Abstract
Traditional Chinese medicine (TCM), as a vital component of traditional healthcare systems, relies heavily on its chemical constituents, which serve as a bridge between ancient therapeutic theories and modern biomedical science. Efficient access to compound-related information is crucial for promoting the modernization and scientific understanding of TCM. However, existing approaches primarily rely on fragmented databases and literature-based retrieval methods, which suffer from low intelligence, poor data integration, and limited retrieval efficiency.This study presents a novel AI agent-based retrieval system tailored for compound information in TCM. The core innovation of the system lies in its hybrid retrieval-augmented generation framework, which seamlessly combines structured database queries with semantic vector retrieval. Furthermore, it integrates knowledge from three complementary sources—locally built knowledge bases, domain-specific APIs, and open web search—allowing for comprehensive coverage and adaptive handling of diverse natural language queries. Experiments conducted on a benchmark dataset of 150 compound-related queries demonstrate that the system achieves a peak accuracy of 96.67% across multiple mainstream LLMs. Ablation studies further reveal that removing either the hybrid RAG or multi-source knowledge module leads to a notable accuracy decline, while the full system outperforms typical RAG baselines by over 25%. These results confirm the effectiveness and robustness of the proposed architecture in TCM compound retrieval, and highlight the advantage of combining structured matching with dynamic knowledge access in specialized biomedical applications.
1. Introduction
Traditional Chinese medicine (TCM), as an integral part of China’s medical heritage, has played a vital role in disease treatment and health preservation throughout history [1,2]. The chemical compounds found in TCM form the pharmacological basis of its therapeutic practices. These compounds reflect the structural and functional principles of traditional formulations and support modern pharmacological investigations. In addition, they serve as a critical bridge connecting traditional medical theories with contemporary drug discovery [3,4,5]. A notable example is artemisinin, derived from the TCM herb Artemisia annua, which illustrates the significant therapeutic potential of TCM in modern medicine [6].
As the modernization of TCM continues to progress, the systematic and efficient retrieval of compound-related information has become a core requirement for both basic research and practical development [7]. Currently, such information is distributed mainly in the traditional literature, academic journals, and various online databases. Despite the richness of these sources, challenges such as fragmentation of data, structural heterogeneity, and low retrieval efficiency persist, significantly hindering integration and deep utilization [8].
However, despite the emergence of a number of specialized databases for TCM, such as HERB 2.0 [9], TCMID 2.0 [10], and ETCM [11], which provide valuable data resources, these platforms remain largely static and isolated, relying primarily on keyword matching search methods. Data models are inconsistent across platforms, and cross-database integration using identifiers such as CAS numbers or InChIKeys often requires manual intervention. Furthermore, most systems lack support for natural language understanding or tolerance for synonyms and misspellings, which limits accessibility for nonexpert users and impedes intelligent interaction [12]. Therefore, there is an urgent need to develop an efficient and intelligent TCM compound information retrieval system to further advance the modernization of Chinese medicine.
Recent advances in LLM-based AI agents have significantly enhanced the performance of knowledge retrieval and question answering systems [13,14]. When augmented with retrieval-augmented generation (RAG) techniques, these agents can dynamically retrieve external evidence and generate concise, interpretable responses [15]. Building on this paradigm, several recent studies have demonstrated domain-specific improvements: Feng et al. improved factual accuracy in biomedical QA by integrating an LLM with comprehensive cancer-related resources [16]; Duan et al. improved the relevance of the answer in TCM queries using specialized domain data [17]. CodeQA combines an LLM with RAG to address programming questions, effectively reducing hallucinations compared to non-RAG baselines [18]. Vakayil et al. developed an RAG chatbot based on LLaMA-2 that retrieves counseling information and achieves a response accuracy of 95% [19], while Suresh et al. used a collider physics literature vector database to produce summaries supported by citations [20]. While these studies affirm the practical utility of LLM-based AI agents for answering questions centered on retrieval, they typically rely on a single static knowledge source and a fixed retrieval strategy.
In contrast, our system advances this line of research by integrating structured field matching with semantic vector retrieval in a hybrid RAG architecture, and by coordinating three complementary knowledge channels: local databases, domain-specific APIs, and real-time web content. This design offers broader coverage, more timely information, and improved accuracy compared to previous approaches.
This study proposes an AI agent-based intelligent information retrieval system for TCM compounds. The system leverages an LLM as its core module and integrates structured database querying with semantic vector retrieval. A hybrid retrieval-augmented generation (hybrid RAG) mechanism is designed in conjunction with a dynamic multi-source knowledge scheduling and fusion strategy. The primary scientific contribution of this work lies in the design of a hybrid retrieval-augmented generation (hybrid RAG) mechanism that integrates structured field matching with semantic vector retrieval, enabling a more accurate interpretation of and response to complex natural language queries. In addition, the system introduces a hierarchical multi-source retrieval strategy that combines local databases, domain-specific APIs, and web search, thereby enhancing the comprehensiveness, timeliness, and robustness of information acquisition.
2. Materials and Methods
2.1. System Overview
The overall architecture of the proposed AI agent-based system for TCM compound information retrieval is illustrated in Figure 1. Centered on an LLM, the system integrates three core modules—intent recognition, dynamic multi-source retrieval, and knowledge fusion—to support a fully automated workflow from user query to final response.
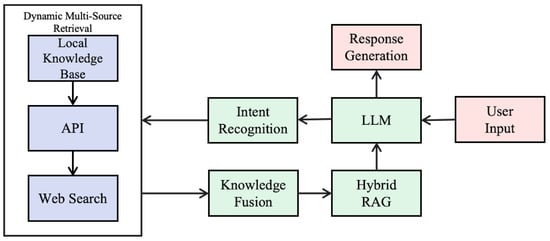
Figure 1.
System architecture of the AI agent-based retrieval system for TCM compounds.
Upon receiving a query, the agent first classifies the input as either identifier-based or natural language-based. It then initiates a hierarchical retrieval process, sequentially querying local structured databases, external domain-specific APIs (for example, PubChem [21]), and, if necessary, open web resources. The retrieved data are subsequently standardized and integrated through the knowledge fusion module and the consolidated information is delivered to the LLM to generate a coherent response in natural language.
This architecture offers a comprehensive solution for intelligent information querying and interactive response generation in the domain of TCM compound retrieval.
2.2. Knowledge Base Construction and Data Sources
The local knowledge base of the system is based on information related to traditional Chinese medicinal materials (TCM) and their constituent chemical compounds. Primary data sources include the Pharmacopoeia of the People’s Republic of China (2020 edition) and the HERB 2.0 database platform [22]. As the official national drug standard, the Chinese Pharmacopoeia provides authoritative data on commonly used TCM materials, including quality standards and clinically verified efficacy. HERB 2.0 is an integrated TCM database that aggregates evidence from systematic reviews, clinical trials, literature mining, and high-throughput experimental studies, offering comprehensive coverage and structured data organization.
During the data integration phase, information on 614 medicinal materials was extracted from the Chinese Pharmacopoeia, of which 404 entries were annotated with specific chemical compounds. To enhance the coverage and structural representation of compound data, additional records were incorporated from the HERB 2.0 platform. After performing deduplication, field mapping, and consistency checks, a local structured knowledge base was constructed, comprising 582 TCM materials and 7695 unique chemical compounds. This knowledge base supports precise matching using various fields, including compound names, CAS numbers, and InChIKeys.
To supplement potential information gaps in the local repository, the system integrates dynamic augmentation mechanisms through external APIs and web search engines. Two primary external data sources are incorporated into this process.
The PubChem API [23], managed by the US National Center for Biotechnology Information (NCBI), provides access to over 150 million chemical compound records. These include molecular structures, physicochemical properties, biological activities, drug targets, and the associated literature. Using PubChem significantly improves the coverage and credibility of the system data in the field of natural product research. When local retrieval fails to yield results, the system automatically invokes the PubChem API. The retrieved data are then normalized and incorporated into the knowledge fusion pipeline for validation and enrichment.
The Tavily API enables a real-time semantic web search and provides access to recent research developments [24], patent filings, and industry reports that are not indexed in structured databases. Tavily draws from platforms such as Google Scholar and PubMed and applies natural language processing techniques to improve query matching accuracy. Within the system, Tavily is primarily used to address open-ended queries involving newly discovered compounds or molecular structures that are not yet formally registered.
To improve the robustness and reliability of API calls, the system adopts a caching mechanism based on the in-memory key-value store Redis. When a user submits a query, the system first computes a unique hash value derived from the normalized query content and checks whether a corresponding recent response exists in the Redis cache. If a match is found, the cached result is returned directly, reducing external API calls and mitigating latency. If not, the system invokes the appropriate external API, stores the returned response in the cache along with a timestamp, and then delivers the result to the user. Each cached entry is configured to expire eight hours after its creation, ensuring a balance between response freshness and storage efficiency. Furthermore, the cache employs a Least Recently Used (LRU) eviction policy to manage memory usage under high-load conditions. This mechanism significantly improves system availability and fault tolerance, particularly during periods of network instability or temporary service outages.
The entire knowledge supplementation process follows the principle of “local-first, external-supplement”, ensuring broad coverage without compromising data reliability. Through this integrated knowledge infrastructure and dynamic enrichment strategy, the system supports structured and semantic-level queries, offering a high-quality, scalable, and adaptive foundation for intelligent information retrieval.
2.3. Dynamic Multi-Source Retrieval Mechanism
To address challenges such as semantic ambiguity and incomplete data in the retrieval of compound information in TCM, the system adopts a progressive hierarchical multi-source strategy. This approach follows a “local-first, progressively extended” principle, enabling efficient handling of complex queries and comprehensive information completion by leveraging local knowledge bases, authoritative external database APIs, and web-based semantic search engines in a coordinated manner.
In the initial retrieval stage, the system analyzes the structure of the user query through the intent recognition module to determine the appropriate retrieval pathway. Queries containing standardized identifiers (e.g., CAS numbers or InChIKeys [25,26]) are processed using a structured retrieval approach, which involves querying predefined fields in a local MySQL relational database. In contrast, natural language queries are converted into 1024-dimensional semantic embeddings using the BGE-large-zh-v1.5 model [27], and similarity-based retrieval is performed against a preconstructed local vector index. The results obtained from both structured and semantic retrieval processes are merged to form an initial set of candidates.
In the semantic retrieval module, we employ the BGE-large-zh-v1.5 embedding model to encode both user queries and candidate entries in dense vectors. This model represents a state-of-the-art Chinese language encoder optimized for retrieval tasks, demonstrating strong performance across multiple Chinese-language benchmarks. Its ability to capture domain-specific semantics makes it particularly suitable for applications in the TCM domain.
If this initial set of results falls below the predefined confidence threshold () or lacks essential information, the system activates the external database retrieval layer. The threshold value was empirically determined based on preliminary experiments, which showed that values below 0.6 often indicated incomplete or low-quality results in our test. At this stage, the PubChem API is invoked to obtain standardized chemical structure information. All retrieved data undergo field normalization before being integrated into the knowledge base to ensure consistency and compatibility with the local schema.
If both preceding stages fail to meet the retrieval requirements, the system escalates to a final layer involving open web search. Using the Tavily API, the system accesses up-to-date scientific content from authoritative platforms such as PubMed [28], CNKI [29], and Google Scholar, allowing the retrieval of real-time knowledge, newly identified compounds, and unstructured information not yet present in curated databases.
This dynamic and layered retrieval mechanism ensures both robustness and scalability in handling diverse query types, while offering broad information coverage and maintaining data integrity, as illustrated in Figure 2.
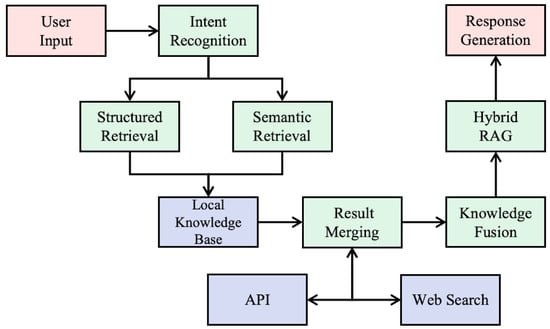
Figure 2.
Workflow of the AI agent-based retrieval system for TCM compounds.
2.4. Knowledge Fusion
Following multi-source retrieval, the system performs data fusion across results obtained from the structured database, semantic vector database, PubChem API, and Tavily API. Due to differences in field definitions, value units, and information granularity between these sources, direct concatenation may result in redundancy, semantic inconsistencies, or data omissions. To ensure consistency, completeness, and reliability for downstream processing, the system applies a unified field normalization and semantic integration strategy.
All retrieved data are first subjected to field mapping and format harmonization. This process includes standardizing field names, unifying measurement units, and converting various data structures into a common JSON Schema. For example, compound entries retrieved from the PubChem API are automatically transformed to match the schema of the local knowledge base, ensuring structural compatibility for subsequent operations.
The fully duplicated information is automatically removed during the deduplication step. In cases where key attributes such as molecular weight or chemical properties diverge across sources, the system employs a credibility-based resolution mechanism that prioritizes values from authoritative sources. Metadata concerning data provenance are preserved and appended to the output to transparently communicate potential discrepancies to the end user.
Furthermore, the fusion module incorporates rule-based consistency checking, validates the completeness of the field, enforces value range constraints, and applies logical coherence rules to cross-validate interdependent fields. A confidence score is assigned to each finalized data record, integrating source reliability and contextual relevance, which is subsequently used to guide response generation.
This module is critical to the integration of heterogeneous information, ensuring that contextual data fed into the LLM are semantically coherent, structurally standardized, and verifiably sourced. As a result, it significantly improves the accuracy, robustness, and trustworthiness of the system’s responses, particularly in handling ambiguous or open-ended TCM compound queries.
2.5. Hybrid Retrieval-Augmented Generation Technique
To improve the accuracy and domain relevance of generated answers in complex query scenarios, the system integrates a hybrid RAG mechanism built upon its multi-source retrieval framework. This mechanism coordinates structured data with semantic vector information and dynamically merges both local and external data sources to generate interpretable, high-quality natural language responses.
Unlike conventional RAG methods that typically depend on a single knowledge source, the hybrid RAG approach is specifically designed to accommodate the structural heterogeneity and semantic richness inherent to TCM compound data. By combining various retrieval strategies with flexible context construction techniques, the system enhances its ability to adapt to various types of query and information demands. Initially, candidate results aggregated through the retrieval strategy described in Section 2.3 are processed via the knowledge fusion module in Section 2.4, where field alignment, format standardization, and conflict resolution are applied to produce a unified semantic input.
During the answer generation stage, the fused knowledge is embedded into predefined prompt templates. The key semantic fields are formatted using a lightweight markup syntax and integrated with system-level instructions and the user’s original query to construct the final input. The LLM then generates domain-specific natural language responses tailored for scientific and technical users. These outputs span a range of formats for presenting knowledge, including named entity recognition, structured relationship explanation, and pharmacological mechanism inference.
By effectively integrating structured and unstructured information across multiple retrieval layers, this hybrid RAG mechanism significantly enhances the system’s performance in handling complex, multi-hop, and ambiguous queries. It ensures that generated content remains semantically coherent, factually accurate, and linguistically professional, making it particularly suitable for domain-intensive, multiturn scientific question-answering tasks.
2.6. System Implementation
The system is implemented based on the Dify platform [30], an open-source framework for developing large LLM-based applications. Dify provides essential functionalities such as visual workflow design, model orchestration, and knowledge base integration. This section elaborates on how the key modules described in the previous sections are translated into functional system components.
LLMs constitute the core component for natural language understanding and generation in this system. While the architecture remains compatible with various LLM backbones, four mainstream models were selected during evaluation based on their performance, robustness, and ease of integration. Details regarding the selected models and their usage are provided in Section 3.
The system employs a dual channel knowledge access strategy to build its local knowledge infrastructure. For structured queries, a local MySQL database is connected to support standardized field-level retrieval. For semantic queries, a vector-based knowledge base is constructed using paragraph embeddings derived from the literature, enabling semantic similarity matching. External data sources such as PubChem API and Tavily API are used to supplement the system when local retrieval does not produce results, thus expanding the system’s overall information coverage.
Upon receiving a user query, the system first determines the query type through an intent recognition module. If the query contains standardized identifiers, the structured retrieval module is triggered; otherwise, a semantic vector retrieval is performed. When local retrieval proves insufficient, external APIs are automatically invoked, enabling a confidence-driven multi-source retrieval workflow that balances coverage and accuracy.
The data returned from these sources is then unified through the knowledge fusion module, which performs field mapping, data format normalization, conflict resolution, and consistency checks. In this process, conflicting values are resolved based on the credibility of the data source to ensure logical and semantic consistency in the fused knowledge.
Consolidated knowledge is then injected into the LLM through a standardized prompt template, organized into a three-part input structure: system instruction, contextual information, and user query. The response generation module applies the hybrid RAG strategy, leveraging both structured field data and semantic knowledge fragments to produce accurate, coherent, and professionally expressed natural language responses tailored to the needs of compound-specific TCM queries.
Through tightly integrated module coordination and dynamic execution flow, the system successfully implements a complete processing loop—from query understanding to knowledge retrieval, data fusion, and answer generation—effectively operationalizing the methodological designs introduced in Section 2.2 through Section 2.5.
This execution loop is summarized in Figure 3, which illustrates the linear processing pipeline from user input to final response generation. It provides a concise view of how the system components are orchestrated to handle compound-specific TCM queries in a unified manner.
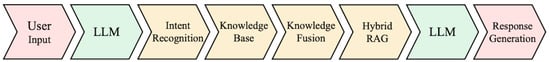
Figure 3.
Linear pipeline of the system’s core modules.
To demonstrate the interactive capabilities of the system and its practical effectiveness, Figure 4 displays the system interface and typical usage scenarios. It includes: (a) the main interface with natural language query support, (b) an example of compound identification results, and (c) a semantic explanation of pharmacological effects based on integrated knowledge.
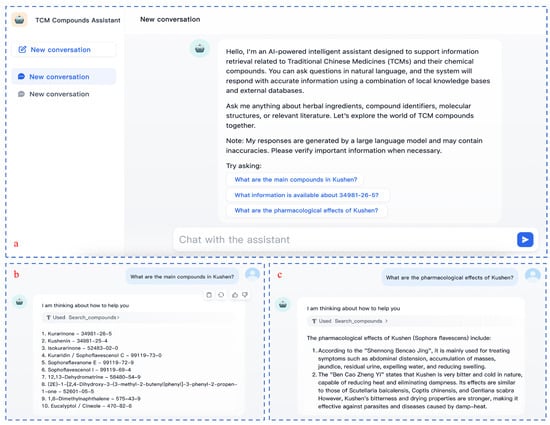
Figure 4.
Interface and usage examples of the AI agent-based retrieval system for TCM compounds.
3. Results
To systematically evaluate the accuracy and effectiveness of the proposed AI agent-based system for retrieving compound information in TCM during real-world question-answering tasks, an experimental framework was designed around practical query scenarios. Under a unified system architecture, the influence of the LLM was isolated by substituting only the core model within the agent. This design enables a comparative analysis of how different LLMs affect the overall performance of the system and facilitates the identification of the most suitable model for this application.
Within the system framework, the LLM plays a pivotal role in intent interpretation, query semantic modeling, and response generation. It serves as the central intelligence module that enables the interactive capabilities of the AI agent. To evaluate the system’s adaptability to different models and examine their performance boundaries in practical deployment, four state-of-the-art LLMs were selected as agent backbones: DeepSeek V3 [31], Qwen-Max-202501 [32], GLM-4-Plus [33], and Doubao v1.5-Pro [34]. All models were accessed via their respective official APIs using standardized protocols, ensuring procedural consistency and comparability of the results.
3.1. Evaluation Dataset
To rigorously assess the system performance in practical query tasks, a domain-specific benchmark dataset was constructed for compound-related information retrieval in the context of TCM. The dataset comprises 150 natural language questions covering commonly used TCM herbs and their representative chemical constituents. The question formats, content structures, and expected answer types were designed with reference to real-world research settings, ensuring strong representativeness and practical applicability.
The dataset was derived primarily from the local knowledge base of the system and was curated by manual sampling. Each test instance consists of a natural language question written in authentic Chinese and a corresponding reference answer. These questions simulate typical user interactions with the system under realistic usage scenarios.
The core evaluation task focuses on TCM-to-compound mapping and evaluates system performance across three key dimensions:
- The recognition of herb names;
- The accuracy of compound-level information retrieval and matching;
- The completeness and fault tolerance of responses under semantically ambiguous expressions.
The test questions cover a broad range of herbs from TCM and reference more than 600 compounds, featuring structural diversity and cross-entry distribution. This design effectively simulates the complexity of real-world information retrieval tasks.
3.2. Experimental Setup and Evaluation Metrics
To objectively evaluate the performance of the TCM compound retrieval system under different LLM configurations, we established a standardized experimental procedure based on the unified benchmark dataset. Two core evaluation metrics were adopted: accuracy and average response time. Accuracy quantifies the alignment between system-generated answers and reference answers, while the average response time reflects the system’s operational efficiency and responsiveness in practical usage scenarios.
All experiments were conducted in a controlled runtime environment. The system was deployed on a unified hardware platform, and all LLMs were accessed through their official APIs using identical hyperparameter settings. This ensured that hardware and configuration differences did not influence the results. Each model completed all 150 benchmark queries in a full evaluation cycle. The system automatically recorded the output and response time for each query. All test questions were provided in natural language, and the full agent workflow—intent recognition, multi-source retrieval, and generation—was invoked.
Accuracy was used to evaluate whether the system generated correct or reasonable answers for input queries.To ensure fairness and reproducibility, all reference answers were curated in advance by two domain experts with experience in TCM and cheminformatics. The correctness of each system output was independently evaluated by both reviewers based on predefined task-specific criteria. Disagreements were resolved through discussion or adjudication by a third evaluator. A response was marked as correct if it accurately matched the reference answer or provided an acceptable synonym or equivalent representation. Let denote the total number of test questions, and the number of correctly answered instances. Then, the accuracy is computed as:
The correctness of each output was manually assessed against the annotated reference responses. The criteria varied depending on the type of question and were defined as follows:
- For compound identification questions: whether the system correctly listed the major chemical constituents of the queried herb;
- For field-specific extraction tasks: whether the system returned accurate attribute values, such as molecular formula, CAS number, or Chinese compound name;
- For logical reasoning questions: whether the system’s output exhibited factual correctness and reasonable inference based on known data.
To evaluate runtime efficiency, average response time was calculated as the mean time required to complete the full processing loop for all test queries. Let denote the response time (in seconds) for the i-th query. Then, the average response time is given by:
This metric reflects the overall latency of the system in intent recognition, multi-source retrieval, knowledge fusion, and LLM-based response generation. All measurements were carried out under consistent execution conditions to ensure comparability between model configurations.
3.3. Experimental Results
The experimental results indicate that the system achieved high accuracy in both compound identification and information retrieval tasks, with an average accuracy exceeding 94%. These findings demonstrate that the proposed AI agent-based system for retrieving TCM compound information exhibits strong stability and practical utility in handling semantic queries, retrieving knowledge from structured sources, and integrating data from external APIs. Among the 150 representative queries, most were correctly interpreted and answered by the system, reflecting strong natural language understanding and effective multi-source data fusion.
The adaptability of the system to different LLMs was further evaluated by testing four main LLMs as the agent core. As shown in Figure 5, all models achieved accuracy scores above 0.94 under identical system configurations, indicating strong model compatibility. DeepSeek V3 achieved the highest accuracy at 96.67%, while GLM-4-Plus demonstrated the fastest average response time of 7.82 s, making it well suited for latency-sensitive applications.
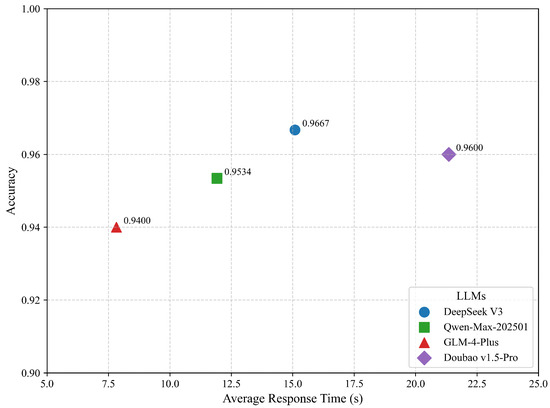
Figure 5.
Performance comparison of different large language models on TCM compound QA tasks.
Qwen-Max-202501 and Doubao v1.5-Pro exhibited comparable performance in both accuracy and response time, suggesting that the system remains stable across different model configurations and demonstrates strong generalization across models. The observed performance differences were primarily attributed to variations in the depth of semantic understanding and response latency, rather than inconsistencies in the core retrieval and fusion mechanisms of the system.
3.4. Ablation Study
To further assess the functional contributions of core system modules, we conducted a two-tier ablation study. The first tier compared the complete agent-based configuration (A0) with a baseline that relies solely on a large language model (LLM-only), disabling all retrieval and fusion mechanisms. In this simplified setting, the LLM generates responses directly from the input prompt without access to structured fields, vector indices, or external databases. As shown in Figure 6, the full system significantly outperformed the LLM-only baseline in terms of accuracy. For instance, DeepSeek V3 achieved 96.67% accuracy under the full configuration, while performance dropped to 44.67% in the LLM-only setting. Similar trends were observed across other tested models, confirming the essential role of hybrid retrieval, knowledge fusion, and dynamic Multi-Source Knowledge Retrieval in delivering accurate and reliable outputs.
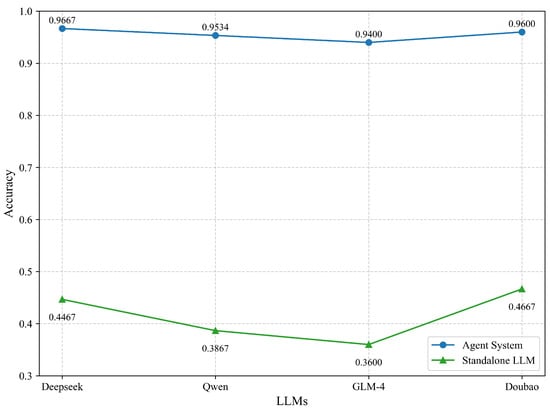
Figure 6.
Accuracy comparison between the full agent system and standalone LLMs on TCM compound QA tasks.
In the second tier, we conducted module-level ablations using DeepSeek V3 as the fixed backbone. Three reduced configurations were tested:
- A1: Disables intent recognition, directing all queries to the hybrid RAG pipeline without identifier-based structured routing.
- A2: Disables dynamic multi-source knowledge retrieval, restricting retrieval to the local knowledge base.
- A3: Disables both A1 and A2, simulating a minimal RAG setup based solely on vector retrieval from the local database.
As summarized in Table 1, removing either module resulted in a moderate accuracy decline—85.33% for A1 and 84.00% for A2. However, when both modules were disabled in A3, accuracy dropped more sharply to 71.33%. These results indicate that structured intent routing and dynamic multi-source knowledge retrieval each contribute independently to overall system performance, and their combined presence is critical for handling diverse and complex natural language queries with high precision.

Table 1.
Accuracy of each ablation setting on the 150-query benchmark (DeepSeek V3).
4. Discussion
The system exhibits strong functional coupling and synergistic effects in all its core modules. The hybrid RAG mechanism integrates structured field matching with semantic vector retrieval, effectively balancing precision with the flexibility to interpret open-ended semantics. This approach addresses the limitations of traditional methods in both structured data coverage and semantic adaptability. It is particularly well-suited for the TCM domain, where queries often involve high semantic ambiguity and dense domain-specific terminology.
To enhance knowledge completeness and adaptability, the system employs a layered multi-source retrieval strategy following a “local-first, progressive expansion” policy. It sequentially invokes the local knowledge base, the domain-specific API, and the web search. This strategy not only broadens the scope of knowledge coverage, but also mitigates the risk of blind spots caused by the dependence on a single data source, thus improving the fault tolerance of the system.
In terms of the adaptability of the model, the system shows strong compatibility between models. Comparative experiments show that regardless of whether the underlying LLM is Qwen, DeepSeek, or GLM-4, the system consistently achieves high response accuracy with minimal performance variation. These findings highlight the independence and portability of the architecture model, providing a solid foundation for future upgrades and system evolution.
The knowledge fusion module plays a pivotal role in harmonizing outputs from diverse data sources. To address heterogeneity among structured databases, local semantic embeddings, and external API responses, the system applies techniques such as field mapping, confidence-based weighting, and conflict resolution to generate contextualized standardized inputs for response generation.
An extended ablation study was conducted to assess the contribution of key modules, including intent recognition, structured retrieval, and dynamic source scheduling. In the baseline comparison between the full agent system and an LLM-only configuration, removing all retrieval and fusion components led to a substantial accuracy drop—from 96.67% to 44.67%—demonstrating the necessity of integrating knowledge-grounded retrieval strategies.
To further isolate the impact of specific modules, additional ablation settings were evaluated using DeepSeek V3. When intent recognition was removed, but hybrid RAG and multi-source knowledge retrieval were retained (A1), accuracy declined to 85.33%. Disabling the dynamic multi-source knowledge retrieval while preserving structured retrieval (A2) resulted in a similar decrease to 83.87%. Notably, when both modules were removed, leaving only a minimal vector-based RAG pipeline with local knowledge (A3), performance dropped further to 71.33%. These results suggest that the two modules contribute independently to system robustness, and that their synergy is essential for maintaining high retrieval accuracy in complex natural language queries.
The ablation results confirm that the hybrid RAG and dynamic multi-source knowledge retrieval are not merely optional enhancements but essential components. Their absence leads to notable declines in output completeness, semantic precision, and domain-specific relevance, particularly in scenarios involving ambiguous query intent or compound attribute resolution.
Compared to prior works such as CodeQA, RAGS4EIC, and the systems by [16,17], our approach advances the field by integrating hybrid RAG with structured and semantic retrieval across multi-source knowledge: local databases, external domain-specific APIs, and the open web. Although these earlier systems typically rely on a single retrieval method and fixed corpus, our design enables broader coverage, more dynamic knowledge access, and improved robustness for complex open-ended TCM queries.
Despite its overall effectiveness, the system has certain limitations in real-world deployment. First, the reliance on external APIs introduces potential issues related to network latency and third-party service availability. Second, the local knowledge base update process currently requires manual intervention, which may limit the adaptability of the system in rapidly evolving research contexts.
5. Conclusions
This study presents an AI agent-based system designed for the intelligent retrieval of compound information in TCM. The proposed system integrates natural language understanding, structured and unstructured data fusion, and automated response generation within a modular architecture. A key innovation lies in the integration of hybrid RAG that combines structured field matching with semantic vector retrieval, enhancing both the precision and flexibility of information access. Complementing this is a dynamic multi-source knowledge retrieval strategy that enables hierarchical access to local knowledge bases, external domain-specific APIs, and open web search, thereby ensuring comprehensive knowledge coverage and robust system adaptability.
Comprehensive experiments conducted on a 150-query benchmark dataset demonstrate that the system consistently achieves high accuracy and robustness across multiple LLMs. These results confirm the effectiveness of each core module and validate the system’s architectural design in delivering reliable and scalable TCM compound information retrieval. Additional ablation studies further reveal the independent and synergistic contributions of structured querying and multi-source scheduling, emphasizing the superiority of the proposed hybrid framework over conventional LLM-only approaches.
Future work will focus on several key directions. First, the system will incorporate automated pipelines for data update and knowledge reconstruction, thus improving its capacity for dynamic knowledge maintenance. Second, the system’s functionality will be extended to support tasks such as pharmacological mechanism analysis, molecular target identification, and compound synergy evaluation within the context of TCM. Furthermore, integrating this system with existing research platforms or knowledge graph infrastructures could further promote its application in intelligent decision support and contribute to the ongoing modernization of TCM.
Author Contributions
Conceptualization, F.Z. and X.X.; methodology, F.Z. and Q.L.; software, F.Z. and M.W.; validation, F.Z. and Q.L.; formal analysis, F.Z.; investigation, F.Z., Q.L. and M.W.; resources, X.X.; data curation, F.Z. and Q.L.; writing—original draft preparation, F.Z.; writing—review and editing, X.X.; visualization, F.Z.; supervision, X.X.; project administration, X.X. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Science & Technology Fundamental Resources Investigation Program, grant number 2022FY101200.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors on request.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| TCM | Traditional Chinese medicine |
| LLM | Large Language Model |
| RAG | Retrieval-augmented generation |
| CAS | Chemical Abstracts Service |
| API | Application Programming Interface |
References
- Li, L.; Yao, H.; Wang, J.; Li, Y.; Wang, Q. The Role of Chinese Medicine in Health Maintenance and Disease Prevention: Application of Constitution Theory. Am. J. Chin. Med. 2019, 47, 495–506. [Google Scholar] [CrossRef]
- Luo, Y.; Wang, C.Z.; Hesse-Fong, J.; Lin, J.G.; Yuan, C.S. Application of Chinese Medicine in Acute and Critical Medical Conditions. Am. J. Chin. Med. 2019, 47, 1223–1235. [Google Scholar] [CrossRef]
- Zhang, N.D.; Han, T.; Huang, B.K.; Rahman, K.; Jiang, Y.P.; Xu, H.T.; Qin, L.P.; Xin, H.L.; Zhang, Q.Y.; Li, Y.m. Traditional Chinese medicine formulas for the treatment of osteoporosis: Implication for antiosteoporotic drug discovery. J. Ethnopharmacol. 2016, 189, 61–80. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liu, J.; Luo, X.; Hu, L.; Lu, H. Functional metabolomics innovates therapeutic discovery of traditional Chinese medicine derived functional compounds. Pharmacol. Ther. 2021, 224, 107824. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Guo, Z.; Zhu, P.; Chen, J.; Huang, Y. Traditional Chinese medicine as a cancer treatment: Modern perspectives of ancient but advanced science. Cancer Med. 2019, 8, 1958–1975. [Google Scholar] [CrossRef]
- Tu, Y. The discovery of artemisinin (qinghaosu) and gifts from Chinese medicine. Nat. Med. 2011, 17, 1217–1220. [Google Scholar] [CrossRef] [PubMed]
- Li, W.F.; Jiang, J.G.; Chen, J. Chinese Medicine and Its Modernization Demands. Arch. Med. Res. 2008, 39, 246–251. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, Y.; Lin, W.; Wu, Q. How Can We Design a Standardized and Efficient Health Data Management System for Large-Scale Heterogeneous TCM Data? In Proceedings of the 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Lisbon, Portugal, 3–6 December 2024; pp. 4848–4853. [Google Scholar] [CrossRef]
- Gao, K.; Liu, L.; Lei, S.; Li, Z.; Huo, P.; Wang, Z.; Dong, L.; Deng, W.; Bu, D.; Zeng, X.; et al. HERB 2.0: An updated database integrating clinical and experimental evidence for traditional Chinese medicine. Nucleic Acids Res. 2025, 53, D1404–D1414. [Google Scholar] [CrossRef]
- Huang, L.; Xie, D.; Yu, Y.; Liu, H.; Shi, Y.; Shi, T.; Wen, C. TCMID 2.0: A comprehensive resource for TCM. Nucleic Acids Res. 2018, 46, D1117–D1120. [Google Scholar] [CrossRef]
- Xu, H.Y.; Zhang, Y.Q.; Liu, Z.M.; Chen, T.; Lv, C.Y.; Tang, S.H.; Zhang, X.B.; Zhang, W.; Li, Z.Y.; Zhou, R.R.; et al. ETCM: An encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019, 47, D976–D982. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, X.; Bai, H.; Ning, K. Network Pharmacology Databases for Traditional Chinese Medicine: Review and Assessment. Front. Pharmacol. 2019, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Xi, Z.; Chen, W.; Guo, X.; He, W.; Ding, Y.; Hong, B.; Zhang, M.; Wang, J.; Jin, S.; Zhou, E.; et al. The rise and potential of large language model based agents: A survey. Sci. China Inf. Sci. 2025, 68, 121101. [Google Scholar] [CrossRef]
- Zhang, A.; Deng, Y.; Lin, Y.; Chen, X.; Wen, J.R.; Chua, T.S. Large Language Model Powered Agents for Information Retrieval. In Proceedings of the 47th International ACM SIGIR Conference on Research and Development in Information Retrieval, 2024; SIGIR ’24, Washington, DC, USA, 14–18 July 2024; ACM: New York, NY, USA; pp. 2989–2992. [Google Scholar] [CrossRef]
- Singh, A.; Ehtesham, A.; Kumar, S.; Khoei, T.T. Agentic Retrieval-Augmented Generation: A Survey on Agentic RAG. arXiv 2025, arXiv:2501.09136. [Google Scholar] [CrossRef]
- Feng, Y.; Zhou, L.; Ma, C.; Zheng, Y.; He, R.; Li, Y. Knowledge graph-based thought: A knowledge graph-enhanced LLM framework for pan-cancer question answering. GigaScience 2025, 14, giae082. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhou, Q.; Li, Y.; Qin, C.; Wang, Z.; Kan, H.; Hu, J. Research on a traditional Chinese medicine case-based question-answering system integrating large language models and knowledge graphs. Front. Med. 2024, 11, 1512329. [Google Scholar] [CrossRef]
- Ahmed, M.; Dorrah, M.; Ashraf, A.; Adel, Y.; Elatrozy, A.; Mohamed, B.E.; Gomaa, W. CodeQA: Advanced programming question-answering using LLM agent and RAG. In Proceedings of the 2024 6th Novel Intelligent and Leading Emerging Sciences Conference (NILES), Giza, Egypt, 19–21 October 2024; pp. 494–499. [Google Scholar] [CrossRef]
- Vakayil, S.; Juliet, D.S.; J, A.; Vakayil, S. RAG-based LLM chatbot using llama-2. In Proceedings of the 2024 7th International Conference on Devices, Circuits and Systems (ICDCS), Coimbatore, India, 19–20 April 2024; pp. 1–5. [Google Scholar] [CrossRef]
- Suresh, K.; Kackar, N.; Schleck, L.; Fanelli, C. Towards a RAG-based summarization for the electron ion collider. J. Instrum. 2024, 19, C07006. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef]
- Commission, C.P. Pharmacopoeia of the People’s Republic of China, Volume I; China Medical Science and Technology Press: Beijing, China, 2020. [Google Scholar]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Bryant, S.H. PUG-SOAP and PUG-REST: Web services for programmatic access to chemical information in PubChem. Nucleic Acids Res. 2015, 43, W605–W611. [Google Scholar] [CrossRef]
- Tavily Search API. 2025. Available online: https://tavily.com/ (accessed on 15 March 2025).
- Chemical Abstracts Service. CAS REGISTRY. 2025. Available online: https://www.cas.org/cas-data/cas-registry (accessed on 13 May 2025).
- Heller, S.R.; McNaught, A.; Pletnev, I.; Stein, S.; Tchekhovskoi, D. InChI, the IUPAC International Chemical Identifier. J. Cheminform. 2015, 7, 23. [Google Scholar] [CrossRef]
- Xiao, S.; Liu, Z.; Zhang, P.; Muennighoff, N.; Lian, D.; Nie, J.Y. C-Pack: Packed Resources For General Chinese Embeddings. arXiv 2024, arXiv:2309.07597. [Google Scholar] [CrossRef]
- PubMed. 2025. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 15 March 2025).
- CNKI (China National Knowledge Infrastructure). 2025. Available online: https://www.cnki.net/ (accessed on 15 March 2025).
- Inc, LangGenius Dify: Open-Source LLM Application Development Platform. 2025. Available online: https://github.com/langgenius/dify (accessed on 15 March 2025).
- DeepSeek-AI; Liu, A.; Feng, B.; Xue, B.; Wang, B.; Wu, B.; Lu, C.; Zhao, C.; Deng, C.; Zhang, C.; et al. DeepSeek-V3 Technical Report. arXiv 2025, arXiv:2412.19437. [Google Scholar] [CrossRef]
- Yang, A.; Yang, B.; Zhang, B.; Hui, B.; Zheng, B.; Yu, B.; Li, C.; Liu, D.; Huang, F.; Wei, H.; et al. Qwen2.5 Technical Report. arXiv 2025, arXiv:2412.15115. [Google Scholar] [CrossRef]
- GLM Team; Zeng, A.; Xu, B.; Wang, B.; Zhang, C.; Yin, D.; Zhang, D.; Rojas, D.; Feng, G.; Zhao, H.; et al. ChatGLM: A Family of Large Language Models from GLM-130B to GLM-4 All Tools. arXiv 2024, arXiv:2406.12793. [Google Scholar] [CrossRef]
- ByteDance. Doubao v1.5-Pro. 2025. Available online: https://seed.bytedance.com/en/special/doubao_1_5_pro/ (accessed on 15 March 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).