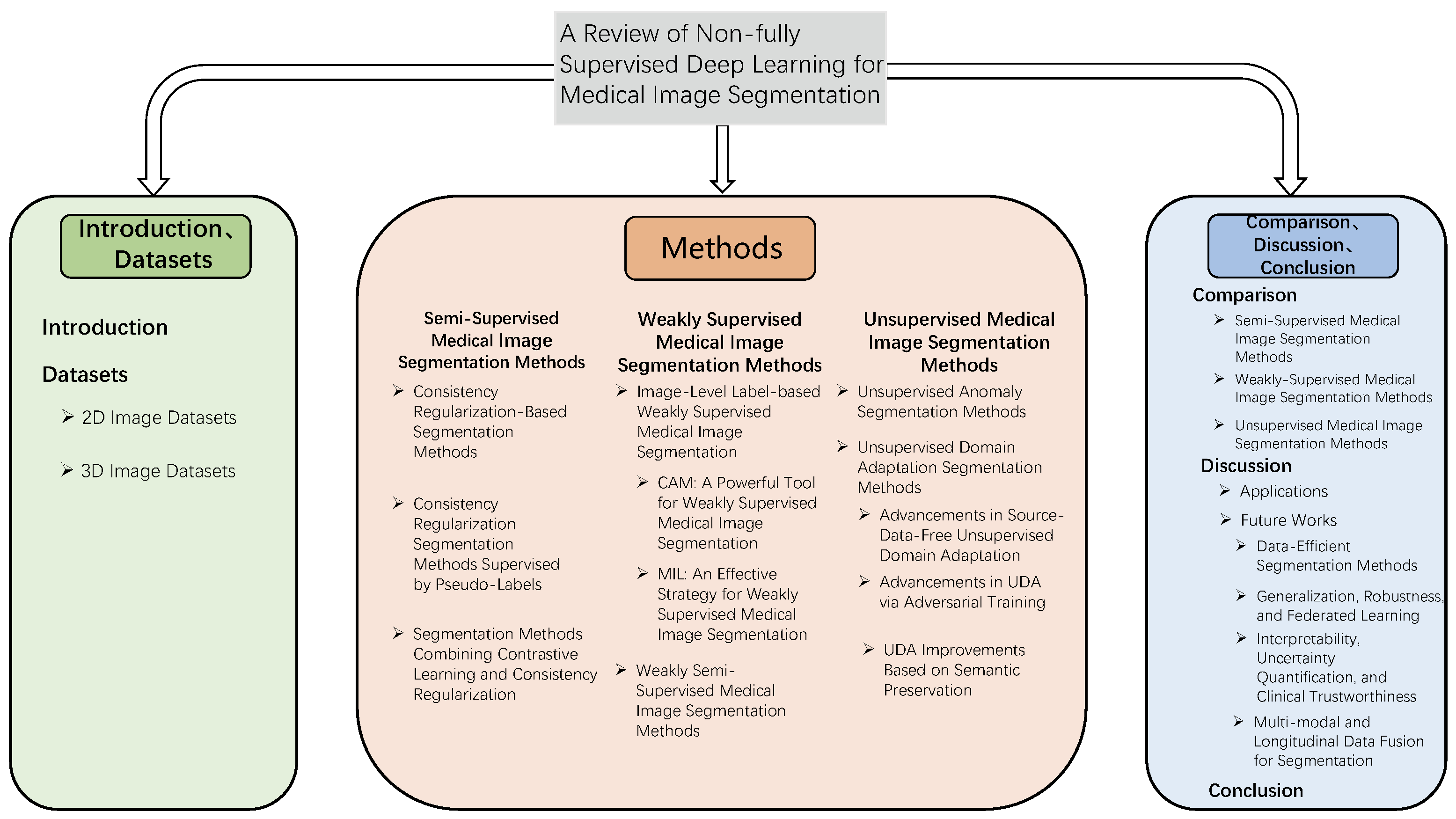
A Review of Non-Fully Supervised Deep Learning for Medical Image Segmentation
Abstract
1. Introduction
2. Datasets
2.1. Two-Dimensional Image Datasets
2.2. Three-Dimensional Image Datasets
3. Semi-Supervised Medical Image Segmentation Methods
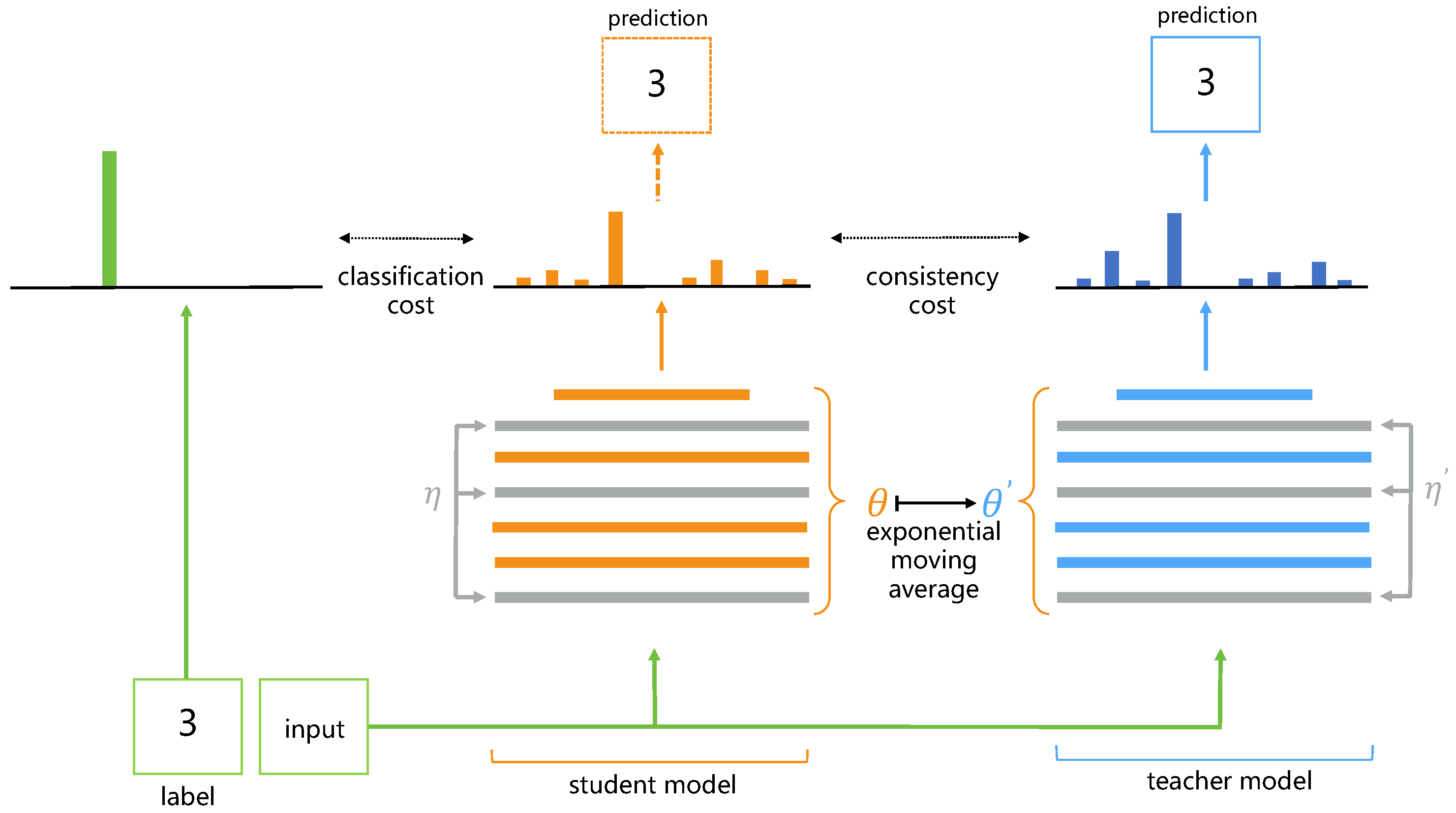
3.1. Consistency Regularization-Based Segmentation Methods
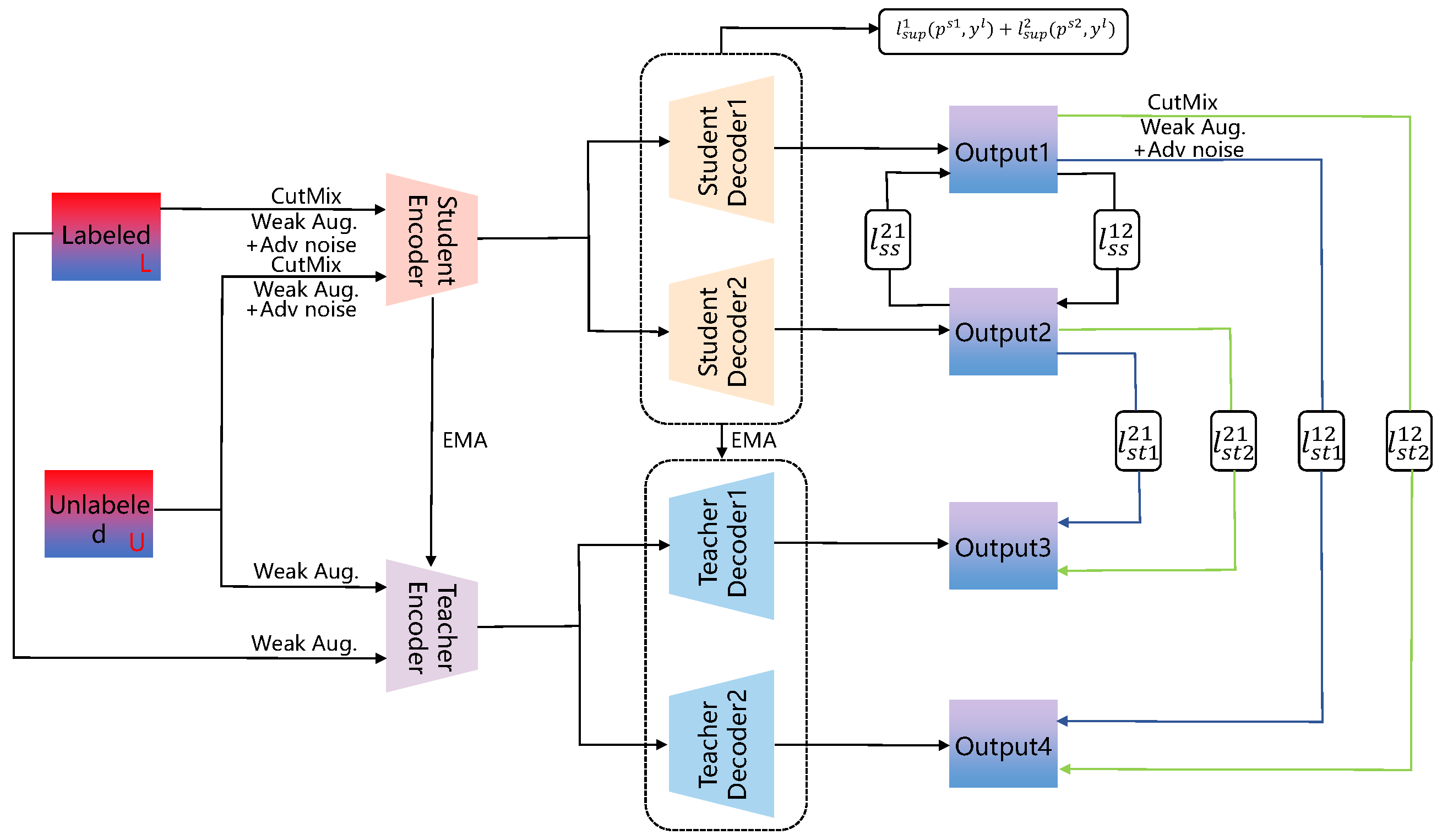
3.2. Consistency Regularization Segmentation Methods Supervised by Pseudo-Labels
3.3. Segmentation Methods Combining Contrastive Learning and Consistency Regularization
- Context-aware consistency path (green path): Two overlapping patches, and , cropped from the unlabeled image are passed through the shared backbone network. Their resulting features are mapped through a projection head (Φ) to obtain embeddings and . A contrastive loss, , is employed to enforce feature consistency under differing contextual views.
- Cross-consistency training path (brown path): Features extracted from the complete unlabeled image are fed into the main classifier to yield prediction . Concurrently, these features, subjected to perturbation (P), are input to multiple auxiliary classifiers, producing predictions . A cross-consistency loss, , enforces consistency between the outputs of the main and auxiliary classifiers.
4. Weakly Supervised Medical Image Segmentation Methods
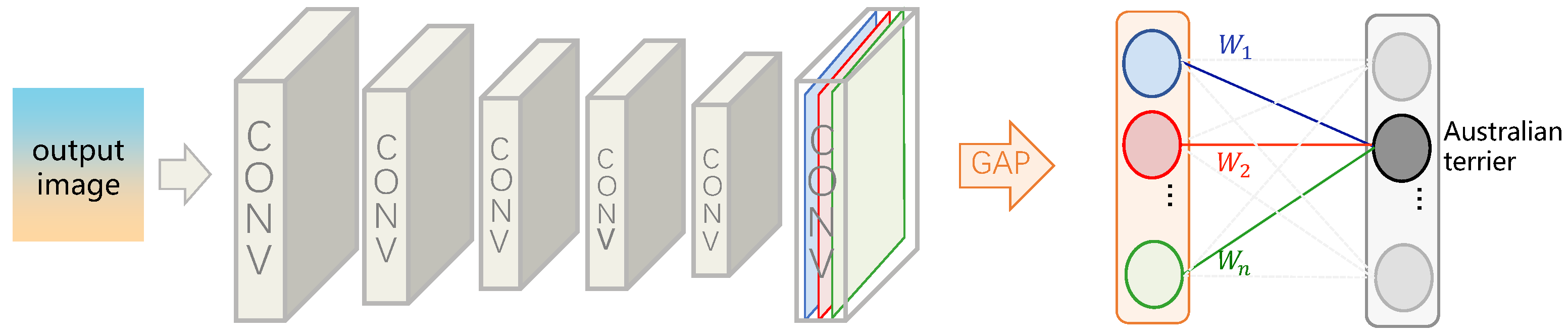
4.1. Image-Level Label-Based Weakly Supervised Medical Image Segmentation
4.1.1. CAM: A Powerful Tool for Weakly Supervised Medical Image Segmentation
| Algorithm 1 Training algorithm |
|
4.1.2. MIL: An Effective Strategy for Weakly Supervised Medical Image Segmentation
4.2. Weakly Semi-Supervised Medical Image Segmentation Methods
5. Unsupervised Medical Image Segmentation Methods
5.1. Unsupervised Anomaly Segmentation Methods
5.2. Unsupervised Domain Adaptation Segmentation Methods
5.2.1. Advancements in Source-Data-Free Unsupervised Domain Adaptation
5.2.2. Advancements in UDA via Adversarial Training
5.2.3. UDA Improvements Based on Semantic Preservation
6. Comparison
6.1. Semi-Supervised Medical Image Segmentation Methods
6.2. Weakly Supervised Medical Image Segmentation Methods
6.3. Unsupervised Medical Image Segmentation Methods
7. Discussion
7.1. Applications
7.2. Future Works
7.2.1. Data-Efficient Segmentation Methods
7.2.2. Generalization, Robustness, and Federated Learning
7.2.3. Interpretability, Uncertainty Quantification, and Clinical Trustworthiness
7.2.4. Multi-Modal and Longitudinal Data Fusion for Segmentation
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Commun. ACM 2012, 60, 84–90. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. arXiv 2015, arXiv:1411.4038. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. Lect. Notes Comput. Sci. 2015, 9351, 234–241. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention Is All You Need. In Advances in Neural Information Processing Systems; The MIT Press: Cambridge, MA, USA, 2017; pp. 5999–6009. [Google Scholar]
- Kirillov, A.; Mintun, E.; Ravi, N.; Mao, H.; Rolland, C.; Gustafson, L.; Xiao, T.; Whitehead, S.; Berg, A.C.; Lo, W.Y.; et al. Segment Anything. arXiv 2023, arXiv:2304.02643. [Google Scholar]
- Baur, C.; Wiestler, B.; Albarqouni, S.; Navab, N. Deep autoencoding models for unsupervised anomaly segmentation in brain MR images. Lect. Notes Comput. Sci. 2019, 11383, 161–169. [Google Scholar]
- Yarkony, J.; Wang, S. Accelerating Message Passing for MAP with Benders Decomposition. arXiv 2018, arXiv:1805.04958. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning Deep Features for Discriminative Localization. arXiv 2016, arXiv:1512.04150. [Google Scholar]
- Dai, J.; He, K.; Sun, J. BoxSup: Exploiting Bounding Boxes to Supervise Convolutional Networks for Semantic Segmentation. arXiv 2015, arXiv:1503.01640. [Google Scholar]
- Ballan, L.; Castaldo, F.; Alahi, A.; Palmieri, F.; Savarese, S. Knowledge transfer for scene-specific motion prediction. Lect. Notes Comput. Sci. 2016, 9905, 697–713. [Google Scholar]
- Tanaka, K. Minimal networks for sensor counting problem using discrete Euler calculus. Jpn. J. Ind. Appl. Math. 2017, 34, 229–242. [Google Scholar] [CrossRef]
- Wei, Y.; Feng, J.; Liang, X.; Cheng, M.M.; Zhao, Y.; Yan, S. Object Region Mining With Adversarial Erasing: A Simple Classification to Semantic Segmentation Approach. arXiv 2017, arXiv:1703.08448. [Google Scholar]
- Hu, F.; Wang, Y.; Ma, B.; Wang, Y. Emergency supplies research on crossing points of transport network based on genetic algorithm. In Proceedings of the 2015 International Conference on Intelligent Transportation, Big Data and Smart City, ICITBS 2015, Halong Bay, Vietnam, 19–20 December 2015; pp. 370–375. [Google Scholar] [CrossRef]
- Gannon, S.; Kulosman, H. The condition for a cyclic code over Z4 of odd length to have a complementary dual. arXiv 2019, arXiv:1905.12309. [Google Scholar]
- Abraham, N.; Khan, N.M. A novel focal tversky loss function with improved attention u-net for lesion segmentation. In Proceedings of the International Symposium on Biomedical Imaging, Venice, Italy, 8–11 April 2019; pp. 683–687. [Google Scholar] [CrossRef]
- Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Ballester, M.A.G.; et al. Deep Learning Techniques for Automatic MRI Cardiac Multi-Structures Segmentation and Diagnosis: Is the Problem Solved? IEEE Trans. Med. Imaging 2018, 37, 2514–2525. [Google Scholar] [CrossRef]
- Graham, S.; Chen, H.; Gamper, J.; Dou, Q.; Heng, P.A.; Snead, D.; Tsang, Y.W.; Rajpoot, N. MILD-Net: Minimal information loss dilated network for gland instance segmentation in colon histology images. Med. Image Anal. 2019, 52, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Demner-Fushman, D.; Kohli, M.D.; Rosenman, M.B.; Shooshan, S.E.; Rodriguez, L.; Antani, S.; Thoma, G.R.; McDonald, C.J. Preparing a collection of radiology examinations for distribution and retrieval. J. Am. Med. Inform. Assoc. JAMIA 2016, 23, 304–310. [Google Scholar] [CrossRef]
- Johnson, A.E.; Pollard, T.J.; Berkowitz, S.J.; Greenbaum, N.R.; Lungren, M.P.; Deng, C.Y.; Mark, R.G.; Horng, S. MIMIC-CXR, a de-identified publicly available database of chest radiographs with free-text reports. Sci. Data 2019, 6, 317. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, D.; Xu, C.; Wang, W.; Hong, Q.; Li, Q.; Tian, J. TFCNs: A CNN-Transformer Hybrid Network for Medical Image Segmentation. Lect. Notes Comput. Sci. 2022, 13532, 781–792. [Google Scholar] [CrossRef]
- Bannur, S.; Hyland, S.; Liu, Q.; Pérez-García, F.; Ilse, M.; Castro, D.C.; Boecking, B.; Sharma, H.; Bouzid, K.; Thieme, A.; et al. Learning to Exploit Temporal Structure for Biomedical Vision-Language Processing. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Juan, PR, USA, 17–19 June 1997; pp. 15016–15027. [Google Scholar] [CrossRef]
- McCollough, C.H.; Bartley, A.C.; Carter, R.E.; Chen, B.; Drees, T.A.; Edwards, P.; Holmes, D.R.; Huang, A.E.; Khan, F.; Leng, S.; et al. Low-dose CT for the detection and classification of metastatic liver lesions: Results of the 2016 Low Dose CT Grand Challenge. Med. Phys. 2017, 44, e339–e352. [Google Scholar] [CrossRef]
- Leuschner, J.; Schmidt, M.; Baguer, D.O.; Maass, P. LoDoPaB-CT, a benchmark dataset for low-dose computed tomography reconstruction. Sci. Data 2021, 8, 109. [Google Scholar] [CrossRef]
- Moen, T.R.; Chen, B.; Holmes, D.R.; Duan, X.; Yu, Z.; Yu, L.; Leng, S.; Fletcher, J.G.; McCollough, C.H. Low-dose CT image and projection dataset. Med. Phys. 2021, 48, 902–911. [Google Scholar] [CrossRef]
- Xiong, Z.; Xia, Q.; Hu, Z.; Huang, N.; Bian, C.; Zheng, Y.; Vesal, S.; Ravikumar, N.; Maier, A.; Yang, X.; et al. A global benchmark of algorithms for segmenting the left atrium from late gadolinium-enhanced cardiac magnetic resonance imaging. Med. Image Anal. 2021, 67, 101832. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Vendt, B.; Smith, K.; Freymann, J.; Kirby, J.; Koppel, P.; Moore, S.; Phillips, S.; Maffitt, D.; Pringle, M.; et al. The cancer imaging archive (TCIA): Maintaining and operating a public information repository. J. Digit. Imaging 2013, 26, 1045–1057. [Google Scholar] [CrossRef]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef]
- Liew, S.Q.; Ngoh, G.C.; Yusoff, R.; Teoh, W.H. Acid and Deep Eutectic Solvent (DES) extraction of pectin from pomelo (Citrus grandis (L.) Osbeck) peels. Biocatal. Agric. Biotechnol. 2018, 13, 1–11. [Google Scholar] [CrossRef]
- Petzsche, M.R.H.; de la Rosa, E.; Hanning, U.; Wiest, R.; Valenzuela, W.; Reyes, M.; Meyer, M.; Liew, S.L.; Kofler, F.; Ezhov, I.; et al. ISLES 2022: A multi-center magnetic resonance imaging stroke lesion segmentation dataset. Sci. Data 2022, 9, 762. [Google Scholar] [CrossRef] [PubMed]
- Maier, O.; Menze, B.H.; von der Gablentz, J.; Häni, L.; Heinrich, M.P.; Liebrand, M.; Winzeck, S.; Basit, A.; Bentley, P.; Chen, L.; et al. ISLES 2015—A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Med. Image Anal. 2017, 35, 250–269. [Google Scholar] [CrossRef]
- Hakim, A.; Christensen, S.; Winzeck, S.; Lansberg, M.G.; Parsons, M.W.; Lucas, C.; Robben, D.; Wiest, R.; Reyes, M.; Zaharchuk, G. Predicting Infarct Core From Computed Tomography Perfusion in Acute Ischemia With Machine Learning: Lessons From the ISLES Challenge. Stroke 2021, 52, 2328–2337. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.; Han, K.; Li, X.; Cheng, X.; Li, Y.; Wang, Y.; Yu, Y. Symmetry-Enhanced Attention Network for Acute Ischemic Infarct Segmentation with Non-contrast CT Images. Lect. Notes Comput. Sci. 2021, 12907, 432–441. [Google Scholar] [CrossRef]
- Campello, V.M.; Gkontra, P.; Izquierdo, C.; Martin-Isla, C.; Sojoudi, A.; Full, P.M.; Maier-Hein, K.; Zhang, Y.; He, Z.; Ma, J.; et al. Multi-Centre, Multi-Vendor and Multi-Disease Cardiac Segmentation: The MMs Challenge. IEEE Trans. Med. Imaging 2021, 40, 3543–3554. [Google Scholar] [CrossRef]
- Heller, N.; Isensee, F.; Maier-Hein, K.H.; Hou, X.; Xie, C.; Li, F.; Nan, Y.; Mu, G.; Lin, Z.; Han, M.; et al. The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: Results of the KiTS19 challenge. Med. Image Anal. 2021, 67, 101821. [Google Scholar] [CrossRef]
- Littlejohns, T.J.; Holliday, J.; Gibson, L.M.; Garratt, S.; Oesingmann, N.; Alfaro-Almagro, F.; Bell, J.D.; Boultwood, C.; Collins, R.; Conroy, M.C.; et al. The UK Biobank imaging enhancement of 100,000 participants: Rationale, data collection, management and future directions. Nat. Commun. 2020, 11, 2624. [Google Scholar] [CrossRef] [PubMed]
- Bilic, P.; Christ, P.; Li, H.B.; Vorontsov, E.; Ben-Cohen, A.; Kaissis, G.; Szeskin, A.; Jacobs, C.; Mamani, G.E.H.; Chartrand, G.; et al. The Liver Tumor Segmentation Benchmark (LiTS). Med. Image Anal. 2023, 84, 102680. [Google Scholar] [CrossRef] [PubMed]
- Kavur, A.E.; Gezer, N.S.; Barış, M.; Aslan, S.; Conze, P.H.; Groza, V.; Pham, D.D.; Chatterjee, S.; Ernst, P.; Özkan, S.; et al. CHAOS Challenge-combined (CT-MR) healthy abdominal organ segmentation. Med. Image Anal. 2021, 69, 101950. [Google Scholar] [CrossRef]
- Zhang, P.; Zhang, B.; Chen, D.; Yuan, L.; Wen, F. Cross-domain correspondence learning for exemplar-based image translation. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Juan, PR, USA, 17–19 June 1997; pp. 5142–5152. [Google Scholar] [CrossRef]
- Sohn, K.; Berthelot, D.; Li, C.L.; Zhang, Z.; Carlini, N.; Cubuk, E.D.; Kurakin, A.; Zhang, H.; Raffel, C. FixMatch: Simplifying Semi-Supervised Learning with Consistency and Confidence. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Chen, H.; Tao, R.; Fan, Y.; Wang, Y.; Wang, J.; Schiele, B.; Xie, X.; Raj, B.; Savvides, M. SoftMatch: Addressing the Quantity-Quality Trade-off in Semi-supervised Learning. In Proceedings of the 11th International Conference on Learning Representations, ICLR, Kigali, Rwanda, 1–5 May 2023. [Google Scholar]
- Wang, X.; Tang, F.; Chen, H.; Cheung, C.Y.; Heng, P.A. Deep semi-supervised multiple instance learning with self-correction for DME classification from OCT images. Med. Image Anal. 2023, 83, 102673. [Google Scholar] [CrossRef]
- Tarvainen, A.; Valpola, H. Mean teachers are better role models: Weight-averaged consistency targets improve semi-supervised deep learning results. Adv. Neural Inf. Process. Syst. 2017, 2017, 1196–1205. [Google Scholar]
- Yang, L.; Qi, L.; Feng, L.; Zhang, W.; Shi, Y. Revisiting Weak-to-Strong Consistency in Semi-Supervised Semantic Segmentation. arXiv 2023, arXiv:2208.09910. [Google Scholar]
- Lyu, F.; Ye, M.; Carlsen, J.F.; Erleben, K.; Darkner, S.; Yuen, P.C. Pseudo-Label Guided Image Synthesis for Semi-Supervised COVID-19 Pneumonia Infection Segmentation. IEEE Trans. Med. Imaging 2023, 42, 797–809. [Google Scholar] [CrossRef] [PubMed]
- Bashir, R.M.S.; Qaiser, T.; Raza, S.E.; Rajpoot, N.M. Consistency regularisation in varying contexts and feature perturbations for semi-supervised semantic segmentation of histology images. Med. Image Anal. 2024, 91, 102997. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, G.; Zhang, T.; Wang, R.; Su, J. Semi-supervised medical image segmentation via weak-to-strong perturbation consistency and edge-aware contrastive representation. Med. Image Anal. 2025, 101, 103450. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Y.; Wu, J.; Lu, J.; Ye, Y.; Huang, Y.; Dou, X.; Li, K.; Wang, G.; Zhang, S.; et al. A novel one-to-multiple unsupervised domain adaptation framework for abdominal organ segmentation. Med. Image Anal. 2023, 88, 102873. [Google Scholar] [CrossRef]
- V., S.A.; Dolz, J.; Lombaert, H. Anatomically-aware uncertainty for semi-supervised image segmentation. Med. Image Anal. 2024, 91, 103011. [Google Scholar] [CrossRef]
- Yu, L.; Wang, S.; Li, X.; Fu, C.W.; Heng, P.A. Uncertainty-Aware Self-ensembling Model for Semi-supervised 3D Left Atrium Segmentation. Lect. Notes Comput. Sci. 2019, 11765, 605–613. [Google Scholar]
- Luo, X.; Wang, G.; Liao, W.; Chen, J.; Song, T.; Chen, Y.; Zhang, S.; Metaxas, D.N.; Zhang, S. Semi-supervised medical image segmentation via uncertainty rectified pyramid consistency. Med. Image Anal. 2022, 80, 102517. [Google Scholar] [CrossRef]
- Li, W.; Bian, R.; Zhao, W.; Xu, W.; Yang, H. Diversity matters: Cross-head mutual mean-teaching for semi-supervised medical image segmentation. Med. Image Anal. 2024, 97, 103302. [Google Scholar] [CrossRef]
- Wu, Y.; Ge, Z.; Zhang, D.; Xu, M.; Zhang, L.; Xia, Y.; Cai, J. Mutual consistency learning for semi-supervised medical image segmentation. Med. Image Anal. 2022, 81, 102530. [Google Scholar] [CrossRef]
- Bai, Y.; Chen, D.; Li, Q.; Shen, W.; Wang, Y. Bidirectional Copy-Paste for Semi-Supervised Medical Image Segmentation. arXiv 2023, arXiv:2305.00673. [Google Scholar]
- Su, J.; Luo, Z.; Lian, S.; Lin, D.; Li, S. Mutual learning with reliable pseudo label for semi-supervised medical image segmentation. Med. Image Anal. 2024, 94, 103111. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 4th International Conference on 3D Vision, 3DV, Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Wang, Y.; Song, K.; Liu, Y.; Ma, S.; Yan, Y.; Carneiro, G. Leveraging labelled data knowledge: A cooperative rectification learning network for semi-supervised 3D medical image segmentation. Med. Image Anal. 2025, 101, 103461. [Google Scholar] [CrossRef]
- Chaitanya, K.; Erdil, E.; Karani, N.; Konukoglu, E. Local contrastive loss with pseudo-label based self-training for semi-supervised medical image segmentation. Med. Image Anal. 2023, 87, 102792. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, Z.; Ma, J.; Li, Z.; Zhang, S. Correlation-Aware Mutual Learning for Semi-supervised Medical Image Segmentation. Lect. Notes Comput. Sci. 2023, 14220, 98–108. [Google Scholar] [CrossRef]
- Li, S.; Zhang, C.; He, X. Shape-Aware Semi-supervised 3D Semantic Segmentation for Medical Images. Lect. Notes Comput. Sci. 2020, 12261, 552–561. [Google Scholar] [CrossRef]
- Wang, R.; Chen, S.; Ji, C.; Fan, J.; Li, Y. Boundary-aware context neural network for medical image segmentation. Med. Image Anal. 2022, 78, 102395. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Chen, J.; Song, T.; Wang, G. Semi-supervised Medical Image Segmentation through Dual-task Consistency. Proc. AAAI Conf. Artif. Intell. 2021, 35, 8801–8809. [Google Scholar] [CrossRef]
- Shi, J.; Gao, W. Transverse ultimate capacity of U-type stiffened panels for hatch covers used in ship cargo holds. Ships Offshore Struct. 2021, 16, 608–619. [Google Scholar] [CrossRef]
- Peng, J.; Wang, P.; Desrosiers, C.; Pedersoli, M. Self-Paced Contrastive Learning for Semi-supervised Medical Image Segmentation with Meta-labels. Adv. Neural Inf. Process. Syst. 2021, 20, 16686–16699. [Google Scholar]
- Oh, S.J.; Benenson, R.; Khoreva, A.; Akata, Z.; Fritz, M.; Schiele, B. Exploiting saliency for object segmentation from image level labels. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR, Honolulu, HI, USA, 21–26 July 2016; pp. 5038–5047. [Google Scholar] [CrossRef]
- Durieux, G.; Irles, A.; Miralles, V.; Peñuelas, A.; Perelló, M.; Pöschl, R.; Vos, M. The electro-weak couplings of the top and bottom quarks—Global fit and future prospects. J. High Energy Phys. 2019, 2019, 12. [Google Scholar] [CrossRef]
- Kervadec, H.; Dolz, J.D.; Montral, D.; Wang, S.; Granger, E.G.; Montral, G.; Ben, I.; Ayed, A.; Montral, A. Bounding boxes for weakly supervised segmentation: Global constraints get close to full supervision. Proc. Mach. Learn. Res. 2020, 121, 365–380. [Google Scholar]
- Lin, D.; Dai, J.; Jia, J.; He, K.; Sun, J. ScribbleSup: Scribble-Supervised Convolutional Networks for Semantic Segmentation. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Juan, PR, USA, 17–19 June 1997; pp. 3159–3167. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, J.; Kan, M.; Shan, S.; Chen, X. Self-Supervised Equivariant Attention Mechanism for Weakly Supervised Semantic Segmentation. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Juan, PR, USA, 17–19 June 1997; pp. 12272–12281. [Google Scholar] [CrossRef]
- Dietterich, T.G.; Lathrop, R.H.; Lozano-Pérez, T. Solving the multiple instance problem with axis-parallel rectangles. Artif. Intell. 1997, 89, 31–71. [Google Scholar] [CrossRef]
- Chikontwe, P.; Sung, H.J.; Jeong, J.; Kim, M.; Go, H.; Nam, S.J.; Park, S.H. Weakly supervised segmentation on neural compressed histopathology with self-equivariant regularization. Med. Image Anal. 2022, 80, 102482. [Google Scholar] [CrossRef]
- Patel, G.; Dolz, J. Weakly supervised segmentation with cross-modality equivariant constraints. Med. Image Anal. 2022, 77, 102374. [Google Scholar] [CrossRef]
- Chattopadhay, A.; Sarkar, A.; Howlader, P.; Balasubramanian, V.N. Grad-CAM++: Generalized gradient-based visual explanations for deep convolutional networks. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision, WACV, Lake Tahoe, NV, USA, 12–15 March 2018; pp. 839–847. [Google Scholar] [CrossRef]
- Yang, J.; Mehta, N.; Demirci, G.; Hu, X.; Ramakrishnan, M.S.; Naguib, M.; Chen, C.; Tsai, C.L. Anomaly-guided weakly supervised lesion segmentation on retinal OCT images. Med. Image Anal. 2024, 94, 103139. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Wang, T.; Wu, X.; Hua, X.S.; Zhang, H.; Sun, Q. Class Re-Activation Maps for Weakly-Supervised Semantic Segmentation. arXiv 2022, arXiv:2203.00962. [Google Scholar]
- Zhang, W.; Zhu, L.; Hallinan, J.; Zhang, S.; Makmur, A.; Cai, Q.; Ooi, B.C. BoostMIS: Boosting Medical Image Semi-Supervised Learning With Adaptive Pseudo Labeling and Informative Active Annotation. arXiv 2022, arXiv:2203.02533. [Google Scholar]
- Li, K.; Qian, Z.; Han, Y.; Chang, E.I.; Wei, B.; Lai, M.; Liao, J.; Fan, Y.; Xu, Y. Weakly supervised histopathology image segmentation with self-attention. Med. Image Anal. 2023, 86, 102791. [Google Scholar] [CrossRef] [PubMed]
- Yao, T.; Pan, Y.; Li, Y.; Ngo, C.W.; Mei, T. Wave-ViT: Unifying Wavelet and Transformers for Visual Representation Learning. Lect. Notes Comput. Sci. 2022, 13685, 328–345. [Google Scholar] [CrossRef]
- Cheng, B.; Parkhi, O.; Kirillov, A. Pointly-Supervised Instance Segmentation. arXiv 2022, arXiv:2104.06404. [Google Scholar]
- Seeböck, P.; Orlando, J.I.; Michl, M.; Mai, J.; Schmidt-Erfurth, U.; Bogunović, H. Anomaly guided segmentation: Introducing semantic context for lesion segmentation in retinal OCT using weak context supervision from anomaly detection. Med. Image Anal. 2024, 93, 103104. [Google Scholar] [CrossRef]
- Gao, F.; Hu, M.; Zhong, M.E.; Feng, S.; Tian, X.; Meng, X.; Ni-jia ti, M.; Huang, Z.; Lv, M.; Song, T.; et al. Segmentation only uses sparse annotations: Unified weakly and semi-supervised learning in medical images. Med. Image Anal. 2022, 80, 102515. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, H.; Ji, H.; Liu, H.; Li, Y.; He, N.; Wei, D.; Huang, Y.; Dai, Q.; Wu, J.; et al. A deep weakly semi-supervised framework for endoscopic lesion segmentation. Med. Image Anal. 2023, 90, 102973. [Google Scholar] [CrossRef]
- Ahn, J.; Shin, S.Y.; Shim, J.; Kim, Y.H.; Han, S.J.; Choi, E.K.; Oh, S.; Shin, J.Y.; Choe, J.C.; Park, J.S.; et al. Association between epicardial adipose tissue and embolic stroke after catheter ablation of atrial fibrillation. J. Cardiovasc. Electrophysiol. 2019, 30, 2209–2216. [Google Scholar] [CrossRef]
- Viniavskyi, O.; Dobko, M.; Dobosevych, O. Weakly-Supervised Segmentation for Disease Localization in Chest X-Ray Images. Lect. Notes Comput. Sci. 2020, 12299, 249–259. [Google Scholar] [CrossRef]
- Ma, X.; Ji, Z.; Niu, S.; Leng, T.; Rubin, D.L.; Chen, Q. MS-CAM: Multi-Scale Class Activation Maps for Weakly-Supervised Segmentation of Geographic Atrophy Lesions in SD-OCT Images. IEEE J. Biomed. Health Inform. 2020, 24, 3443–3455. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, J.; Xia, Y. TransWS: Transformer-Based Weakly Supervised Histology Image Segmentation. Lect. Notes Comput. Sci. 2022, 13583, 367–376. [Google Scholar] [CrossRef]
- Wang, T.; Niu, S.; Dong, J.; Chen, Y. Weakly Supervised Retinal Detachment Segmentation Using Deep Feature Propagation Learning in SD-OCT Images. Lect. Notes Comput. Sci. 2020, 12069, 146–154. [Google Scholar] [CrossRef]
- Silva-Rodríguez, J.; Naranjo, V.; Dolz, J. Constrained unsupervised anomaly segmentation. Med. Image Anal. 2022, 80, 102526. [Google Scholar] [CrossRef]
- Pinaya, W.H.; Tudosiu, P.D.; Gray, R.; Rees, G.; Nachev, P.; Ourselin, S.; Cardoso, M.J. Unsupervised brain imaging 3D anomaly detection and segmentation with transformers. Med. Image Anal. 2022, 79, 102475. [Google Scholar] [CrossRef]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef]
- Kingma, D.P.; Welling, M. Auto-Encoding Variational Bayes. In Proceedings of the 2nd International Conference on Learning Representations, ICLR 2014—Conference Track Proceedings, Banff, AB, Canada, 14–16 April 2014. [Google Scholar] [CrossRef]
- Schlegl, T.; Seeböck, P.; Waldstein, S.M.; Langs, G.; Schmidt-Erfurth, U. f-AnoGAN: Fast unsupervised anomaly detection with generative adversarial networks. Med. Image Anal. 2019, 54, 30–44. [Google Scholar] [CrossRef]
- Stan, S.; Rostami, M. Unsupervised model adaptation for source-free segmentation of medical images. Med. Image Anal. 2024, 95, 103179. [Google Scholar] [CrossRef]
- Liu, X.; Xing, F.; Fakhri, G.E.; Woo, J. Memory consistent unsupervised off-the-shelf model adaptation for source-relaxed medical image segmentation. Med. Image Anal. 2023, 83, 102641. [Google Scholar] [CrossRef]
- Sun, Y.; Dai, D.; Xu, S. Rethinking adversarial domain adaptation: Orthogonal decomposition for unsupervised domain adaptation in medical image segmentation. Med. Image Anal. 2022, 82, 102623. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Xin, J.; You, C.; Shi, P.; Dong, S.; Dvornek, N.C.; Zheng, N.; Duncan, J.S. Style mixup enhanced disentanglement learning for unsupervised domain adaptation in medical image segmentation. Med. Image Anal. 2025, 101, 103440. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Zhang, R.; Diao, S.; Zhu, J.; Yuan, Y.; Cai, J.; Shao, L.; Li, S.; Qin, W. Dual domain distribution disruption with semantics preservation: Unsupervised domain adaptation for medical image segmentation. Med. Image Anal. 2024, 97, 103275. [Google Scholar] [CrossRef] [PubMed]
- Dou, Q.; Ouyang, C.; Chen, C.; Chen, H.; Glocker, B.; Zhuang, X.; Heng, P.A. PnP-AdaNet: Plug-and-Play Adversarial Domain Adaptation Network with a Benchmark at Cross-modality Cardiac Segmentation. arXiv 2018, arXiv:1812.07907. [Google Scholar] [CrossRef]
- Vu, T.H.; Jain, H.; Bucher, M.; Cord, M.; Perez, P. ADVENT: Adversarial Entropy Minimization for Domain Adaptation in Semantic Segmentation. arXiv 2019, arXiv:1811.12833. [Google Scholar]
- Chen, C.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.A. Unsupervised Bidirectional Cross-Modality Adaptation via Deeply Synergistic Image and Feature Alignment for Medical Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 2494–2505. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhuang, X. CF Distance: A New Domain Discrepancy Metric and Application to Explicit Domain Adaptation for Cross-Modality Cardiac Image Segmentation. IEEE Trans. Med. Imaging 2020, 39, 4274–4285. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhu, Z.; Zheng, S.; Liu, Y.; Zhou, J.; Zhao, Y. Margin Preserving Self-Paced Contrastive Learning Towards Domain Adaptation for Medical Image Segmentation. IEEE J. Biomed. Health Inform. 2022, 26, 638–647. [Google Scholar] [CrossRef]
- Chen, J.; Huang, W.; Zhang, J.; Debattista, K.; Han, J. Addressing inconsistent labeling with cross image matching for scribble-based medical image segmentation. IEEE Trans. Image Process. 2025, 34, 842–853. [Google Scholar] [CrossRef]
- Gao, W.; Wan, F.; Pan, X.; Peng, Z.; Tian, Q.; Han, Z.; Zhou, B.; Ye, Q. TS-CAM: Token Semantic Coupled Attention Map for Weakly Supervised Object Localization. arXiv 2021, arXiv:2103.14862. [Google Scholar]
- Mahapatra, D. Generative Adversarial Networks and Domain Adaptation for Training Data Independent Image Registration. arXiv 2019, arXiv:1910.08593. [Google Scholar] [CrossRef]
- Wang, G.; Li, W.; Zuluaga, M.A.; Pratt, R.; Patel, P.A.; Aertsen, M.; Doel, T.; David, A.L.; Deprest, J.; Ourselin, S.; et al. Interactive Medical Image Segmentation Using Deep Learning with Image-Specific Fine Tuning. IEEE Trans. Med. Imaging 2018, 37, 1562–1573. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Dou, Q.; Chen, H.; Qin, J.; Heng, P.A. Synergistic Image and Feature Adaptation: Towards Cross-Modality Domain Adaptation for Medical Image Segmentation. Proc. AAAI Conf. Artif. Intell. 2019, 33, 865–872. [Google Scholar] [CrossRef]
- Lei, T.; Zhang, D.; Du, X.; Wang, X.; Wan, Y.; Nandi, A.K. Semi-Supervised Medical Image Segmentation Using Adversarial Consistency Learning and Dynamic Convolution Network. IEEE Trans. Med. Imaging 2023, 42, 1265–1277. [Google Scholar] [CrossRef]
- Kalinicheva, E.; Ienco, D.; Sublime, J.; Trocan, M. Unsupervised Change Detection Analysis in Satellite Image Time Series Using Deep Learning Combined with Graph-Based Approaches. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 1450–1466. [Google Scholar] [CrossRef]
- Kamnitsas, K.; Baumgartner, C.; Ledig, C.; Newcombe, V.; Simpson, J.; Kane, A.; Menon, D.; Nori, A.; Criminisi, A.; Rueckert, D.; et al. Unsupervised Domain Adaptation in Brain Lesion Segmentation with Adversarial Networks. Lect. Notes Comput. Sci. 2017, 10265, 597–609. [Google Scholar] [CrossRef]
- Hoffman, J.; Tzeng, E.; Park, T.; Zhu, J.Y.; Isola, P.; Saenko, K.; Efros, A.; Darrell, T. CyCADA: Cycle-Consistent Adversarial Domain Adaptation. arXiv 2018, arXiv:1711.03213. [Google Scholar]
- Zhang, Y.; Miao, S.; Mansi, T.; Liao, R. Task driven generative modeling for unsupervised domain adaptation: Application to X-ray image segmentation. Lect. Notes Comput. Sci. 2018, 11071, 599–607. [Google Scholar]

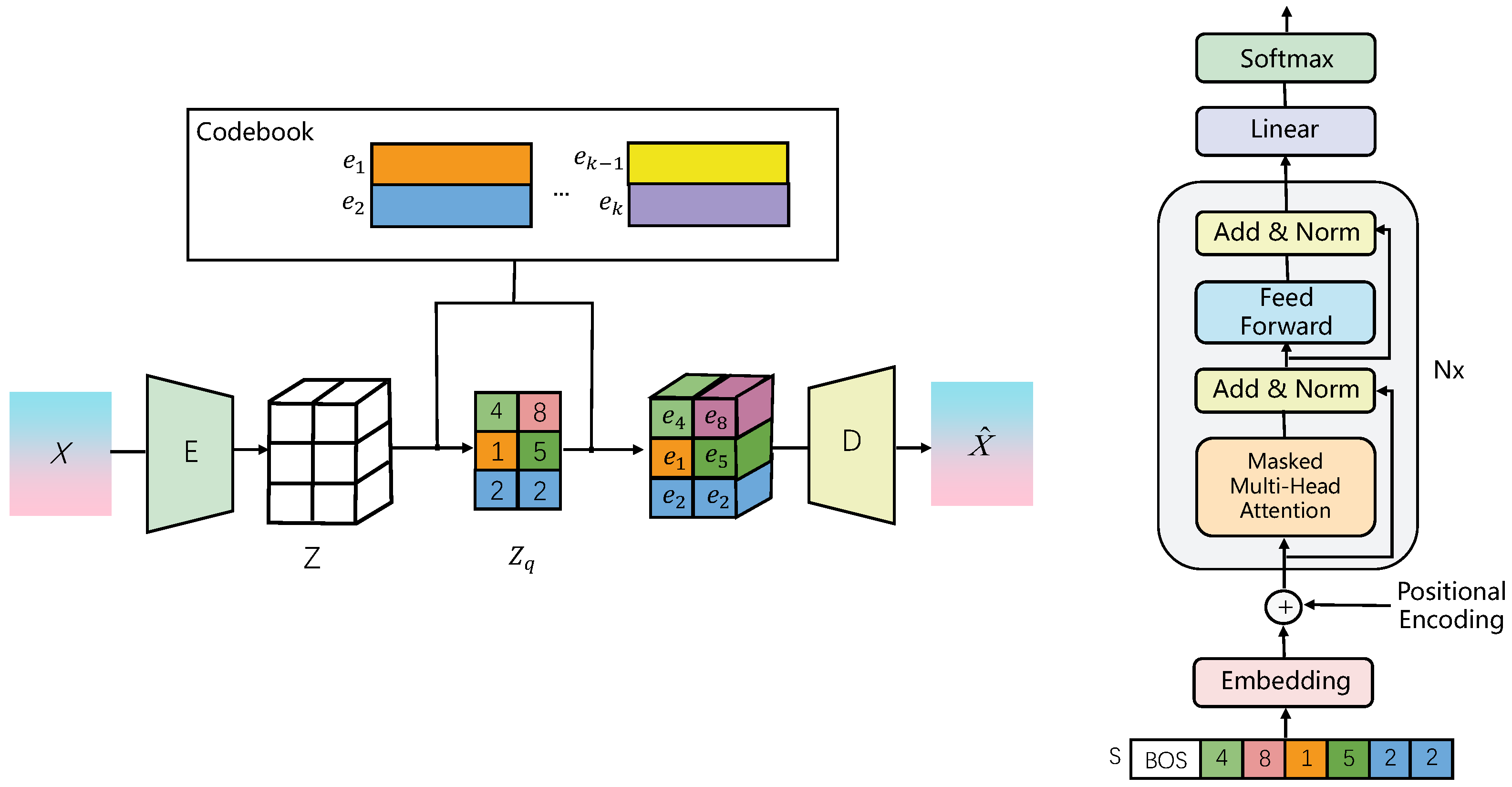
| Dimension | Semi-Supervised Learning (SSL) | Weakly Supervised Learning (WSL) | Unsupervised Learning (UL) |
|---|---|---|---|
| Supervision Source | Small amount of precise labels + large amount of unlabeled data | Coarse-grained/indirect labels (e.g., image-level tags, bounding boxes, points/scribbles) | No direct segmentation labels; relies on inherent data structure or priors |
| Annotation Cost | Moderate | Low | Very low/none (for the target task) |
| Core Mechanism | Leverages unlabeled data to boost performance (e.g., consistency regularization, pseudo-labeling) | Infers strong segmentation from weak signals (e.g., CAMs, MIL) | Discovers inherent data patterns/anomalies (e.g., clustering, reconstruction, UDA) |
| Performance Potential | High; can approach fully supervised | Moderate to high; depends on weak label quality | Generally lower than supervised; valuable for specific tasks (e.g., anomaly detection) |
| Application Scenarios | Few precise labels but abundant unlabeled data; enhancing robustness | Precise labeling difficult but coarse information available; large-scale screening | No labels available; exploratory analysis; domain adaptation |
| Dataset | Modality | Anatomical Area | Application Scenarios | Supervision Type |
|---|---|---|---|---|
| ACDC [16] | MRI | Heart (left and right ventricles) | Cardiac function analysis, ventricular segmentation | Fully supervised |
| Colorectal adenocarcinoma glands [17] | Pathology Sections (H&E staining) | Colorectal tissue | Segmentation of the glandular structure | Fully supervised |
| IU Chest X-ray [18] | X-ray (chest X-ray) | Chest (cardiopulmonary area) | Classification of lung diseases | Weakly supervised |
| MIMIC-CXR [19] | X-ray (chest X-ray) + clinical report | Chest | Automatic diagnosis of multiple diseases | Weakly supervised |
| COV-CTR [20] | CT (chest) | Lung | COVID-19 severity rating | Weakly supervised |
| MS-CXR-T [21] | X-ray (chest X-ray) | Chest | Multilingual report generation | Weakly supervised |
| NIH-AAPM-Mayo Clinical LDCT [22] | Low-dose CT (chest) | Lung | Lung nodule detection | Fully supervised |
| LoDoPaB [23] | Low-dose CT (simulation) | Body | CT reconstruction algorithm development | Fully supervised |
| LDCT [24] | Low-dose CT | Chest/abdomen | Radiation dose reduction studies | Fully supervised |
| Dataset | Modality | Anatomical Area | Application Scenarios | Supervision Type |
|---|---|---|---|---|
| LA [25] | MRI | Heart (left atrium) | Surgical planning for atrial fibrillation | Fully supervised |
| Pancreas-CT [26] | CT (abdomen) | Pancreas | Pancreatic tumor segmentation | Fully supervised |
| BraTS [27] | Multiparametric MRI | Brain (glioma) | Brain tumor segmentation | Fully supervised |
| ATLAS [28] | MRI (T1) | Brain (stroke lesions) | Stroke analysis | Fully supervised |
| ISLES [29,30,31] | MRI (multiple sequences) | Brain | Ischemic stroke segmentation | Fully supervised |
| AISD [32] | Ultrasonic | Abdominal organs | Organ boundary segmentation | Fully supervised |
| Cardiac [33] | MRI | Heart | Ventricular division | Fully supervised |
| KiTS19 [34] | CT (abdomen) | Kidney | Segmentation of kidney tumors | Fully supervised |
| UKB [35] | MRI/CT/X-ray | Body | Multi-organ phenotypic analysis | Weakly supervised |
| LiTS [36] | CT (abdomen) | Liver | Segmentation of liver tumors | Fully supervised |
| CHAOS [37] | CT/MRI (abdomen) | Multi-organ | Cross-modal organ segmentation | Fully supervised |
| Method | % Labeled | 2017 ACDC (2D) | |||
|---|---|---|---|---|---|
| Scans | DSC (%) | Jaccard (%) | 95HD (mm) | ASD (mm) | |
| Using 5% labeled scans | |||||
| UAMT [49] | 5 | 51.23 (1.96) | 41.82 (1.62) | 17.13 (2.82) | 7.76 (2.01) |
| SASSNet [59] | 5 | 58.47 (1.74) | 47.04 (2.02) | 18.04 (3.63) | 7.31 (1.53) |
| Tri-U-MT [60] | 5 | 59.15 (2.01) | 47.37 (1.82) | 17.37 (2.77) | 7.34 (1.31) |
| DTC [61] | 5 | 57.09 (1.57) | 45.61 (1.23) | 20.63 (2.61) | 7.05 (1.94) |
| CoraNet [62] | 5 | 59.91 (2.08) | 48.37 (1.75) | 15.53 (2.23) | 5.96 (1.42) |
| SPCL [63] | 5 | 81.82 (1.24) | 70.62 (1.04) | 5.96 (1.62) | 2.21 (0.29) |
| MC-Net+ [52] | 5 | 63.47 (1.75) | 53.13 (1.41) | 7.38 (1.68) | 2.37 (0.32) |
| URPC [50] | 5 | 62.57 (1.18) | 52.75 (1.36) | 7.79 (1.85) | 2.64 (0.36) |
| PLCT [57] | 5 | 78.42 (1.45) | 67.43 (1.25) | 6.54 (1.62) | 2.48 (0.24) |
| DGCL [41] | 5 | 80.57 (1.12) | 68.74 (0.96) | 6.04 (1.73) | 2.17 (0.30) |
| CAML [58] | 5 | 79.04 (0.83) | 68.45 (0.97) | 6.28 (1.79) | 2.24 (0.26) |
| DCNet [40] | 5 | 71.57 (1.58) | 61.12 (1.19) | 8.37 (1.92) | 4.08 (0.84) |
| SFPC [43] | 5 | 80.52 (1.03) | 68.73 (0.88) | 6.08 (1.47) | 2.14 (0.22) |
| Using 10% labeled scans | |||||
| UAMT [49] | 10 | 81.86 (1.25) | 71.07 (1.43) | 12.92 (1.68) | 3.49 (0.64) |
| SASSNet [59] | 10 | 84.61 (1.97) | 74.53 (1.78) | 6.02 (1.54) | 1.71 (0.35) |
| Tri-U-MT [60] | 10 | 84.06 (1.69) | 74.32 (1.77) | 7.41 (1.63) | 2.59 (0.51) |
| DTC [61] | 10 | 82.91 (1.65) | 71.61 (1.81) | 8.69 (1.84) | 3.04 (0.59) |
| CoraNet [62] | 10 | 84.56 (1.53) | 74.41 (1.49) | 6.11 (1.15) | 2.35 (0.44) |
| SPCL [63] | 10 | 87.57 (1.15) | 78.63 (0.89) | 4.87 (0.79) | 1.31 (0.27) |
| MC-Net+ [52] | 10 | 86.78 (1.41) | 77.31 (1.27) | 6.92 (0.95) | 2.04 (0.37) |
| URPC [50] | 10 | 85.18 (0.98) | 74.65 (0.83) | 5.01 (0.79) | 1.52 (0.26) |
| PLCT [57] | 10 | 86.83 (1.17) | 77.04 (0.83) | 6.62 (0.86) | 2.27 (0.42) |
| DGCL [41] | 10 | 87.74 (1.06) | 78.82 (1.22) | 4.74 (0.73) | 1.56 (0.24) |
| CAML [58] | 10 | 87.67 (0.83) | 78.70 (0.91) | 4.97 (0.62) | 1.35 (0.17) |
| DCNet [40] | 10 | 87.81 (0.88) | 78.96 (0.94) | 4.84 (0.81) | 1.23 (0.21) |
| SFPC [43] | 10 | 87.76 (0.92) | 78.94 (0.83) | 4.90 (0.74) | 1.28 (0.23) |
| Method | % Labeled | BraTS2020 (3D) | |||
|---|---|---|---|---|---|
| Scans | DSC (%) | Jaccard (%) | 95HD (mm) | ASD (mm) | |
| Using 5% labeled scans | |||||
| UAMT [49] | 5 | 49.46 (2.51) | 38.46 (1.86) | 19.57 (3.28) | 6.54 (0.86) |
| SASSNet [59] | 5 | 51.82 (1.74) | 43.93 (1.42) | 23.47 (2.83) | 7.47 (1.09) |
| Tri-U-MT [60] | 5 | 53.95 (1.97) | 44.33 (2.18) | 19.68 (3.06) | 7.29 (0.84) |
| DTC [61] | 5 | 56.72 (2.04) | 45.78 (1.67) | 17.38 (4.31) | 6.28 (1.22) |
| CoraNet [62] | 5 | 57.97 (1.83) | 46.40 (1.64) | 19.52 (2.80) | 5.83 (0.85) |
| SPCL [63] | 5 | 78.73 (1.54) | 67.90 (1.29) | 16.26 (1.68) | 4.47 (1.08) |
| MC-Net+ [52] | 5 | 58.91 (1.47) | 47.24 (1.36) | 20.82 (3.35) | 7.14 (1.12) |
| URPC [50] | 5 | 60.48 (2.01) | 50.69 (1.99) | 18.21 (3.27) | 7.12 (0.95) |
| PLCT [57] | 5 | 65.74 (2.17) | 55.40 (1.85) | 16.61 (3.04) | 6.85 (1.39) |
| DGCL [41] | 5 | 80.21 (0.75) | 68.86 (0.63) | 14.91 (1.53) | 4.63 (1.16) |
| CAML [58] | 5 | 77.86 (0.96) | 66.42 (1.37) | 15.21 (1.74) | 5.10 (1.12) |
| DCNet [40] | 5 | 78.52 (1.21) | 67.81 (1.07) | 17.37 (1.48) | 4.32 (0.96) |
| SFPC [43] | 5 | 80.76 (0.74) | 69.18 (0.83) | 14.87 (1.92) | 4.02 (0.75) |
| Using 10% labeled scans | |||||
| UAMT [49] | 10 | 81.04 (1.46) | 68.88 (1.57) | 17.27 (3.35) | 6.25 (1.63) |
| SASSNet [59] | 10 | 82.36 (2.08) | 71.03 (2.35) | 14.80 (3.72) | 4.11 (1.54) |
| Tri-U-MT [60] | 10 | 82.83 (1.35) | 71.52 (1.21) | 15.19 (2.86) | 3.57 (1.30) |
| DTC [61] | 10 | 81.98 (2.41) | 70.41 (2.73) | 16.27 (3.62) | 3.62 (1.71) |
| CoraNet [62] | 10 | 81.38 (1.68) | 70.01 (1.83) | 13.94 (2.72) | 3.95 (1.26) |
| SPCL [63] | 10 | 84.65 (1.16) | 73.91 (1.19) | 12.24 (1.47) | 3.28 (0.42) |
| MC-Net+ [52] | 10 | 83.93 (1.73) | 72.34 (1.69) | 13.52 (2.74) | 3.37 (1.13) |
| URPC [50] | 10 | 84.23 (1.41) | 72.37(1.26) | 11.52 (1.79) | 3.26 (1.14) |
| PLCT [57] | 10 | 83.66 (1.82) | 71.99 (1.67) | 13.68 (1.29) | 3.59 (1.02) |
| DGCL [41] | 10 | 84.02 (1.24) | 72.16 (1.07) | 12.98 (1.28) | 3.02 (0.96) |
| CAML [58] | 10 | 84.34 (1.03) | 73.84 (0.92) | 12.02 (1.84) | 3.31 (0.58) |
| DCNet [40] | 10 | 83.39 (0.97) | 71.94 (0.88) | 11.93 (1.24) | 3.50 (0.33) |
| SFPC [43] | 10 | 85.01 (0.89) | 74.67 (1.14) | 10.73 (1.36) | 3.03 (0.31) |
| Dataset | RESC | Duke | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Lesion | BG | SRF | PED | BG | Fluid | |||||
| Metric | DSC | mIoU | DSC | mIoU | DSC | mIoU | DSC | mIoU | DSC | mIoU |
| IRNet [82] | 98.88% | 97.78% | 49.18% | 33.75% | 22.98% | 14.66% | 99.02% | 98.10% | 17.79% | 20.45% |
| SEAM [68] | 98.69% | 97.43% | 46.44% | 34.13% | 28.09% | 10.71% | 98.48% | 97.03% | 25.48% | 17.87% |
| ReCAM [74] | 98.81% | 97.66% | 31.19% | 14.23% | 31.99% | 19.11% | 98.16% | 96.41% | 18.91% | 11.67% |
| WSMIS [83] | 96.90% | 95.64% | 45.91% | 24.64% | 10.34% | 2.96% | 98.16% | 96.41% | 0.42% | 0.42% |
| MSCAM [84] | 98.59% | 97.25% | 18.52% | 10.14% | 17.03% | 11.97% | 98.98% | 98.00% | 29.93% | 17.98% |
| TransWS [85] | 99.07% | 98.18% | 52.44% | 34.88% | 30.28% | 17.22% | 99.06% | 98.15% | 37.58% | 27.01% |
| DFP [86] | 98.83% | 97.72% | 20.39% | 6.40% | 31.39% | 15.64% | 99.10% | 98.24% | 27.53% | 15.14% |
| AGM [73] | 99.15% | 98.34% | 57.84% | 43.94% | 34.03% | 22.33% | 99.13% | 98.29% | 40.17% | 30.06% |
| Methods | Cardiac MRI → Cardiac CT | Cardiac CT → Cardiac MRI | ||
|---|---|---|---|---|
| AA | AA | |||
| Dice (%) | ASSD (mm) | Dice (%) | ASSD (mm) | |
| Supervised training | ||||
| (upper bound) | 92.0 ± 7.2 | 1.5 ± 0.8 | 80.12 ± 4.0 | 4.2 ± 1.9 |
| Without adaptation | ||||
| (lower bound) | 0.1 ± 0.1 | 51.0 ± 9.1 | 18.1 ± 13.7 | 32.9 ± 4.7 |
| One-shot Finetune | 46.2 ± 9.2 | 10.7 ± 2.1 | 39.9 ± 11.2 | 8.2 ± 1.5 |
| Five-shot Finetune | 73.1 ± 3.4 | 8.6 ± 1.7 | 39.5 ± 10.3 | 8.5 ± 1.2 |
| PnP-AdaNet [97] | 74.0 ± 21.1 | 24.9 ± 6.7 | 43.7 ± 6.2 | 3.1 ± 2.2 |
| AdvEnt [98] | 84.2 ± 3.0 | 9.1 ± 4.1 | 53.0 ± 5.9 | 6.9 ± 1.7 |
| SIFA [99] | 81.3 ± 5.7 | 7.9 ± 2.7 | 65.3 ± 10.9 | 7.3 ± 5.0 |
| VarDA [100] | 81.9 ± 9.1 | 8.1 ± 5.0 | 54.6 ± 9.3 | 15.5 ± 4.5 |
| BMCAN [101] | 83.0 ± 6.8 | 5.8 ± 4.1 | 72.2 ± 4.3 | 3.7 ± 2.6 |
| DAAM [74] | 87.0 ± 2.1 | 5.4 ± 3.0 | 76.0 ± 7.3 | 6.8 ± 3.2 |
| ADR [94] | 87.9 ± 3.6 | 5.9 ± 4.4 | 69.7 ± 4.2 | 5.1 ± 2.1 |
| MPSCL [101] | 86.8 ± 2.6 | 7.7 ± 3.9 | 64.6 ± 4.7 | 4.5 ± 2.3 |
| SMEDL [95] | 88.3 ± 3.5 | 4.3 ± 2.3 | 80.12 ± 4.0 | 4.2 ± 1.9 |
| Method | Authors (Year) | Key Feature | Application Domain(s) | Strengths |
|---|---|---|---|---|
| AC-MT [47] | Xu et al. (2023) | Ambiguity recognition module selectively calculates consistency loss | Medical image segmentation | High-ambiguity-pixel screening with entropy and selective consistency learning improves segmentation index |
| AAU-Net [48] | Adiga V. et al. (2024) | Uncertainty estimation of anatomical prior (DAE) | Abdominal CT multi-organ segmentation | Denoising Autoencoder optimizes prediction anatomy rationality and improves DSC/HD |
| CMMT-Net [51] | Li et al. (2024) | Cross-head mutual-aid mean teaching and multi-level perturbations | Medical image segmentation on LA, Pancreas-CT, ACDC | Multi-head decoder enhances prediction diversity and improves Dice |
| MLRPL [54] | Su et al. (2024) | Collaborative learning framework with dual reliability evaluation | Medical image segmentation (e.g., Pancreas-CT) | Dual decoders with mutual comparison strategy, achieves near-fully supervised performance |
| CRLN [56] | Wang et al. (2025) | Prototype learning and dynamic interaction correction for pseudo-labeling | 3D medical image segmentation (LA, Pancreas-CT, BraTS19) | Multi-prototype learning captures intra-class diversity to enhance generalization |
| CRCFP [45] | Bashir et al. (2024) | Context-aware contrast and cross-consistency training | Histopathology image segmentation (BCSS, MoNuSeg) | Dual-path unsupervised learning with lightweight classifier, achieves near-fully supervised performance |
| AGM [73] | Yang et al. (2024) | Iterative refinement learning stage | Handling small size, low contrast, and multiple co-existing lesions in medical images | Enhances lesion localization accuracy |
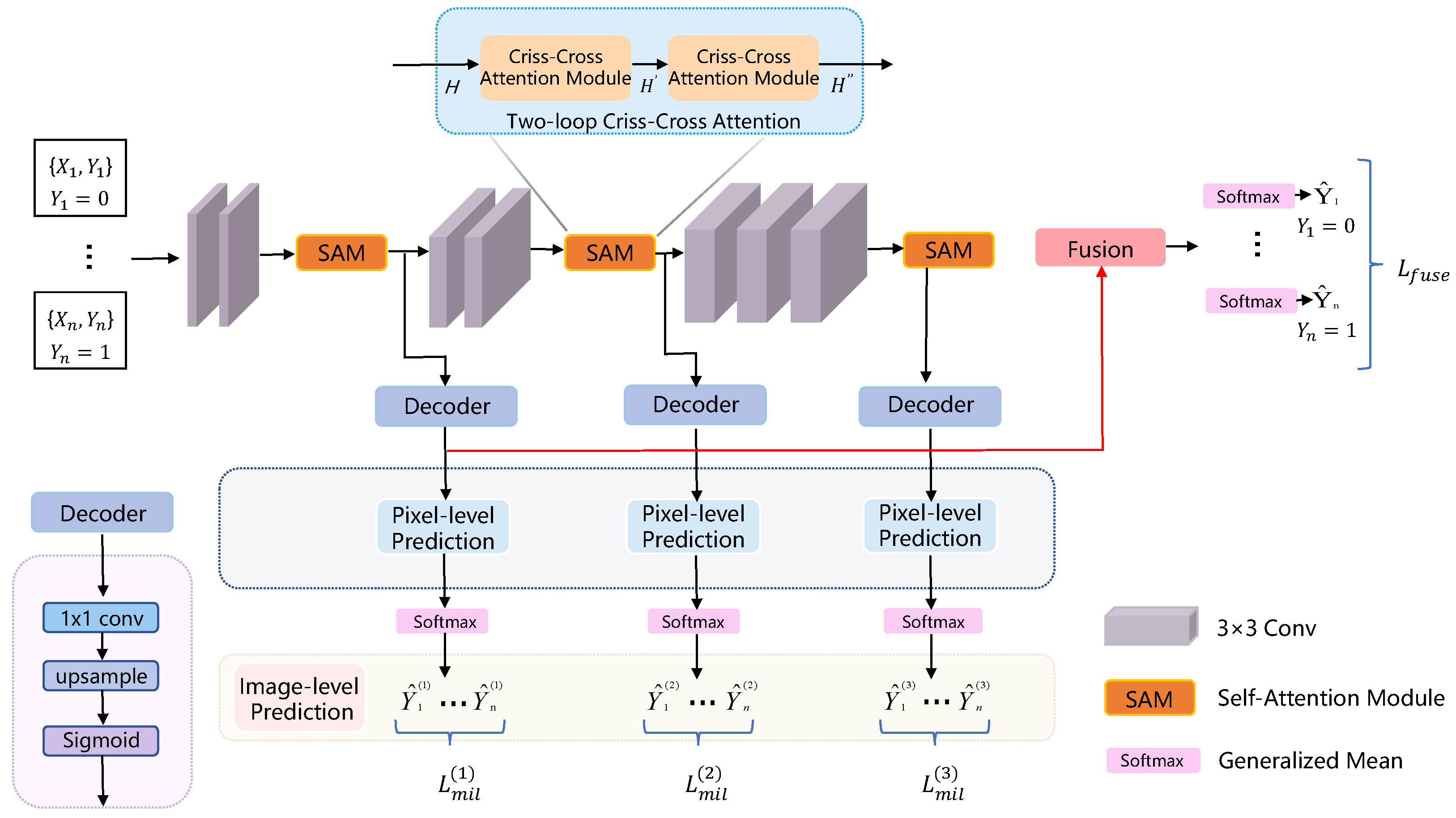
| SA-MIL [76] | Li et al. (2023) | Criss-Cross Attention | Better differentiation between foreground (e.g., cancerous regions) and background | Enhances feature representation capability |
| Method | Authors (Year) | Key Feature | Application Domain(s) | Strengths |
|---|---|---|---|---|
| SOUSA [80] | Gao et al. (2022) | Multi-angle projection reconstruction loss | More accurate segmentation boundaries, fewer false positive regions | Significantly improves segmentation accuracy |
| Point SEGTR [81] | Shi et al. (2023) | Fuses limited pixel-level annotations with abundant point-level annotations | Endoscopic image analysis | Significantly reduces dependency on pixel-level annotations |
| VAE [87] | Silva-Rodríguez et al. (2022) | Attention mechanism (Grad-CAM) + extended log-barrier method | Unsupervised anomaly detection and segmentation; lesion detection and localization | Effectively separates activation distributions of normal and abnormal patterns |
| OSUDA [93] | Liu et al. (2023) | Exponential momentum decay; High-order BN Statistics Consistency Loss | Source-free unsupervised domain adaptation (SFUDA); privacy-preserving knowledge transfer | Improves performance and stability in the target domain |
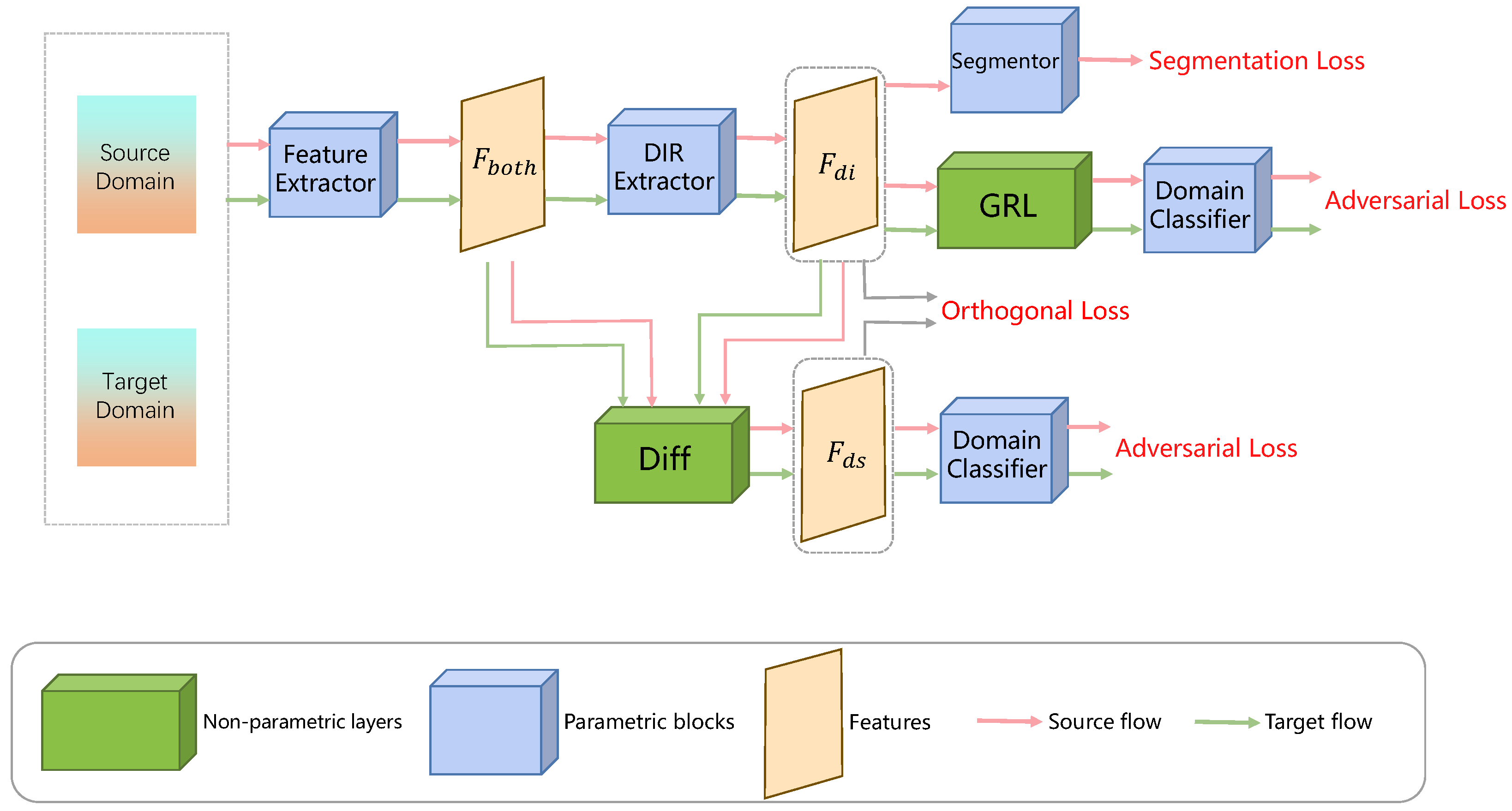
| ODADA [94] | Sun et al. (2022) | Domain-invariant representation and domain-specific representation decomposition | Scenarios with significant domain shift; unsupervised domain adaptation | Learns purer and more effective domain-invariant features |
| SMEDL [95] | Cai et al. (2025) | Disentangled Style Mixup (DSM) strategy | Cross-modal medical image segmentation tasks | Leverages both intra-domain and inter-domain variations to learn robust representations |
| DDSP [96] | Zheng et al. (2024) | Dual domain distribution disruption strategy; Inter-channel Feature Alignment (IFA) mechanism | Scenarios with complex domain shift; unsupervised domain adaptation tasks | Significantly improves shared classifier accuracy for target domains |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Wang, J.; Wei, J.; Yuan, X.; Wu, M. A Review of Non-Fully Supervised Deep Learning for Medical Image Segmentation. Information 2025, 16, 433. https://doi.org/10.3390/info16060433
Zhang X, Wang J, Wei J, Yuan X, Wu M. A Review of Non-Fully Supervised Deep Learning for Medical Image Segmentation. Information. 2025; 16(6):433. https://doi.org/10.3390/info16060433
Chicago/Turabian StyleZhang, Xinyue, Jianfeng Wang, Jinqiao Wei, Xinyu Yuan, and Ming Wu. 2025. "A Review of Non-Fully Supervised Deep Learning for Medical Image Segmentation" Information 16, no. 6: 433. https://doi.org/10.3390/info16060433
APA StyleZhang, X., Wang, J., Wei, J., Yuan, X., & Wu, M. (2025). A Review of Non-Fully Supervised Deep Learning for Medical Image Segmentation. Information, 16(6), 433. https://doi.org/10.3390/info16060433






