Focal Cosine-Enhanced EfficientNetB0: A Novel Approach to Classifying Breast Histopathological Images
Abstract
1. Introduction
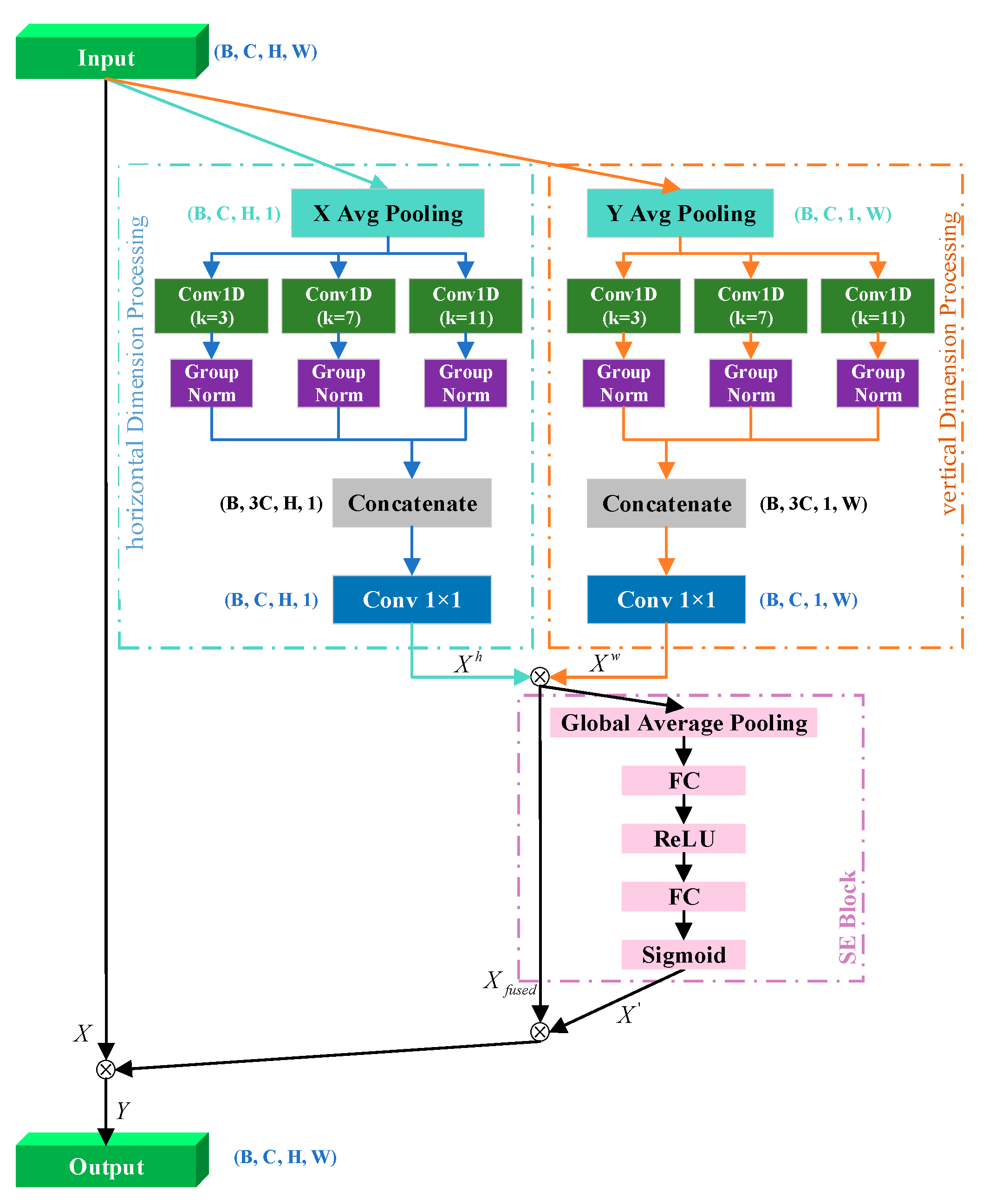
- Traditional CNNs are constrained by fixed receptive fields, limiting their ability to collaboratively extract cellular and tissue structural features from low- and high-magnification microscopy images. Therefore, we propose the multiscale ELA module, which integrates multi-branch convolutions with attention mechanisms to automatically focus on critical lesion regions across scales, significantly enhancing the feature extraction capabilities for key pathological characteristics under varying magnifications.
- To address the shortcomings in terms of small interclass differences and large intraclass variations in histopathological breast images, the model is trained using an improved focal cosine loss. This loss function integrates modified focal loss and enhanced cosine loss mechanisms, which enhance the model’s sensitivity to minority class samples by adaptively adjusting the weights for hard-to-classify samples and optimizing the interclass angular discrepancies, thereby improving the overall classification performance.
- By adopting EfficientNetB0 as the backbone network and integrating transfer learning techniques, the model applies ImageNet pre-trained weights to the breast cancer image classification task. Through structural optimization and feature fine-tuning, both the classification accuracy and generalization capabilities are significantly improved.
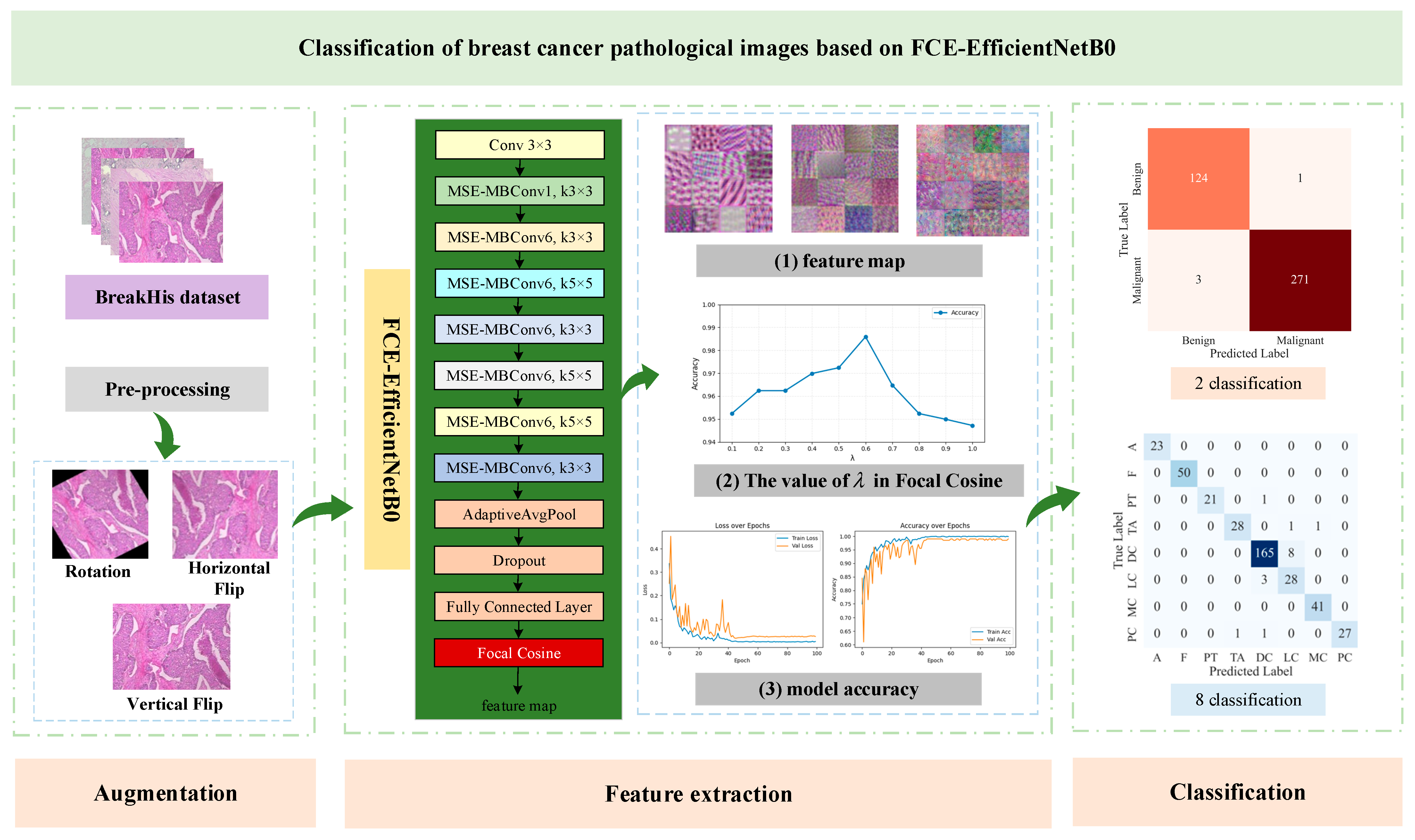
2. Proposed Approach
2.1. Multiscale ELA
2.2. Focal Cosine Loss Function
- 1.
- Optimized Focal Loss
- 2.
- Optimized Cosine Embedding Loss Function
- 3.
- Focal Cosine Loss Function
3. Results
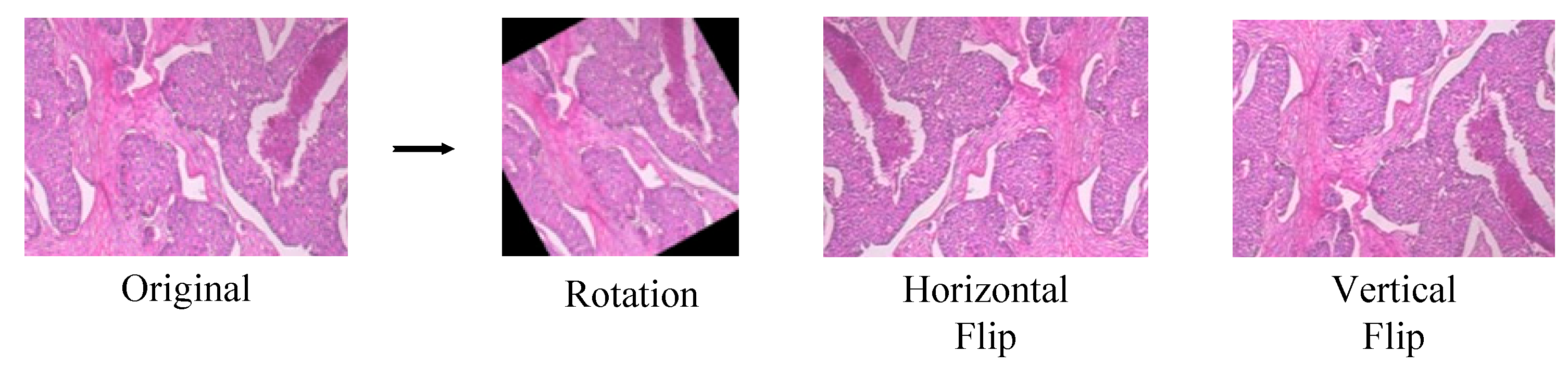
3.1. Data Description and Augmentation
3.2. Evaluation Metrics and Experimental Settings
- True Positives (): Correctly predicted instances of class .
- False Positives (): Instances predicted as class but belonging to other classes.
- False Negatives (): Instances of class incorrectly predicted as other classes.
3.3. Classification Results of FCE-EfficientNetB0 at Different Magnifications
3.4. Contrast Test
3.5. Comparison with Other Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Huang, S.; Yang, J.; Fong, S.; Zhao, Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020, 471, 61–71. [Google Scholar] [CrossRef]
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics 2024. CA A Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Massafra, R.; Bove, S.; Lorusso, V.; Biafora, A.; Comes, M.C.; Didonna, V.; Diotaiuti, S.; Fanizzi, A.; Nardone, A.; Nolasco, A.; et al. Radiomic feature reduction approach to predict breast cancer by contrast-enhanced spectral mammography images. Diagnostics 2021, 11, 684. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef]
- Senousy, Z.; Abdelsamea, M.M.; Gaber, M.M.; Abdar, M.; Acharya, U.R.; Khosravi, A.; Nahavandi, S. MCUa: Multi-level context and uncertainty aware dynamic deep ensemble for breast cancer histology image classification. IEEE Trans. Biomed. Eng. 2021, 69, 818–829. [Google Scholar] [CrossRef]
- Venugopal, A.; Sreelekshmi, V.; Nair, J.J. Ensemble Deep Learning Model for Breast Histopathology Image Classification. In ICT Infrastructure and Computing: Proceedings of ICT4SD 2022; Springer Nature: Singapore, 2022; pp. 499–509. [Google Scholar]
- Man, R.; Yang, P.; Xu, B. Classification of breast cancer histopathological images using discriminative patches screened by generative adversarial networks. IEEE Access 2020, 8, 155362–155377. [Google Scholar] [CrossRef]
- Seo, H.; Brand, L.; Barco, L.S.; Wang, H. Scaling multi-instance support vector machine to breast cancer detection on the BreaKHis dataset. Bioinformatics 2022, 38 (Suppl. 1), i92–i100. [Google Scholar] [CrossRef]
- Ortiz, S.; Rojas-Valenzuela, I.; Rojas, F.; Valenzuela, O.; Herrera, L.J.; Rojas, I. Novel methodology for detecting and localizing cancer area in histopathological images based on overlapping patches. Comput. Biol. Med. 2024, 168, 107713. [Google Scholar] [CrossRef]
- Bakshi, A.A.; Joarder, R.H.; Tasmi, S.T. Wavelet-Infused U-Net for Breast Ultrasound Image Segmentation. Ph.D. Thesis, Islamic University of Technology (IUT), Board Bazar, Gazipur, Bangladesh, 2024. [Google Scholar]
- Kang, D.U.; Chun, S.Y. Multi-Scale Curriculum Learning for Efficient Automatic Whole Slide Image Segmentation. In Proceedings of the 2022 IEEE International Conference on Big Data and Smart Computing (BigComp), Daegu, Republic of Korea, 17–20 January 2022; pp. 366–367. [Google Scholar]
- Tong, L.; Sha, Y.; Wang, M.D. Improving Classification of Breast Cancer by Utilizing the Image Pyramids of Whole-Slide Imaging and Multi-Scale Convolutional Neural Networks. In Proceedings of the 2019 IEEE 43rd Annual Computer Software and Applications Conference (COMPSAC), Milwaukee, WI, USA, 15–19 July 2019; Volume 1, pp. 696–703. [Google Scholar]
- Xie, P.; Li, T.; Li, F.; Zuo, K.; Zhou, J.; Liu, J. Multi-Scale Convolutional Neural Network for Melanoma Histopathology Image Classification. In Proceedings of the 2021 IEEE 3rd International Conference on Frontiers Technology of Information and Computer (ICFTIC), Qingdao, China, 12–14 November 2021; pp. 551–554. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-Excitation Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Li, X.; Wang, W.; Hu, X.; Yang, J. Selective Kernel Networks. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 510–519. [Google Scholar]
- Yu, D.; Lin, J.; Cao, T.; Chen, Y.; Li, M.; Zhang, X. SECS: An effective CNN joint construction strategy for breast cancer histopathological image classification. J. King Saud Univ.-Comput. Inf. Sci. 2023, 35, 810–820. [Google Scholar] [CrossRef]
- Hou, Q.; Zhou, D.; Feng, J. Coordinate Attention for Efficient Mobile Network Design. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 13713–13722. [Google Scholar]
- Xu, W.; Wan, Y. ELA: Efficient local attention for deep convolutional neural networks. arXiv 2024, arXiv:2403.01123. [Google Scholar]
- Reza, M.S.; Ma, J. Imbalanced Histopathological Breast Cancer Image Classification with Convolutional Neural Network. In Proceedings of the 2018 14th IEEE International Conference on Signal Processing (ICSP), Beijing, China, 12–16 August 2018; pp. 619–624. [Google Scholar]
- Larrazabal, A.J.; Nieto, N.; Peterson, V.; Milone, D.H.; Ferrante, E. Gender imbalance in medical imaging datasets produces biased classifiers for computer-aided diagnosis. Proc. Natl. Acad. Sci. USA 2020, 117, 12592–12594. [Google Scholar] [CrossRef]
- Mosquera, C.; Ferrer, L.; Milone, D.H.; Luna, D.; Ferrante, E. Class imbalance on medical image classification: Towards better evaluation practices for discrimination and calibration performance. Eur. Radiol. 2024, 34, 7895–7903. [Google Scholar] [CrossRef]
- Salmi, M.; Atif, D.; Oliva, D.; Abraham, A.; Ventura, S. Handling imbalanced medical datasets: Review of a decade of research. Artif. Intell. Rev. 2024, 57, 273. [Google Scholar] [CrossRef]
- Edward, J.; Rosli, M.M.; Seman, A. A new multi-class rebalancing framework for imbalance medical data. IEEE Access 2023, 11, 92857–92874. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Lin, T.Y.; Goyal, P.; Girshick, R.; He, K.; Dollár, P. Focal Loss for Dense Object Detection. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2980–2988. [Google Scholar]
- Meng, Z.; Zhao, Z.; Su, F. Multi-Classification of Breast Cancer Histology Images by Using Gravitation Loss. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, Great Britain, 12–17 May 2019; pp. 1030–1034. [Google Scholar]
- Cao, B.; Li, L.; Ma, Y.; Ye, S.; Li, S.; He, X. RAANet: Residual Aggregation Attention Network for Classification of Small Intestinal Endoscopic Images. In Proceedings of the 2023 IEEE 12th Data Driven Control and Learning Systems Conference (DDCLS), Xiangtan, China, 12–14 May 2023; pp. 1014–1019. [Google Scholar]
- Hu, S.; Zhang, Z.; Yang, J. Handling Intra-Class Dissimilarity and Inter-Class Similarity for Imbalanced Skin Lesion Image Classification. In Proceedings of the International Joint Conference on Rough Sets, Krakow, Poland, 5–8 October 2023; Springer Nature: Cham, Switzerland, 2023; pp. 565–579. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]
- Spanhol, F.A.; Oliveira, L.S.; Petitjean, C.; Heutte, L. A dataset for breast cancer histopathological image classification. IEEE Trans. Biomed. Eng. 2015, 63, 1455–1462. [Google Scholar] [CrossRef]
- Abdulaal, A.H.; Yassin, R.A.; Valizadeh, M.; Abdulwahhab, A.H.; Jasim, A.M.; Mohammetd, A.J.; Jabir, H.J.; Albaker, B.M.; Dheyaa, N.H.; Amirani, M.C. Cutting-Edge CNN Approaches for Breast Histopathological Classification: The Impact of Spatial Attention Mechanisms. ShodhAI J. Artif. Intell. 2024, 1, 109–130. [Google Scholar] [CrossRef]
- Dihin, R.A. Breast Cancer Detection and Diagnosis Using Gabor Features and EfficientNetV2 Model. J. Al-Qadisiyah Comput. Sci. Math. 2024, 16, 290–299. [Google Scholar]
- Kaur, A.; Kaushal, C.; Sandhu, J.K.; Damaševičius, R.; Thakur, N. Histopathological image diagnosis for breast cancer diagnosis based on deep mutual learning. Diagnostics 2023, 14, 95. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. Mobilenetv2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 4510–4520. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Bardou, D.; Zhang, K.; Ahmad, S.M. Classification of breast cancer based on histology images using convolutional neural networks. IEEE Access 2018, 6, 24680–24693. [Google Scholar] [CrossRef]
- Sharma, S.; Mehra, R. Conventional machine learning and deep learning approach for multi-classification of breast cancer histopathology images—a comparative insight. J. Digit. Imaging 2020, 33, 632–654. [Google Scholar] [CrossRef]
- Taheri, S.; Golrizkhatami, Z.; Basabrain, A.A.; Hazzazi, M.S. A comprehensive study on classification of breast cancer histopathological images: Binary versus multi-category and magnification-specific versus magnification-independent. IEEE Access 2024, 12, 50431–50443. [Google Scholar] [CrossRef]







| Class | Subtype | 40× | 100× | 200× | 400× | Total |
|---|---|---|---|---|---|---|
| Benign (B) | Adenosis (A) | 114 | 113 | 111 | 106 | 444 |
| Fibroadenoma (F) | 253 | 260 | 264 | 237 | 1014 | |
| Phyllodes Tumor (PT) | 149 | 150 | 140 | 130 | 569 | |
| Tubular Adenoma (TA) | 109 | 121 | 108 | 115 | 453 | |
| Malignant (M) | Ductal Carcinoma (DC) | 864 | 903 | 896 | 788 | 3451 |
| Lobular Carcinoma (LC) | 156 | 170 | 163 | 137 | 626 | |
| Mucinous Carcinoma (MC) | 205 | 222 | 196 | 169 | 792 | |
| Papillary Carcinoma (PC) | 145 | 142 | 135 | 138 | 560 | |
| Total | 1995 | 2081 | 2013 | 1820 | 7909 |
| Magnification | Method | Metric (%) | |||
|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1-Score | ||
| 40× | EfficientNetB0 | 89.22 ± 1.08 | 87.84 ± 1.88 | 87.08 ± 0.62 | 87.38 ± 1.0 |
| EfficientNetB0-MSE | 90.47 ± 1.23 | 89.65 ± 0.94 | 88.35 ± 2.62 | 88.85 ± 1.70 | |
| EfficientNetB0-MSE + FC Loss | 96.54 ± 0.50 | 96.69 ± 0.90 | 94.27 ± 0.15 | 94.61 ± 0.50 | |
| FCE-EfficientNetB0 + TL | 98.71 ± 0.63 | 97.06 ± 0.67 | 98.57 ± 0.27 | 97.06 ± 0.97 | |
| 100× | EfficientNetB0 | 89.21 ± 2.41 | 87.30 ± 2.32 | 87.55 ± 3.89 | 87.34 ± 3.13 |
| EfficientNetB0-MSE | 91.37 ± 0.98 | 89.32 ± 1.51 | 91.18 ± 2.77 | 90.14 ± 1.12 | |
| EfficientNetB0-MSE + FC Loss | 95.33 ± 1.80 | 94.71 ± 1.77 | 95.57 ± 0.45 | 95.66 ± 1.69 | |
| FCE-EfficientNetB0 + TL | 98.65 ± 0.21 | 97.28 ± 0.66 | 97.51 ± 0.63 | 97.26 ± 0.78 | |
| 200× | EfficientNetB0 | 92.81 ± 1.53 | 91.85 ± 1.79 | 91.33 ± 2.09 | 91.54 ± 1.85 |
| EfficientNetB0-MSE | 93.14 ± 0.62 | 91.99 ± 0.68 | 92.75 ± 1.22 | 92.09 ± 0.76 | |
| EfficientNetB0-MSE + FC Loss | 96.40 ± 0.87 | 95.21 ± 1.01 | 97.06 ± 0.52 | 95.67 ± 0.89 | |
| FCE-EfficientNetB0 + TL | 98.72 ± 0.42 | 98.63 ± 0.63 | 98.26 ± 0.76 | 98.67 ± 0.46 | |
| 400× | EfficientNetB0 | 91.85 ± 1.44 | 90.82 ± 2.14 | 90.96 ± 1.92 | 90.74 ± 1.60 |
| EfficientNetB0-MSE | 92.03 ± 0.99 | 90.96 ± 1.48 | 91.53 ± 1.95 | 91.03 ± 1.20 | |
| EfficientNetB0-MSE + FC Loss | 96.35 ± 1.15 | 96.01 ± 1.70 | 97.24 ± 0.67 | 97.14 ± 1.13 | |
| FCE-EfficientNetB0 + TL | 98.74 ± 0.28 | 98.16 ± 0.59 | 98.52 ± 0.23 | 98.53 ± 0.22 | |
| Mixed | EfficientNetB0 | 93.78 ± 0.66 | 93.45 ± 0.17 | 91.99 ± 1.44 | 92.64 ± 0.89 |
| EfficientNetB0-MSE | 94.22 ± 0.60 | 93.94 ± 0.84 | 93.08 ± 0.52 | 93.25 ± 0.68 | |
| EfficientNetB0-MSE + FC Loss | 95.50 ± 0.56 | 96.85 ± 0.81 | 96.31 ± 0.49 | 97.57 ± 0.52 | |
| FCE-EfficientNetB0 + TL | 98.59 ± 0.40 | 98.56 ± 0.32 | 98.36 ± 0.41 | 98.26 ± 0.56 | |
| Magnification | Method | Metric (%) | |||
|---|---|---|---|---|---|
| Accuracy | Precision | Recall | F1-Score | ||
| 40× | EfficientNetB0 | 68.84 ± 0.82 | 64.25 ± 2.91 | 59.63 ± 2.22 | 59.63 ± 3.32 |
| EfficientNetB0-MSE | 76.94 ± 0.89 | 72.21 ± 1.83 | 69.87 ± 1.16 | 70.33 ± 1.13 | |
| EfficientNetB0-MSE + FC Loss | 86.23 ± 0.23 | 90.98 ± 0.38 | 88.26 ± 0.65 | 89.31 ± 0.18 | |
| FCE-EfficientNetB0 + TL | 95.17 ± 0.80 | 95.52 ± 0.34 | 95.78 ± 0.17 | 94.43 ± 0.37 | |
| 100× | EfficientNetB0 | 65.83 ± 2.07 | 62.13 ± 1.67 | 63.78 ± 1.65 | 62.11 ± 2.26 |
| EfficientNetB0-MSE | 74.18 ± 1.37 | 69.90 ± 1.22 | 71.38 ± 1.43 | 69.88 ± 1.23 | |
| EfficientNetB0-MSE + FC Loss | 90.31 ± 0.86 | 90.37 ± 0.24 | 89.63 ± 0.62 | 90.20 ± 0.72 | |
| FCE-EfficientNetB0 + TL | 92.07 ± 0.74 | 92.42 ± 0.21 | 92.05 ± 0.70 | 92.68 ± 0.95 | |
| 200× | EfficientNetB0 | 68.65 ± 1.10 | 61.48 ± 1.49 | 62.29 ± 1.31 | 60.72 ± 1.35 |
| EfficientNetB0-MSE | 73.20 ± 0.88 | 67.14 ± 1.05 | 65.60 ± 0.97 | 65.75 ± 0.98 | |
| EfficientNetB0-MSE + FC Loss | 88.44 ± 0.89 | 86.44 ± 0.24 | 86.74 ± 0.08 | 86.88 ± 0.03 | |
| FCE-EfficientNetB0 + TL | 90.45 ± 0.87 | 88.67 ± 1.06 | 88.50 ± 0.79 | 90.35 ± 0.75 | |
| 400× | EfficientNetB0 | 66.48 ± 1.20 | 59.04 ± 1.10 | 56.76 ± 1.60 | 57.05 ± 1.33 |
| EfficientNetB0-MSE | 69.69 ± 1.13 | 63.51 ± 1.40 | 64.93 ± 2.75 | 63.93 ± 1.77 | |
| EfficientNetB0-MSE + FC Loss | 89.17 ± 0.54 | 89.38 ± 0.94 | 85.64 ± 0.45 | 87.61 ± 0.43 | |
| FCE-EfficientNetB0 + TL | 90.26 ± 0.12 | 90.45 ± 0.37 | 91.24 ± 0.21 | 92.05 ± 0.31 | |
| Mixed | EfficientNetB0 | 78.49 ± 1.59 | 74.69 ± 2.52 | 75.18 ± 1.13 | 74.44 ± 1.78 |
| EfficientNetB0-MSE | 79.58 ± 0.62 | 75.93 ± 0.85 | 75.21 ± 0.96 | 76.26 ± 0.89 | |
| EfficientNetB0-MSE + FC Loss | 90.49 ± 0.99 | 90.14 ± 0.75 | 90.48 ± 0.02 | 89.63 ± 0.94 | |
| FCE-EfficientNetB0 + TL | 92.55 ± 0.69 | 92.33 ± 0.22 | 91.38 ± 0.85 | 91.53 ± 0.83 | |
| Model | Bottleneck | Parameters (M) | FLOPs (G) |
|---|---|---|---|
| ResNet50 | Residual | 23.51 | 4.13 |
| Inceptionv3 | Conv | 21.79 | 2.85 |
| DenseNet121 | Conv | 49.84 | 2.90 |
| EfficientNetB0 | MBConv | 4.01 | 0.41 |
| FCE-EfficientNetB0 | MSE-MBConv | 49.85 | 0.82 |
| Loss Function | 40× | 100× | 200× | 400× | Mean |
|---|---|---|---|---|---|
| Cross-Entropy Loss | 96.49 | 96.64 | 98.01 | 97.8 | 97.24 |
| Focal Loss | 95.36 | 94.72 | 95.25 | 94.49 | 94.96 |
| Focal Cosine loss | 98.71 | 98.65 | 98.72 | 98.74 | 98.70 |
| Reference | Method | Magnification | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|---|---|
| Abdulaal A H, et al. [32] | VGG19 + SAM | 40× | 97.49 | 96.80 | 95.28 | 96.03 |
| 100× | 96.71 | 95.68 | 94.33 | 95 | ||
| 200× | 96.03 | 94.40 | 92.91 | 93.65 | ||
| 400× | 97.53 | 95.76 | 96.58 | 96.17 | ||
| Dihin R A, et al. [33] | Gabor- EfficientNetV2 | 40× | 87 | 85.6 | 87.01 | 86.88 |
| 100× | 93.5 | 90.30 | 95 | 93.8 | ||
| 200× | 94.1 | 92.40 | 94.79 | 94.78 | ||
| 400× | 96.3 | 92.90 | 97.3 | 98.52 | ||
| Kaur A, et al. [34] | DML | 40× | 97.87 | 97.56 | 92.56 | 97.89 |
| 100× | 98.56 | 95.38 | 95.45 | 98.34 | ||
| 200× | 98.34 | 98.65 | 98.31 | 97.89 | ||
| 400× | 96.54 | 99.71 | 96.44 | 99.44 | ||
| He K, et al. [35] | ResNet50 | 40× | 84.71 | 82.20 | 82.34 | 82.27 |
| 100× | 87.5. | 86.44 | 83.70 | 84.86 | ||
| 200× | 89.33 | 89.12 | 85.44 | 86.95 | ||
| 400× | 88.74 | 88.33 | 85.49 | 86.68 | ||
| Sandler M, et al. [36] | MobileNetv2 | 40× | 97.24 | 97.44 | 97.93 | 97.50 |
| 100× | 97.84 | 97.20 | 97.80 | 97.49 | ||
| 200× | 97.02 | 97.40 | 95.64 | 96.46 | ||
| 400× | 98.08 | 97.35 | 98.36 | 97.83 | ||
| Huang G, et al. [37] | DenseNet121 | 40× | 84.71 | 82.20 | 82.34 | 82.27 |
| 100× | 87.5. | 86.44 | 83.70 | 84.86 | ||
| 200× | 89.33 | 89.12 | 85.44 | 86.95 | ||
| 400× | 88.74 | 88.33 | 85.49 | 86.68 | ||
| Szegedy C, et al. [38] | Inceptionv3 | 40× | 95.24 | 94.93 | 93.92 | 94.40 |
| 100× | 94.96 | 93.88 | 94.43 | 94.14 | ||
| 200× | 96.38 | 95.39 | 95.98 | 95.68 | ||
| 400× | 95.33 | 94.43 | 94.00 | 94.71 | ||
| This paper | FCE-EfficientNetB0 | 40× | 98.71 ± 0.63 | 97.06 ± 0.67 | 98.57 ± 0.27 | 97.06 ± 0.97 |
| 100× | 98.65 ± 0.21 | 97.28 ± 0.66 | 97.51 ± 0.63 | 97.26 ± 0.78 | ||
| 200× | 98.72 ± 0.42 | 98.63 ± 0.63 | 98.26 ± 0.76 | 98.67 ± 0.46 | ||
| 400× | 98.74 ± 0.28 | 98.16 ± 0.59 | 98.52 ± 0.23 | 98.53 ± 0.22 |
| Reference | Method | Magnification | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|---|---|
| Bardou et al. [39] | Ensemble CNN model | 40× | 88.23 | 84.27 | 83.79 | 83.74 |
| 100× | 84.64 | 84.29 | 84.48 | 84.31 | ||
| 200× | 83.31 | 81.85 | 80.83 | 80.48 | ||
| 400× | 83.98 | 80.84 | 81.03 | 80.63 | ||
| Sharma et al. [40] | VGG19 + SVM (L, 1) (balanced + augmented data) | 40× | 92.64 | 92.00 | 92.00 | 92.00 |
| 100× | 91.25 | 91.00 | 91.00 | 91.00 | ||
| 200× | 81.42 | 82.00 | 82.00 | 82.00 | ||
| 400× | 80.84 | 82.00 | 81.00 | 82.00 | ||
| Taheri et al. [41] | MLF2-CNN | 40× | 90.14 | 88.57 | 82.76 | 86 |
| 100× | 91.38 | 88.02 | 86.96 | 86 | ||
| 200× | 91.45 | 88.1 | 87.14 | 90 | ||
| 400× | 89.9 | 88.57 | 82.76 | 86 | ||
| He K, et al. [35] | ResNet50 | 40× | 83.16 | 79.21 | 86.61 | 79.92 |
| 100× | 86.83 | 79.66 | 83.04 | 85.01 | ||
| 200× | 80.79 | 80.62 | 83.79 | 86.69 | ||
| 400× | 80.71 | 81.55 | 80.23 | 79.33 | ||
| Sandler M, et al. [36] | MobileNetv2 | 40× | 89.22 | 85.84 | 86.42 | 87.37 |
| 100× | 85.61 | 84.69 | 84.17 | 84.06 | ||
| 200× | 87.34 | 86.90 | 86.10 | 85.25 | ||
| 400× | 85.23 | 83.11 | 84.83 | 83.68 | ||
| Huang G, et al. [37] | DenseNet121 | 40× | 87.97 | 85.31 | 86.41 | 86.43 |
| 100× | 85.61 | 82.03 | 82.53 | 83.23 | ||
| 200× | 85.16 | 84.83 | 82.65 | 81.96 | ||
| 400× | 84.20 | 83.64 | 83.79 | 83.15 | ||
| Szegedy C, et al. [38] | Inceptionv3 | 40× | 85.46 | 83.03 | 82.23 | 82.14 |
| 100× | 83.21 | 81.77 | 80.35 | 80.15 | ||
| 200× | 81.89 | 88.76 | 87.25 | 87.02 | ||
| 400× | 88.02 | 83.52 | 83.23 | 83.29 | ||
| This paper | FCE-EfficientNetB0 | 40× | 95.17 ± 0.80 | 95.52 ± 0.34 | 95.78 ± 0.17 | 94.43 ± 0.37 |
| 100× | 92.07 ± 0.74 | 92.42 ± 0.21 | 92.05 ± 0.70 | 92.68 ± 0.95 | ||
| 200× | 90.45 ± 0.87 | 88.67 ± 1.06 | 88.50 ± 0.79 | 90.35 ± 0.75 | ||
| 400× | 90.26 ± 0.12 | 90.45 ± 0.37 | 91.24 ± 0.21 | 92.05 ± 0.31 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.; Pei, Y.; Wu, M.; Wang, J. Focal Cosine-Enhanced EfficientNetB0: A Novel Approach to Classifying Breast Histopathological Images. Information 2025, 16, 444. https://doi.org/10.3390/info16060444
Liu M, Pei Y, Wu M, Wang J. Focal Cosine-Enhanced EfficientNetB0: A Novel Approach to Classifying Breast Histopathological Images. Information. 2025; 16(6):444. https://doi.org/10.3390/info16060444
Chicago/Turabian StyleLiu, Min, Yuzhen Pei, Minghu Wu, and Juan Wang. 2025. "Focal Cosine-Enhanced EfficientNetB0: A Novel Approach to Classifying Breast Histopathological Images" Information 16, no. 6: 444. https://doi.org/10.3390/info16060444
APA StyleLiu, M., Pei, Y., Wu, M., & Wang, J. (2025). Focal Cosine-Enhanced EfficientNetB0: A Novel Approach to Classifying Breast Histopathological Images. Information, 16(6), 444. https://doi.org/10.3390/info16060444






