of many supervised NLP tasks [18,26,31]. Our results imply the similar conclusion. We observed that when the word representation feature is added, the recall of both the training and development sets improved, leading to an increase of F-score by 0.01. After manual analysis, the improvement can be attributed to the fact that the word representation feature enables the supervised learning algorithm to utilize the similarity between known ADR-related words and unknown words determined from the unlabeled data. An example of this is found in the 19th created word cluster, in which 49% of the tagged tokens are ADR-related. Another example can be observed in the development set. The token “eye” occurs only once in the training set. Both “eye” and the token “worse”, which can compose the ADR mention “eyes worse”, are not annotated as ADR-related terms in the training set. Fortunately, they are clustered into two clusters which contain ADR-related tokens in our word clusters. The token “eye” is within the cluster containing “dry” and “nose”, while “worse” is in the cluster that consists of tokens like “teeth” and “reactions”, which are known to be ADR-related terms in the training set. Therefore, the supervised learning algorithm is able to recognize the unseen mention as an ADR with this information.
The word clusters created by this study was based on a relatively small corpus compared with some publicly available word representation models trained on Twitter data. For example, the clusters generated by Nikfarjam et al. [18] were learned from one million tweets. Pennington et al. [31] released a pre-training model learned from two billion tweets by the global vector algorithm. It raises an interesting question about how well the performance of the developed model will be if the cluster information used by our word representation feature is replaced with the information from the two larger pre-trained clusters and vectors. We conducted an additional experiment to study the effect of this replacement, and the results are displayed in Table 10. In the configuration 2, the 150 clusters generated by Nikfarjam et al. was directly used. For the vectors created by Pennington et al., we applied the K-mean algorithm to create 150, 200, and 400 clusters and listed the results for each cluster in configuration 3, 4, and 5, respectively.

Table 10.
ADR-R performance on the test set with different word clusters. The best PRF-scores are highlighted in bold.
Similar to the observation in our previous work [48], it might be surprising to see that the replacement of larger clusters did not significantly improve the F-scores as shown in Table 10. The model with our clusters can achieve compatible performance in comparison to configuration 2. After examining the generated clusters and the manually annotated ADRs in the test set, we believe that it is owing to that the domain of our clusters is more relevant to ADR events because it was compiled using ADR-related keywords. The relevance of the corpus to the domain is more important than the size of the corpus [49]. It is noteworthy that clusters created by Nikfarjam et al. occasionally overlooked common ADR-related words such as “slept” and “forgetting”. On the other hand, after checking Pennington et al.’s clusters used in configuration 3 and 4, we found that most of the ADR-related words such as “depression” are falling into the cluster consisting of words like “the”, “for”, and “do”, implying that the number of pre-determined clusters may be insufficient to separate them from stop words. Therefore, we increased the number of clusters to 400 in configuration 5, and the results indicate an improvement in both R and F-scores. Several other studies have attempted to determine the optimal numbers of clusters or word embedding algorithms for implementing the word representation features, and it is beyond the scope of this study. Instead, we state that the number of clusters generated from vectors based on huge dataset is important, and we would like to further investigate this in our future work.
5.2. ADR Post Classification
As demonstrated in Table 6, although we included the output of ADR-R as a feature, which has shown to be an advantage over using the lexicon matching-based feature in a preliminary experiment, the performance of ADR-C is not satisfactory. We observed that the large number of error cases in the training and development set are due to the large class imbalance. SVM based classifier tends to be biased towards the majority class in an imbalanced dataset. Although the concern of class imbalance was addressed by assigning weights to the class based on the class distribution to a certain extent, applying more sophisticated class imbalance techniques, such as ensemble based classifiers, would further improve the ADR-C performance [50]. In addition, several issues remained despite various approaches during the preprocessing step have been exploited to reduce the noise in the data. For instance, several ill-formed special characters still remained after applying spelling check, which resulted in sparse feature space. Another major issue we noticed is the disambiguation of abbreviations. Many of the tweets included abbreviations or acronyms for ADRs and drug names that are ambiguous with general terms. Considering that there is an entirely different vocabulary of abbreviations and slang words adopted by Twitter users, we believe that a custom-built lexicon of abbreviations and acronyms for ADRs and drug names should mitigate the effect of these terms.
The performance of our ADR post classifier has increased with the addition of topic modeling based features. The improvement due to the addition of topic distribution weights per instance is consistent with the findings from previous studies in automatic text classification [51,52]. However, as shown in Figure 6, the performance varies, depending on the different number of extracted topics in the topic modeling features. The results indicate that when the topics are increased to five from three, the classification performance decreased in both the tenfold CV of the training set and the development set. This may be due to the fact that tweets are short, and extracting large number of topic related information creates sparse and noisy data. Moreover, the topic modeling features used only included per-tweet topic distribution weights. Topic modeling generates large amounts of useful information on a given dataset such as the number of terms in each topic and the weight of each term in a topic. Incorporating information of such might improve the effectiveness of the classifier.
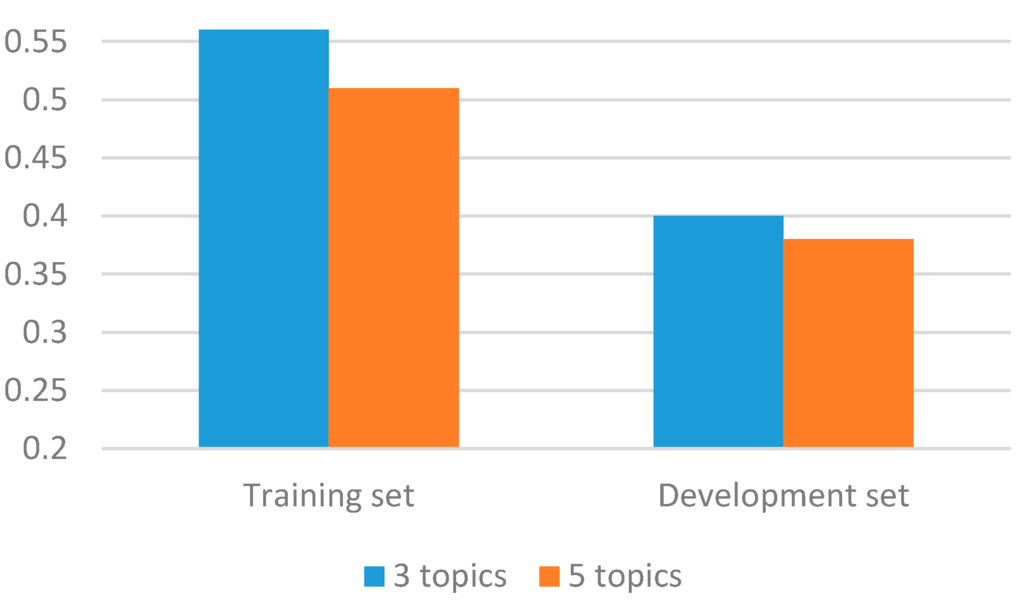
Figure 6.
Comparison of F-scores with different number of topics.
6. Conclusions
In conclusion, this study presented methods to mine ADRs from Twitter posts using an integrated text mining system that utilizes supervised machine learning algorithms to recognize ADR mentions and to classify whether a tweet reports an event of an ADR. We implemented several features proposed for NER including local contextual features, external knowledge features, and the word representation features, and discussed their impact on ADR-R. After applying a feature selection algorithm, the best features included the current token, its surrounding tokens within the three context window, the prefix and suffix, the PoS of the current token, the lexicon feature, and the word representation features. In ADR-C, we proposed a method to automatically classify ADRs using SVM with the topic modeling, polarity, ADR-R, and linguistic features. The results demonstrated that the performance of the classifier could be improved by adding the topic modeling features, but would decline when the number of topics are increased. In the future, we aim to continually improve the performance of our methods by exploiting new features and ensemble based classifiers. In addition, the proposed methods for identifying ADRs will be evaluated in other social media platforms, as well as electronic health records.
Acknowledgments
This work was supported by the Ministry of Science and Technology of Taiwan (MOST-104-2221-E-143-005).
Author Contributions
Hong-Jie Dai conceived and designed the experiments; Musa Touray and Hong-Jie Dai performed the experiments; Jitendra Jonnagaddala and Hong-Jie Dai analyzed the data; Jitendra Jonnagaddala and Hong-Jie Dai developed the systems; Hong-Jie Dai, Jitendra Jonnagaddala, Musa Touray and Shabbir Syed-Abdul wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| ADR | Adverse Drug Reaction |
| BSFS | Backward Sequential Feature Selection |
| CRF | Conditional Random Field |
| F | F-measure |
| FN | False Negative |
| FP | False Positive |
| FSFS | Forward Sequential Feature Selection |
| NER | Name Entity Recognition |
| NLP | Natural Language Processing |
| OOV | Out-Of-Vocabulary |
| P | Precision |
| PSB | Pacific Symposium on Biocomputing |
| PoS | Part of Speech |
| R | Recall |
| SMM | Social Media Mining |
| SVM | Support Vector Machine |
| TP | True Positive |
| UMLS | Unified Medical Language System |
References
- Lardon, J.; Abdellaoui, R.; Bellet, F.; Asfari, H.; Souvignet, J.; Texier, N.; Jaulent, M.C.; Beyens, M.N.; Burgun, A.; Bousquet, C. Adverse Drug Reaction Identification and Extraction in Social Media: A Scoping Review. J. Med. Internet Res. 2015, 17, e171. [Google Scholar] [CrossRef] [PubMed]
- Sarker, A.; Ginn, R.; Nikfarjam, A.; O’Connor, K.; Smith, K.; Jayaraman, S.; Upadhaya, T.; Gonzalez, G. Utilizing social media data for pharmacovigilance: A review. J. Biomed. Inform. 2015, 54, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Blenkinsopp, A.; Wilkie, P.; Wang, M.; Routledge, P.A. Patient reporting of suspected adverse drug reactions: a review of published literature and international experience. Br. J. Clin. Pharmacol. 2007, 63, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Cieliebak, M.; Egger, D.; Uzdilli, F. Twitter can Help to Find Adverse Drug Reactions. Available online: http://ercim-news.ercim.eu/en104/special/twitter-can-help-to-find-adverse-drug-reactions (accessed on 20 May 2016).
- Benton, A.; Ungar, L.; Hill, S.; Hennessy, S.; Mao, J.; Chung, A.; Leonard, C.E.; Holmes, J.H. Identifying potential adverse effects using the web: A new approach to medical hypothesis generation. J. Biomed. Inform. 2011, 44, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Lafferty, J.; McCallum, A.; Pereira, F. Conditional random fields: Probabilistic models for segmenting and labeling sequence data. In Proceedings of the 18th International Conference on Machine Learning (ICML), Williamstown, MA, USA, 28 June 2001.
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Liu, S.; Tang, B.; Chen, Q.; Wang, X.; Fan, X. Feature engineering for drug name recognition in biomedical texts: Feature conjunction and feature selection. Comput. Math. Methods Med. 2015, 2015, 913489. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.J.; Lai, P.T.; Chang, Y.C.; Tsai, R.T. Enhancing of chemical compound and drug name recognition using representative tag scheme and fine-grained tokenization. J. Cheminform. 2015, 7, S14. [Google Scholar] [CrossRef] [PubMed]
- Tkachenko, M.; Simanovsky, A. Named entity recognition: Exploring features. In Proceedings of The 11th Conference on Natural Language Processing (KONVENS 2012), Vienna, Austria, 19–21 September 2012; pp. 118–127.
- Gui, Y.; Gao, Z.; Li, R.; Yang, X. Hierarchical Text Classification for News Articles Based-on Named Entities. In Advanced Data Mining and Applications, Proceedings of the 8th International Conference, ADMA 2012, Nanjing, China, 15–18 December 2012; Zhou, S., Zhang, S., Karypis, G., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 318–329. [Google Scholar]
- Tsai, R.T.-H.; Hung, H.-C.; Dai, H.-J.; Lin, Y.-W. Protein-protein interaction abstract identification with contextual bag of words. In Proceedings of the 2nd International Symposium on Languages in Biology and Medicine (LBM 2007), Singapore, 6–7 December 2007.
- Sarker, A.; Nikfarjam, A.; Gonzalez, G. Social media mining shared task workshop. In Proceedings of the Pacific Symposium on Biocomputing 2016, Big Island, HI, USA, 4–8 January 2016.
- Gimpel, K.; Schneider, N.; O’Connor, B.; Das, D.; Mills, D.; Eisenstein, J.; Heilman, M.; Yogatama, D.; Flanigan, J.; Smith, N.A. Part-of-speech tagging for Twitter: Annotation, features, and experiments. In Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics: Human Language Technologies, Portland, OR, USA, 19–24 June 2011.
- Ritter, A.; Clark, S.; Etzioni, O. Named entity recognition in tweets: an experimental study. In Proceedings of the Conference on Empirical Methods in Natural Language Processing, Edinburgh, UK, 27–31 July 2011.
- Finkel, J.R.; Grenager, T.; Manning, C. Incorporating non-local information into information extraction systems by Gibbs sampling. In Proceedings of the 43rd Annual Meeting on Association for Computational Linguistics, Ann Arbor, MI, USA, 25–30 June 2005.
- Eisenstein, J. What to do about bad language on the internet. In Proceedings of the North American Chapter of the Association for Computational Linguistics (NAACL), Atlanta, GA, USA, 9–15 June 2013.
- Nikfarjam, A.; Sarker, A.; O’Connor, K.; Ginn, R.; Gonzalez, G. Pharmacovigilance from social media: Mining adverse drug reaction mentions using sequence labeling with word embedding cluster features. J. Am. Med. Inform. Assoc. 2015, 22, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Harpaz, R.; DuMochel, W.; Shah, N.H. Big Data and Adverse Drug Reaction Detection. Clin. Pharmacol. Ther. 2016, 99, 268–270. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.-J.; Syed-Abdul, S.; Chen, C.-W.; Wu, C.-C. Recognition and Evaluation of Clinical Section Headings in Clinical Documents Using Token-Based Formulation with Conditional Random Fields. BioMed Res. Int. 2015. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Yang, Z.; Lin, H.; Li, Y. Drug name recognition in biomedical texts: A machine-learning-based method. Drug Discov. Today 2014, 19, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Kazama, J.I.; Torisawa, K. Exploiting Wikipedia as external knowledge for named entity recognition. In Proceedings of the 2007 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning, Prague, Czech Republic, 28–30 June 2007; pp. 698–707.
- Zhang, T.; Johnson, D. A robust risk minimization based named entity recognition system. In Proceedings of the Seventh Conference on Natural language Learning at HLT-NAACL 2003, Edmonton, AB, Canada, 31 May–1 June 2003.
- Tsai, R.T.-H.; Sung, C.-L.; Dai, H.-J.; Hung, H.-C.; Sung, T.-Y.; Hsu, W.-L. NERBio: Using selected word conjunctions, term normalization, and global patterns to improve biomedical named entity recognition. BMC Bioinform. 2006, 7, S11. [Google Scholar] [CrossRef] [PubMed]
- Cohen, W.W.; Sarawagi, S. Exploiting dictionaries in named entity extraction: combining semi-Markov extraction processes and data integration methods. In Proceedings of the 10th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Seattle, WA, USA, 22–25 August 2004.
- Turian, J.; Ratinov, L.; Bengio, Y. Word representations: A simple and general method for semi-supervised learning. In Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics, Uppsala, Sweden, 11–16 July 2010; pp. 384–394.
- Brown, P.F.; de Souza, P.V.; Mercer, R.L.; Pietra, V.J.D.; Lai, J.C. Class-based n-gram models of natural language. Comput. Linguist. 1992, 18, 467–479. [Google Scholar]
- Ratinov, L.; Roth, D. Design challenges and misconceptions in named entity recognition. In Proceedings of the 19th Conference on Computational Natural Language Learning, Boulder, CO, USA, 4–5 June 2009.
- Lin, W.-S.; Dai, H.-J.; Jonnagaddala, J.; Chang, N.-W.; Jue, T.R.; Iqbal, U.; Shao, J.Y.-H.; Chiang, I.J.; Li, Y.-C. Utilizing Different Word Representation Methods for Twitter Data in Adverse Drug Reactions Extraction. In Proceedings of the 2015 Conference on Technologies and Applications of Artificial Intelligence (TAAI), Tainan, Taiwan, 20–22 November 2015.
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.S.; Dean, J. Distributed representations of words and phrases and their compositionality. In Proceedings of Advances in Neural Information Processing Systems (NIPS 2013), Lake Taheo, NV, USA, 5–10 December 2013; pp. 3111–3119.
- Pennington, J.; Socher, R.; Manning, C.D. Glove: Global vectors for word representation. In Proceedings of the Empiricial Methods in Natural Language Processing (EMNLP 2014), Doha, Qatar, 25–29 October 2014; Volume 12, pp. 1532–1543.
- Yates, A.; Goharian, N.; Frieder, O. Extracting Adverse Drug Reactions from Social Media. In Proceedings of the Twenty-Ninth AAAI Conference on Artificial Intelligence (AAAI-15), Austin, TX, USA, 25–30 Jaunary 2015; pp. 2460–2467.
- Sarker, A.; Gonzalez, G. Portable automatic text classification for adverse drug reaction detection via multi-corpus training. J. Biomed. Inform. 2015, 53, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Sarker, A.; O’Connor, K.; Ginn, R.; Scotch, M.; Smith, K.; Malone, D.; Gonzalez, D. Social Media Mining for Toxicovigilance: Automatic Monitoring of Prescription Medication Abuse from Twitter. Drug Saf. 2016, 39, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Paul, M.J.; Dredze, M. You Are What You Tweet: Analyzing Twitter for Public Health. In Proceedings of the Fifth International AAAI Conference on Weblogs and Social Media (ICWSM-11), Barcelona, Spain, 17–21 July 2011.
- Owoputi, O.; O’Connor, B.; Dyer, C.; Gimpel, K.; Schneider, N.; Smith, N.A. Improved part-of-speech tagging for online conversational text with word clusters. In Proceedings of the Conference of the North American Chapter of the Association for Computational Linguistics, Atlanta, GA, USA, 9–14 June 2013.
- Leaman, R.; Wojtulewicz, L.; Sullivan, R.; Skariah, A.; Yang, J.; Gonzalez, G. Towards internet-age pharmacovigilance: extracting adverse drug reactions from user posts to health-related social networks. In Proceedings of the 2010 Workshop on Biomedical Natural Language Processing, Uppsala, Sweden, 15 July 2010; pp. 117–125.
- Bodenreider, O. The unified medical language system (UMLS): Integrating biomedical terminology. Nucleic Acids Res. 2004, 32, D267–D270. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Campillos, M.; Letunic, I.; Jensen, L.J.; Bork, P. A side effect resource to capture phenotypic effects of drugs. Mol. Syst. Biol. 2010, 6. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Zhu, X.; Li, J.; Hirst, G. Analysis of Polarity Information in Medical Text. AMIA Ann. Symp. Proc. 2005, 2005, 570–574. [Google Scholar]
- Tsai, R.T.-H.; Wu, S.-H.; Chou, W.-C.; Lin, C.; He, D.; Hsiang, J.; Sung, T.-Y.; Hsu, W.-L. Various criteria in the evaluation of biomedical named entity recognition. BMC Bioinform. 2006, 7. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-D.; Ohta, T.; Tsuruoka, Y.; Tateisi, Y. Introduction to the bio-entity recognition task at JNLPBA. In Proceedings of the International Workshop on Natural Language Processing in Biomedicine and its Applications (JNLPBA-04), Geneva, Switzerland, 28–29 August 2004; pp. 70–75.
- Tsuruoka, Y.; Tateishi, Y.; Kim, J.D.; Ohta, T.; McNaught, J.; Ananiadou, S.; Tsujii, J.I. Developing a robust part-of-speech tagger for biomedical text. In Advances in Informatics, Proceedings of the 10th Panhellenic Conference on Informatics, PCI 2005, Volas, Greece, 11–13 November 2005; Bozanis, P., Houstis, E.N., Eds.; Lecture Notes in Computer Science. Springer: Berlin/Heidelberg, Germany, 2005; Volume 3746, pp. 382–392. [Google Scholar]
- Aha, D.W.; Bankert, R.L. A comparative evaluation of sequential feature selection algorithms. In Learning from Data: Artificial Intelligence and Statistics V; Fisher, D., Lenz, H.-J., Eds.; Springer: New York, NY, USA, 1995; pp. 199–206. [Google Scholar]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar]
- Klinger, R.; Friedrich, C.M. Feature Subset Selection in Conditional Random Fields for Named Entity Recognition. In Proceedings of the International Conference RANLP 2009, Borovets, Bulgaria, 14–16 September 2009.
- Brody, S.; Diakopoulos, N. Cooooooooooooooollllllllllllll!!!!!!!!!!!!!!: Using word lengthening to detect sentiment in microblogs. In Proceedings of the Conference on Empirical Methods in Natural Language Processing, Edinburgh, UK, 27–29 July 2011.
- Wang, C.-K.; Singh, O.; Dai, H.-J.; Jonnagaddala, J.; Jue, T.R.; Iqbal, U.; Su, E.C.-Y.; Abdul, S.S.; Li, J.Y.-C. NTTMUNSW system for adverse drug reactions extraction in Twitter data. In Proceedings of the Social Media Mining Shared Task Workshop at the Pacific Symposium on Biocomputing, Big Island, HI, USA, 4–8 January 2016.
- Lai, S.; Liu, K.; Xu, L.; Zhao, J. How to Generate a Good Word Embedding? 2015; arXiv:1507.05523. [Google Scholar]
- Galar, M.; Fernandez, A.; Barrenechea, E.; Bustince, H.; Herrera, F. A review on ensembles for the class imbalance problem: Bagging-, boosting-, and hybrid-based approaches. In IEEE Transactions on Systems, Man, and Cybernetics, Part C: Applications and Reviews; IEEE: New York, NY, USA, 2012; Volume 42, pp. 463–484. [Google Scholar]
- Jonnagaddala, J.; Dai, H.-J.; Ray, P.; Liaw, S.-T. A preliminary study on automatic identification of patient smoking status in unstructured electronic health records. ACL-IJCNLP 2015, 2015, 147–151. [Google Scholar]
- Jonnagaddala, J.; Jue, T.R.; Dai, H.-J. Binary classification of Twitter posts for adverse drug reactions. In Proceedings of the Social Media Mining Shared Task Workshop at the Pacific Symposium on Biocomputing, Big Island, HI, USA, 4–8 January 2016.
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).